Abstract
Alcohol consumption causes disruptions in a variety of daily rhythms, including the circadian free-running rhythm. A previous study conducted in our laboratories has shown that larval ethanol exposure alters the free-running period in adult Canton-S Drosophila melanogaster. Few studies, however, have explored the effect of alcohol exposure on organisms exhibiting circadian periods radically different than (normal) 24-hours. We reared Canton-S, period long, and period short Drosophilamelanogaster larvae on 10%-ethanol supplemented food, and assessed their adult free-running locomotor activity and period transcript at ZT 12. We demonstrate that in Canton-S larval ethanol exposure shortens the adult free-running locomotor activity but does not significantly alter period mRNA levels at ZT 12. Period long mutants exposed to larval ethanol had significantly shortened adult free-running locomotor activity rhythms and decreased period mRNA levels, while period short mutants lengthened their free-running rhythm and showed increased period mRNA levels at ZT 12 after being exposed to larval ethanol. These results indicate that the effects of ethanol on the circadian clock might depend upon the baseline circadian period of the organism or that period mutant gene expression is sensitive to developmental ethanol treatment.
Keywords: ethanol, circadian, period, Drosophila, mutant, rhythm
Alcohol intake can lead to disruptions of daily behavioral and molecular rhythms in both humans and animal models [1]. Previous investigations using rodent models illustrate that chronic alcohol treatment can alter the free-running rhythm, a fundamental parameter of circadian clocks [2–5]. Chronic ethanol can alter molecular rhythms as well, including period (per) mRNA expression within the Suprachiasmatic Nucleus (SCN), the location of the mammalian light-entrainable circadian clock [6]. Not only does ethanol intake affect the adult circadian clock, perinatal or neonatal ethanol exposure also causes disruptions of per mRNA expression rhythms [7,8], and free-running rhythms [7,9]. These studies show clear connections between ethanol consumption and alterations of the circadian clock, at both behavioral and molecular levels.
While Drosophila melanogaster has been a widely used model in studying the behavior and genetics of both the circadian clock [10] and ethanol exposure [11], few studies have investigated effects of ethanol on the free-running rhythm in fruit flies. A recent study from our laboratories [12] shows that chronic developmental ethanol exposure alters the adult circadian free-running locomotor rhythm in wild-type Canton-S (CS) fruit flies in a dose dependent manner, even after ethanol treatment has been suspended. These previous investigations (including the aforementioned mammalian studies) have used only “wild-type” animal models, all with free-running periods approximating 24 hours. There appears to be a link between ethanol consumption and genes regulating the circadian clock, as mutations in per2 lead to significantly increased ethanol drinking in mice and are associated with human alcoholics [13]. Per2 mutant mice exhibit increased drinking bouts compared to wild-type mice, and acamprosate (a drug known to reduce ethanol drinking) reduces the number of drinking bouts in an LD cycle [14]. It is currently unknown, however, how chronic ethanol affects the free-running rhythm in circadian mutants which do not exhibit wild-type length circadian periods. Thus, the present experiments aim to uncover effects of chronic ethanol ingestion on circadian behavior (locomotor activity rhythm) and its molecular mechanism (per mRNA) in Drosophila melanogaster, using the Long (perL) and Short (perS) variants of the period mutants, which have circadian periods of approximately 28.5 and 19.5 hours, respectively [10].
For the behavioral assays, CS, perS, and perL Drosophila melanogaster, were reared and the activity and free-running periods were calculated using the protocols as previously described [12]. Flies were considered entrained only with LD periods of 24.00 ± 0.05 h. Composite actograms were generated by copying the raw numbers within each individual channel file (produced by FileScan) into a single column within a spreadsheet. Each individual fly received its own column. As each row constitutes the activity during a single 10-min bin for each fly, the numbers were averaged across each row and copied into a blank text file. The newly formed averaged activity file was imported into ClockLab to generate the actogram. A bout analysis for both LD and DD was conducted for all genotypes and ethanol treatments. The mean length of time (minutes), beam crosses per bout, and number of bouts per day were analyzed. An activity bout was defined as being greater than or equal to the average size of activity counts across the day, separated by at least 10-minutes of inactivity.
Six separate groups of these fly strains were also raised on either 0%- or 10%-ethanol supplemented food (same as in the behavioral analyses), and after eclosion, the flies were tested to determine the relative per mRNA level at ZT 12, using quantitative real time – Polymerase Chain Reaction (qRT-PCR). Total RNA (with DNAse treatment) from 25–30 heads of appropriate genotype and treatment was isolated using RNeasy miniprep kit (Qiagen, Valencia, CA). RNA quality and yield was measured using nanodrop spectrometer (Thermo Scientific, Wilmington, DE). qRT-PCR was performed in duplicates on 50–100 ng total RNA using one step Quantifast SYBR Green RT-PCR kit (Qiagen) on StepONE Real-Time PCR system (Applied Biosystems, Foster City, CA). Dissociation curve was analyzed to ensure primer specificity. Relative normalized transcript level was determined by delta-delta Ct method. RP49 was used as the normalizing gene. Mean ± SEM per transcript was calculated from six independent experiments. Welch’s t-tests with the Bonferroni correction were performed to determine differences between ethanol receiving flies and controls, among the three genotypes. Per primers were 5′-GACCGAATCCCTGCTCAATA-3′ and 5′-GTGTCATTGGCGGACTTCTT-3′, and RP49 primers were 5′-CGGTTACGGATCGAACAAGC-3′ and 5′-CTTGCGCTTCTTGGAGGAGA-3′.
The mean activity level (number of beam crosses per 10 min bin) in LD and DD across all genotypes and ethanol treatments are listed in Table 1. CS receiving larval 10%-ethanol were not significantly different from CS control receiving 0%-ethanol, regarding the mean activity level in LD (t1,48= 1.378=; p=0.18) or DD (t1,48= 0.048; p=0.96). Regardless of ethanol treatment, perS flies had significantly greater mean activity than perL, in both LD (Two-way ANOVA; F1,110=88.99; p<0.001) and DD (F1,110=66.82; p<0.001). In addition, there was no genotype by ethanol interaction for mean activity for perS and perL, in LD (p=0.37) or DD (p=0.57). These results are consistent with our previous study in flies [12], where no differences in activity level in DD were found between larval-ethanol treated animals versus controls.
Table 1.
Adult locomotor activity levels (average beam crosses per 10 min bin ± SEM) and bout analysis of Canton-S, perS, and perL, in LD and DD, under the differing ethanol treatment conditions. On average, perS exhibit higher locomotor activity than perL, manifesting itself in more beam crosses per bin and longer bouts in both LD and DD. There are no differences in the number of bouts between perS and perL in LD, but perL showed increased number of bouts per day in DD compared to perS. There is no effect of ethanol on the activity amongst individual genotypes for overall activity levels, length of bout, beam crosses per bout, or number of bouts per day in LD or DD, excepting that perL receiving larval 10% ethanol showed increased number of bouts per day compared to control perL in DD only. Values with differing letters (a, b, c) are significantly different (p<0.05).
| Genotype | EtOH % | N | Activity LD | Activity DD |
|---|---|---|---|---|
| Canton-S | 0% | 26 | 5.84 ± .36a | 5.40 ± .36a |
| 10% | 24 | 5.18 ± .31a | 5.42 ± .31a | |
| perL | 0% | 34 | 4.31 ± .30a | 4.58 ± .25a |
| 10% | 25 | 3.97 ± .27a | 4.95 ± .34a | |
| perS | 0% | 30 | 9.85 ± .89b | 9.24 ± .71b |
| 10% | 25 | 10.66 ± .86b | 10.32 ± .86b |
| LD | DD | ||||||
|---|---|---|---|---|---|---|---|
| Genotype | EtOH % | Length of Bout (min) | Beam Crosses per Bout | Bouts per Day | Length of Bout (min) | Beam Crosses per Bout | Bouts per Day |
| Canton-S | 0% | 48.56 ± 5.82 | 73.86 ± 9.57 | 11.92 ± 1.04 | 44.89 ± 6.73 | 58.33 ± 9.63 | 14.36 ± 1.14 |
| 10% | 46.54 ± 4.17 | 69.75 ± 6.56 | 9.90 ± 0.83 | 40.57 ± 3.61 | 54.51 ± 6.37 | 14.07 ± 0.73 | |
| perL | 0% | 23.67 ± 1.70a | 27.60 ± 2.43a | 12.35 ± 0.92a | 22.84 ± 2.56a | 28.62 ± 4.59a | 15.12 ± 1.00a |
| 10% | 21.04 ± 1.49a | 30.46 ± 2.48a | 10.22 ± 0.91a | 26.78 ± 2.40a | 34.64 ± 3.74a | 19.32 ± 1.15b | |
| perS | 0% | 33.04 ± 4.94b | 74.93 ± 13.98b | 11.84 ± 0.84a | 39.01 ± 6.11b | 79.81 ± 16.10b | 12.42 ± 1.05c |
| 10% | 37.09 ± 4.52b | 81.85 ± 11.90b | 11.97 ± 0.68a | 41.20 ± 7.43b | 82.67 ± 16.62b | 10.69 ± 0.85c | |
The bout analysis showed that perS, regardless of ethanol treatment, had increased bout length (Two-way ANOVA; p=0.001) and beam crosses per bout (p<0.001), but not number of bouts per day (p=0.64), compared to perL in LD. In DD, perS had increased bout length (p=0.005) and beam crosses per bout (p<0.001), but had decreased number of bouts per day (p<0.001) compared to perL. There was no genotype by ethanol interaction for the bout duration, number of counts per bout, and number of bouts per day in LD or DD (all p>0.10), except that perL exposed to larval-ethanol showed more bouts per day than control perL in DD (p=0.028). This result suggests that perL may be more sensitive to the effects of ethanol on activity than perS. There were no differences between CS receiving ethanol and controls for the bout analysis (t-tests, all p>0.10). The results from the bout analyses are different from a previous report using mice, where mean bout duration and quantity was altered during ethanol exposure and withdrawal in a 1-minute light pulse skeleton photoperiod [15]. This implies that there may be species differences in the effects of ethanol on the organization of locomotor activity between Drosophila and mammals.
CS receiving larval 10%-ethanol treatment had a significantly shortened mean free-running period (24.13 ± .04 h) compared to CS controls (24.35 ± .05 h; t1,48=3.59; p=0.002), confirming the results of our previous study (Fig. 1, 2a) [12]. This shortening of the circadian period corresponded with a trending, but statistically non-significant reduction of per mRNA level in larval 10%-ethanol treated flies at ZT 12 (t1,4=2.55; p=0.064, Fig. 2b). Control and 10%-ethanol-treated perL flies had a mean free-running period of 28.73 ± .16 h and 28.15 ± .11 h, respectively (Two-Way ANOVA F1,110=15.31; p<0.001; p=0.029; Fig. 1, 3a), while control and 10% ethanol-treated perS flies had a mean free-running period of 19.33 ± .10 h and 19.90 ± .14 h, respectively (p=0.038; Fig. 3a). Of course, perS have a significantly shorter free-running period than perL, regardless of ethanol exposure (p<0.001). The shortening of the free-running period in ethanol-treated perL flies corresponded to a significant decrease in per mRNA level at ZT 12 (t1,5=3.25; p=0.045; Fig. 3b) compared to control perL flies. In contrast, perS receiving larval 10%-ethanol had a significantly higher per mRNA level (t1,4=4.09; p=0.030; Fig. 3b) than the control perS group.
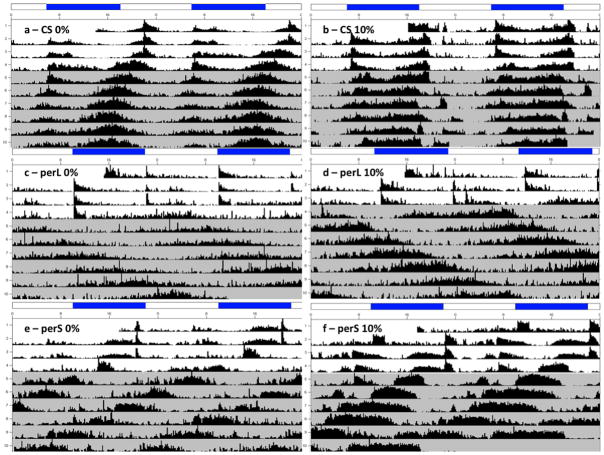
Figure 1.
Composite actograms for each of the genotype/ethanol conditions. Double-plotted composite actograms for (a) CS 0%-ethanol and (b) CS 10%-ethanol (c) perL 0%-ethanol and (d) perL 10%-ethanol (e) perS0%-ethanol (f) perS 10%-ethanol. LD bars are represented on the top of each actogram, and the shaded portion indicated DD.
Figure 2.
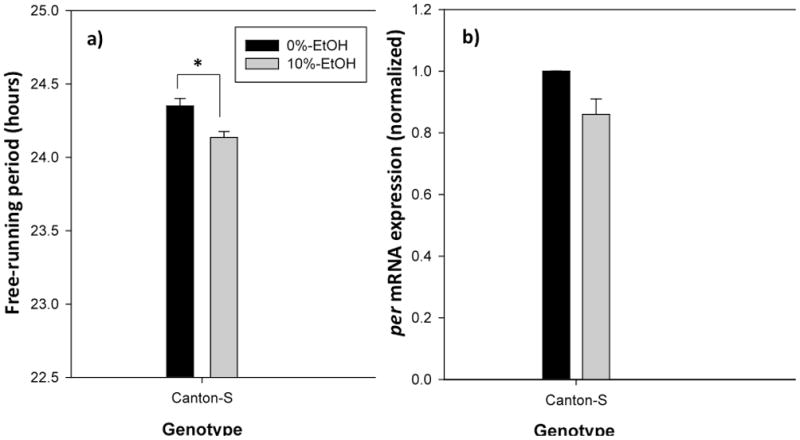
The effects of larval ethanol exposure on CS free-running period and per transcript level. (a) Larval 10%-ethanol treatment significantly shortens the behavioral locomotor activity rhythm, (b) but not the per mRNA level, compared to 0%-ethanol receiving controls. * indicates significantly different from each other (p<0.05)
Figure 3.
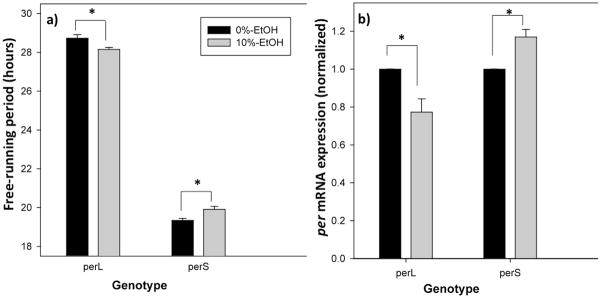
Larval ethanol exposure differently alters both behavioral and molecular rhythms of per mutants. (a) Larval 10%-ethanol treatment leads to lengthening and shortening of the free-running locomotor activity rhythm in perS and perL, respectively, compared to 0%-ethanol receiving controls. (b) perS shows an increase, while perL shows a decrease, in per mRNA level at ZT 12 compared to 0%-ethanol receiving controls. * indicates significantly different from each other (p<0.05).
Although both CS and perL displayed a shorter circadian period when treated with ethanol, only the perL showed a significantly reduced level of per at ZT 12. One possibility for this difference might be the extent of the period difference produced among the wild-type (CS) and two mutant strains. Ethanol produces an alteration of the free-running period of approximately 0.20–0.25 h in CS (similar to what is found within mammalian studies), while producing differences greater than 0.50 h in per mutants. Thus, the smaller decrease in per mRNA levels might be reflective of the smaller change in circadian period by ethanol, when compared to the mutants. The results from these per mutant experiments are some of the first to show that ethanol exposure affects the circadian clock differently between organisms with very short free-running periods compared to long periods, both behaviorally and molecularly. In summary, these results confirm and extend the work on the effects of chronic ethanol consumption on development and the circadian clock, which are consistent between rats and fruit flies. The alterations in overall per mRNA levels by developmental ethanol exposure found in this study provide an explanation for the subsequent shortening or lengthening of the locomotor free-running rhythm found in the Drosophilaper mutants.
Modifying per transcript level is one mechanism that leads to altered behavioral circadian rhythms, as lithium produces both lengthened locomotor activity rhythms and significantly higher per2 levels, in adult mice [16]. Other studies have also documented that ethanol consumption affects per mRNA levels, in both developing animals [7,8] and in adult animals [6]. Perinatal ethanol exposure decreases per1 mRNA levels at ZT 7, and per2 mRNA levels at ZT 15, in LD in SCN of Sprague-Dawley rats [8]. Sprague-Dawley rats exposed to early neonatal ethanol exposure expressed depressed per1 and per2 mRNA levels at CT 6, and an altered free-running period in the adult even after ethanol treatment ended [7]. While the differences between the results of these two studies might be due to the different lighting conditions and/or the timing of ethanol exposure, it is worth noting that both adult and developmental ethanol exposure significantly decreased per1 and per2 levels only at the peak of the mRNA expression rhythm in each of those specific conditions, and at no other circadian time points. As per gene mRNA expression in adult fly heads peaks at or close to ZT 12 [17], our results showing ethanol modulation of the Drosophila per gene in perS and perL at ZT 12 are consistent with that observation.
Our results show that larval ethanol treatment alters both the behavioral and molecular rhythms of per mutants in a baseline free-running rhythm dependent manner. Previous work has illustrated ethanol-consumption-dependent lengthening or shortening of the free-running rhythm based upon the individual’s baseline period in Long-Evans rats [4]. Taken together there appears to be an emerging body of evidence demonstrating that the effects of ethanol on the circadian clock are dependent upon the innate free-running period of the organism, and those effects are mediated through altered per expression levels.
These results also demonstrate that developmental environmental factors can alter the adult circadian clock behavior and molecular state even after the treatment stimulus has ended. Light-pulses during first instar larvae can have lasting effects into adulthood [18]; Drosophila larvae also exhibit cyclical oscillations of PER and other clock proteins [19]. A recent study has shown that mice exposed to differing seasonal photoperiods perinatally show lasting effects on the adult circadian clock. Per1:GFP mice exposed to long-day seasonal photoperiods show significantly earlier peaks in per1 rhythms within individual SCN neurons, in addition to shortened behavioral rhythms, which can persist for several weeks after alternate photoperiodic input [20].
One possibility for the developmental affects seen in this study is through the effects of slowpoke, which encodes BK-channels, is necessary and sufficient to induce ethanol tolerance [21], and upon deletion, leads to arrhythmia in flies [22]. As slowpokein duction leads to ethanol resistance, developmental ethanol exposure should induce high levels of slowpoke expression throughout development, which can lead to differing free-running period lengths. PER and TIM cycling is, however, similar between slowpoke mutants and wild-type flies, indicating that the arrhythmia caused by slowpoke deletion is downstream of the oscillator [22]. Thus, effects of ethanol on slowpoke expression may be affecting the downstream processes, while the current results show that larval ethanol exposure affects the circadian oscillator directly by altering per mRNA expression. Developmental effects on slowpoke in wild-type flies, however, have not been examined and can’t be ruled out in the present study. It is interesting to note that both per and slowpoke mutants show altered courtship songs [23]. If flies reared on ethanol have slightly altered perand slowpoke expression they might also have altered courtship songs. This may promote assortative mating with respect to developmental ethanol exposure, since it should create a slight variation in courtship song frequency which could allow females to recognize males whose courtship song frequencies reflect rearing in ethanol. Future studies could test whether males reared on ethanol have an altered courtship song and whether the ethanol-raised females prefer to mate with them.
The differing effect of ethanol on the circadian period seen in perS and perL may also be partly due to the effects of ethanol on PER degradation. Casein Kinase 1ε (CK1ε) inhibitors prevent relapse-like ethanol consumption in rats, indicating a link between PER degradation and ethanol drinking [24]. Ethanol might be affecting the ortholog of CK1ε, doubletime (dbt), which is responsible for PER degradation in Drosophila. So while our study showed that larval ethanol affects per mRNA levels in per mutants, ethanol might also be affecting the PER turnover rate differently in the per mutants by affecting dbt function, manifesting itself in lengthening or shortening of the behavioral free-running periods in perS and perL, respectively. It is worth noting, however, that CK1ε cannot rescue or replace dbt function in Drosophila [25], so there are differences between the two species that needs to be examined further. Future studies are needed to determine the effects of ethanol exposure on the trafficking of mRNA and proteins that specifically regulate the circadian clock (i.e., the “core-clock” genes), in different species. Regardless of the effects of ethanol PER degradation, as larval-ethanol treatment alters per mRNA levels, our results indicate that ethanol affects the genetic mechanism of the circadian clock itself differently in organisms with different period lengths. Since fruit fly and mammalian circadian systems both appear to have the potential to display a significant degree of plasticity in response to developmental conditions, it would also be interesting to investigate the potential for transgenerational expression of these effects in further experiments.
The lengthening of the free-running rhythm and increases found in per transcript levels in perS may have implications for other sleep and circadian disorders, including familial advanced sleep-phase syndrome (FASPS), which is characterized by having behavioral and physiological rhythms phase advanced by several hours and is associated with a mutation in the human per2 gene. Both perS[2–6] and humans with FASPS [27] exhibit premature nuclear degradation and clearance of dPER and hPER2 (respectively for each species), which is one of the causes of the severely advanced rhythms seen in these organisms. Additionally, the same mutation in human per2 found by the Spanagel lab [13] is significantly associated with both alcoholism and sleep problems, including premature final awakening in adolescent males [28]. Investigating the mechanisms underlying the effects of ethanol on circadian clock function in wild-type and mutant strains may lead to better understanding of how individual differences in circadian period may alter response to chronic or developmental ethanol exposure and its potential relevance to understanding mechanisms involved in addiction to alcohol.
Highlights.
Larval ethanol exposure alters circadian rhythm in D. melanogaster period mutants
Changes in circadian period and per mRNA persist in adults after removal of ethanol
Larval ethanol shortens period and decreases per mRNA levels in perL mutants
Larval ethanol lengthens period and increases per mRNA levels in perS mutants
Acknowledgments
Dr. Michael Rosbash (Brandeis University/HHMI) generously provided the perL and perS fly stocks. This project was supported by the grants from the National Center for Research Resources, INBRE (5P20RR016463-12) and the National Institute of General Medical Sciences (8 P20 GM103423-12) from the National Institutes of Health to Colby College and Science Division Grant, Colby College to STA.
Abbreviations
- CT
Circadian Time
- LD
Light-Dark
- DD
Dark-Dark (i.e., constant darkness)
- ZT
Zeitgeber Time
Footnotes
Competing Financial Interests:
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spanagel R, Rosenwasser AM, Schumann G, Sarkar DK. Alcohol consumption and the body’s biological clock. Alcohol Clin Exp Res. 2005;29:1550–7. doi: 10.1097/01.alc.0000175074.70807.fd. [DOI] [PubMed] [Google Scholar]
- 2.Mistlberger RE, Nadeau J. Ethanol and circadian rhythms in the Syrian hamster: effects on entrained phase, reentrainment rate, and period. Pharmacol Biochem Behav. 1992;43:159–65. doi: 10.1016/0091-3057(92)90652-v. [DOI] [PubMed] [Google Scholar]
- 3.Dwyer SM, Rosenwasser AM. Neonatal clomipramine treatment, alcohol intake and circadian rhythms in rats. Psychopharmacology (Berl) 1998;138:176–83. doi: 10.1007/s002130050660. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwasser AM, Fecteau ME, Logan RW. Effects of ethanol intake and ethanol withdrawal on free-running circadian activity rhythms in rats. Physiol Behav. 2005a;84:537–42. doi: 10.1016/j.physbeh.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Seggio JA, Fixaris MC, Reed JD, Logan RW, Rosenwasser AM. Chronic ethanol intake alters circadian phase shifting and free-running period in mice. J Biol Rhythms. 2009;24:304–12. doi: 10.1177/0748730409338449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CP, Kuhn P, Advis JP, Sarkar DK. Chronic ethanol consumption impairs the circadian rhythm of pro-opiomelanocortin and period genes mRNA expression in the hypothalamus of the male rat. J Neurochem. 2004;88:1547–54. doi: 10.1046/j.1471-4159.2003.02300.x. [DOI] [PubMed] [Google Scholar]
- 7.Farnell YZ, Allen GC, Nahm SS, Neuendorff N, West JR, Chen WJ, et al. Neonatal alcohol exposure differentially alters clock gene oscillations within the suprachiasmatic nucleus, cerebellum, and liver of adult rats. Alcohol Clin Exp Res. 2008;32:544–52. doi: 10.1111/j.1530-0277.2007.00598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CP, Kuhn P, Advis JP, Sarkar DK. Prenatal ethanol exposure alters the expression of period genes governing the circadian function of beta-endorphin neurons in the hypothalamus. J Neurochem. 2006;97:1026–33. doi: 10.1111/j.1471-4159.2006.03839.x. [DOI] [PubMed] [Google Scholar]
- 9.Allen GC, West JR, Chen WJ, Earnest DJ. Neonatal alcohol exposure permanently disrupts the circadian properties and photic entrainment of the activity rhythm in adult rats. Alcohol Clin Exp Res. 2005;29:1845–52. doi: 10.1097/01.alc.0000183014.12359.9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–6. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heberlein U, Wolf FW, Rothenfluh A, Guarnieri DJ. Molecular Genetic Analysis of Ethanol Intoxication in Drosophila melanogaster. Integr Comp Biol. 2004;44:269–74. doi: 10.1093/icb/44.4.269. [DOI] [PubMed] [Google Scholar]
- 12.Seggio JA, Possidente B, Ahmad ST. Larval ethanol exposure alters adult circadian free-running locomotor activity rhythm in Drosophila melanogaster. Chronobiol Int. 2012;29:75–81. doi: 10.3109/07420528.2011.635236. [DOI] [PubMed] [Google Scholar]
- 13.Spanagel R, Pendyala G, Abarca C, Zghoul T, Sanchis-Segura C, Magnone MC, et al. The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat Med. 2005a;11:35–42. doi: 10.1038/nm1163. [DOI] [PubMed] [Google Scholar]
- 14.Brager AJ, Prosser RA, Glass JD. Circadian and acamprosate modulation of elevated ethanol drinking in mPer2 clock gene mutant mice. Chronobiol Int. 2011;28(8):664–672. doi: 10.3109/07420528.2011.601968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brager AJ, Ruby CL, Prosser RA, Glass JD. Chronic ethanol distrups circadian photic entrainment and daily locomotor activity in the mouse. Alcohol Clin Exp Res. 2010;34:1–8. doi: 10.1111/j.1530-0277.2010.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Lu WQ, Beesley S, Loudon AS, Meng QJ. Lithium impacts on the amplitude and period of the molecular circadian clockwork. PLoS ONE. 2012;7:e33292. doi: 10.1371/journal.pone.0033292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardin PE. Analysis of period mRNA cycling in Drosophila head and body tissues indicates that body oscillators behave differently from head oscillators. Mol Cell Biol. 1994;14:7211–8. doi: 10.1128/mcb.14.11.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sehgal A, Price J, Young MW. Ontogeny of a biological clock in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1992;89:1423–7. doi: 10.1073/pnas.89.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko M, Helfrich-Forster C, Hall JC. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J Neurosci. 1997;17:6745–60. doi: 10.1523/JNEUROSCI.17-17-06745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciarleglio CM, Axley JC, Strauss BR, Gamble KL, McMahon DG. Perinatal photoperiod imprints the circadian clock. Nat Neurosci. 2011;14:25–7. doi: 10.1038/nn.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowmeadow RB, Krishnan HR, Ghezzi A, Al’Hasan YM, Wang YZ, Atkinson NS. Ethanol tolerance caused by slowpoke induction in Drosophila. Alcohol Clin Exp Res. 2006;30:745–53. doi: 10.1111/j.1530-0277.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez MP, Chu J, Villella A, Atkinson N, Kay SA, Ceriani MF. Impaired clock output by altered connectivity in the circadian network. Proc Natl Acad Sci U S A. 2007;104:5650–5. doi: 10.1073/pnas.0608260104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peixoto AA, Hall JC. Analysis of temperature-sensitive mutants reveals new genes involved in the courtship song of Drosophila. Genetics. 1998;148:827–38. doi: 10.1093/genetics/148.2.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perreau-Lenz S, Vengeliene V, Noori HR, Merlo-Pich EV, Corsi MA, Corti C, et al. Inhibition of the casein-kinase-1-epsilon/delta prevents relapse-like alcohol drinking. Neuropsychopharmacology. 2012;37:2121–31. doi: 10.1038/npp.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekine T, Yamaguchi T, Hamano K, Young MW, Shimoda M, Saez L. Casein kinase I epsilon does not rescue double-time function in Drosophila despite evolutionarily conserved roles in the circadian clock. J Biol Rhythms. 2008;23:3–15. doi: 10.1177/0748730407311652. [DOI] [PubMed] [Google Scholar]
- 26.Marrus SB, Zeng H, Rosbash M. Effect of constant light and circadian entrainment of perS flies: evidence for light-mediated delay of the negative feedback loop in Drosophila. EMBO J. 1996;15:6877–86. [PMC free article] [PubMed] [Google Scholar]
- 27.Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, et al. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–72. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Comasco E, Nordquist N, Gokturk C, Aslund C, Hallman J, Oreland L, et al. The clock gene PER2 and sleep problems: association with alcohol consumption among Swedish adolescents. Ups J Med Sci. 2010;115:41–8. doi: 10.3109/03009731003597127. [DOI] [PMC free article] [PubMed] [Google Scholar]