Abstract
Phage ϕC31 encodes an integrase that can mediate the insertion of extrachromosomal DNA into genomic DNA1. Here we show ϕC31 integrase can be used to generate transgenic Xenopus laevis embryos. mRNA encoding integrase was co-injected with a reporter plasmid containing a CMV promoter driven GFP into one cell embryos. The reporter plasmid was integrated into the genome. GFP expression, though robust, was in a limited number of tissues and varied among the embryos analyzed. We attributed this restriction to transcriptional silencing by chromosome position effects. Modification of the reporter plasmid by bracketing the CMV-GFP region with tandem copies of the chicken β-globin 5′ HS4 insulator2 relieved position effects. Embryos transgenic with insulated CMV-GFP expressed GFP uniformly. Tissue specific expression was achieved when the CMV promoter was replaced with a 551 base-pair minimal gamma crystallin lens promoter from Xenopus. Embryos transgenic with this plasmid had GFP expression limited to the lens of the eye. We observed that about a third of embryos assayed one week after fertilization were transgenic. These experiments demonstrate that the integration of insulated gene sequences using ϕC31 integrase can be used to efficiently create transgenic embryos in Xenopus laevis and may increase the practical use of ϕC31 integrase in other systems as well.
The ability to generate transgenic animals has revolutionized studies on gene function. Transgenic frogs of the genus Xenopus have been made using methods based on a restriction-enzyme mediated sperm integration approach described by Kroll and Amaya3. This method has been remarkably successful, but can be technically demanding. In addition, many embryos contain multiple copies of the transgene inserted at random insertion sites 3, 4.
We have tried an alternative approach, creating transgenic Xenopus embryos by using ϕC31 integrase mediated recombination. ϕC31 is a bacteriophage that encodes an integrase that mediates sequence directed recombination between a 34 nucleotide long bacterial attachment site (attB) and a 39 base-pair long phage attachment site (attP). ϕC31 integrase has a high efficiency of recombination, requires no accessory factors5, 6 and has been effectively used to integrate genes into plant cells7, mammalian cells1, 8-13, and Drosophila14. The ϕC31 integrase does not require an attP site to have perfect sequence fidelity for it to be recognized and cleaved9. These imperfect attP sites, or psuedo-attP sites, may have a similarity as low as 24% to an attP site and still allow recombination9. It has been estimated that a mammalian genome may contain 100-1000 psuedo attP sites 9. Since the Xenopus laevis genome is approximately the same size as a mammalian genome (3×109 base pairs), we hypothesized that there would also be psuedo-attP sites in its genome that could be used to create transgenic Xenopus embryos. In the experiments that follow all the plasmids injected are derivatives of pEGFPB2, which contains an attB site and the CMV promoter to drive GFP expression.
Injection of plasmids into Xenopus embryos can recapitulate promoter specific temporal and spatial expression; however, their expression is mosaic, randomly failing to transcribe in all of the cells that they should15. In addition, gene expression from plasmid DNA rarely extends to late stages (stage 42 and beyond) of development15. Typically 50-100 pg of plasmid is used in plasmid injection experiments15. An example of a stage 39 embryo injected with 100 pg of pEGFPB2 reporter plasmid demonstrating mosaic GFP expression is shown in figure 1e. The half life of the GFP protein has been estimated to be as long as 80 hours. Most of the embryos initially GFP positive no longer fluoresce by stage 46, but about 10% still glow at this stage in a mosaic pattern (figure 1f).
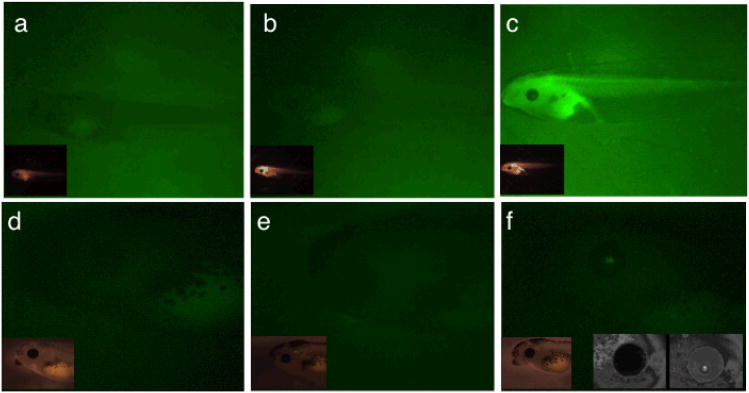
Figure 1.
Co-injection of ϕC31 integrase with CMV-GFP reporter plasmid enables detection of GFP expression in Xenopus embryos in stage 46 embryos. Embryos were anesthetized and the tails curled around toward the head for photo documentation. Images a--f are images of embryos viewed under fluorescent light passing through a 535 nanometer limit GFP emission filter. In every case the insert shows a brightfield image of the embryo. (a) Stage 46 Xenopus embryo injected 5 pg of pEGFPB2. (b-d) Stage 46 Xenopus embryos injected with 1ng of integrase mRNA and 5 pg of pEGFPB2. Depending on the embryo we noted GFP signal in either dorsal muscle (b), eyes and neural tissue (c), or in the gill structures of the embryos (d). (e) Stage 39 Xenopus embryo injected with 100pg pEGFPB2 reporter plasmid. (f) Stage 46 Xenopus embryo injected with 100pg pEGFPB2 reporter plasmid.
We reduced the amount of plasmid injected to levels where we no longer detected CMV-GFP expression in the embryo. We screened hundreds of embryos from three different mating pairs of frogs and all were negative for fluorescence when 5 pg of reporter plasmid was injected. We conclude that this concentration is too low to routinely see expression using pEGFPB2 as an extrachromosomal reporter plasmid. To test whether the ϕC31 integrase can be used to insert DNA into the Xenopus genome, 5 pg of pEGFPB2 was injected into single cell Xenopus embryos along with 1ng of mRNA encoding for the ϕC31 integrase protein. The earliest opportunity for expression of the reporter plasmid coincides with the activation of embryonic transcription at the 4000 cell mid-blastula stage (stage 7.5 about 8 hours after fertilization). We were able to detect green fluorescence as early as stage 14 (about 20 hrs post fertilization) and monitored expression through stage 46 (about 8 days post fertilization, Figure 1).
We focused on embryos that appeared developmentally normal at stage 46. We note that although survivorship varied depending on the specific mating pair of adults, overall there was no deleterious effect of plasmid or integrase mRNA on the health of the embryos when compared to control injections (Table 1).
Table 1.
Assay of GFP fluorescence as an indicator of integrase mediated gene insertion. The table indicates starting numbers of one cell embryos, the number of trials, survivorship to stage 46 and the number of embryos with GFP fluorescence at stage 46.
| Sample | # Embryos Injected (#trials/mothers) | Stage 46 Survival | Stage 46 Fluoresence | Avg % of embryos glowing (Range) |
|---|---|---|---|---|
| Non-Injected | 1194(12/7) | 779 | 0.00E+00 | 0.00E+00 |
| DEPC H2O | 916 (7/5) | 398 | 0.00E+00 | 0.00E+00 |
| Integrase mRNA 1 ng | 125 (1/1) | 44 | 0.00E+00 | 0.00E+00 |
| 5 pg pEGFPB2 | 337 (3/3) | 165 | 0.00E+00 | 0.00E+00 |
| 5 pg insulated pEGFPB2 plasmid | 300 (4/3) | 118 | 0.00E+00 | 0.00E+00 |
| 5 pg pEGFPB2 plasmid + 1 ng Integrase | 452 (3/3) | 258 | 95 | 36.82 (12-49) |
| 5 pg insulated pEGFPB2 plasmid + 1 ng integrase | 300 (4/3) | 148 | 47 | 31.76 (29-44) |
| 5 pg insulated pEGFPB2 plasmid + 500 pg integrase | 100(1/1) | 34 | 4 | 11.76 |
| 5 pg insulated pEGFPB2 plasmid + 50 pg integrase | 100(1/1) | 52 | 2 | 3.85 |
| 5 pg insulated crystallin lens plasmid | 200 (4/2) | 69 | 0.00E+00 | 0.00E+00 |
| 5 pg insulated crystallin lens plasmid + 1 ng Integrase | 200 (4/2) | 73 | 29 | 39.73 (37-44) |
Co-injection of 5 pg of pEGFPB2 with integrase mRNA resulted in approximately one third of the embryos scoring positive for green fluorescence. GFP expression was limited to a subset of tissues despite being driven by a CMV promoter (Figure 1). Embryos could be sorted into three groups based on GFP expression. 57% showed somatic muscle tissue along the tail(fig 1b), 31% the eyes and forebrain (fig 1c), and 7% in gill structures(fig 1d). While the embryos represented in fig 1c and 1d have a very different expression pattern than we have seen with plasmid alone injection, those in 1b are reminiscent of the mosaic expression from unintegrated plasmid. However, without co-injection with integrase mRNA we did not see GFP expression from this concentration of reporter plasmid, and the CMV promoter was not expected to give such limited mosaic expression. Integration of the reporter plasmid (pEGFPB2) was confirmed by Southern blot analysis (Figure 2, lanes 1 and 2). We did not routinely detect extrachromosomal reporter plasmid, indicating that plasmid did not persist in a non-integrated state. It is worth noting that one quarter of non-fluorescing embryos tested by Southern blot analysis still had an integrated copy of the GFP plasmid. These experiments suggest that the ϕC31 integrase can mediate recombination of plasmid carrying an attB site in Xenopus embryos. However, they failed to provide transgenic embryos with the expected expression pattern.
Figure 2.
Southern blot analysis of treated embryos indicates insertion of the GFP reporter plasmid into the embryonic genome. Markers in kilobase pairs are indicted on the left side of the blot. Lane c is linearized pEGFPB2, lane 1 and 2 used DNA isolated from the embryos in figure 1 in panel b and c. Lane 3 and 4 are from embryos transgenic for the insulated crystallin lens-GFP reporter and lane 5 is from an embryo transgenic for insulated CMV-GFP reporter.
The CMV promoter should have been active in every tissue, so we sought to explain why GFP expression was not ubiquitous. As cells differentiate, multicellular eukaryotes render regions of chromosomes transcriptionally inactive by a process called silencing16, 17. Chromatin silencing can spread to neighboring loci and the insertion of a transgene into a silenced region of chromatin prevents the expression of the transgene. The transcriptional silencing (or the inappropriate activation by insertion next to an enhancer) of a transgene based on where it inserts into the genome is referred to as chromosome position effect 18. Insulators are DNA sequences that protect genes from chromosome position effects 16, 17. One of the best characterized insulators is the chicken β-globin 5′ HS4 insulator 2, 19. Experimentally, it has been shown that tandem copies of the 250 base-pair core region of the chicken β-globin 5′HS4 insulator is sufficient to block the spread of chromatin silencing 2, 17, 19.
We inserted duplicated copies of the chicken β-globin 5′HS4 250 base-pair insulator flanking the CMV driven GFP into our reporter plasmid. 5 pg of insulated reporter plasmid along with 1ng of mRNA encoding ϕC31 integrase was injected into single cell Xenopus embryos. Embryos injected with integrase mRNA and insulated reporter plasmid began to express GFP ubiquitously at stage 14 and continued expressing until stage 46 when the experiment was stopped (Figure 3c). Embryos injected with only reporter plasmid did not fluoresce at any stage (Figure 3b).
Figure 3.
Co-injection of ϕC31 integrase with insulated CMV-GFP or crystallin-lens-GFP reporter plasmid generated Xenopus embryos with tissue appropriate expression. . Images (a-f) are images of embryos viewed under fluorescent light passing through a 535 nanometer limit GFP emission filter. In every case the insert shows a brightfield image of the embryo. (a-c) are stage 46 embryos assaying the expression of insulated CMV-GFP reporter (a) is non-injected, (b) is the insulated CMV-GFP reporter without co-injection of ϕC31 integrase (c) is an embryo co-injected with insulated CMV-GFP and ϕC31 integrase. (d-f) are stage 42 embryos assaying the expression of insulated crystallin-lens-GFP reporter (d) is non-injected, (e) is the insulated crystallin lens-GFP reporter without co-injection of ϕC31 integrase (f) is an embryo co-injected with insulated crystallin lens-GFP reporter and ϕC31 integrase. The two grayscale inserts represent photographs of the eye using either brightfield or fluorescence of the embryos eye taken through a 10× objective on a Zeiss Axioplan 2 microscope.
When embryos injected with 1ng of integrase mRNA and insulated reporter plasmid were assayed at stage 46, approximately 34-42% expressed GFP uniformly (Table 1). Integration was confirmed by Southern blot analysis (an example of integration is shown in Figure 2, lane 5). Embryos injected with less integrase mRNA (500 pg and 50 pg respectively) produced tadpoles with the expected GFP expression pattern, just at lower frequencies (table 1). We note that the quality of the integrase mRNA is critical to the success of this technique. Though further testing is needed, the expression patterns of GFP is consistent with integration generally occurring prior to first cleavage. Most of the remaining embryos failed to express GFP, while an occasional embryo expressed GFP in a manner similar to that seen for uninsulated plasmid, or only on one side.
We next asked if a tissue specific promoter derived from Xenopus would behave in an appropriate manner. Using the insulated GFP reporter plasmid we replaced the CMV promoter with 551 base-pairs of the Xenopus gamma crystallin lens promoter 20(Figure 3d-g). Unlike the global expression seen with the CMV promoter, embryos injected with integrase mRNA and the crystallin lens-GFP plasmid expressed GFP only in the lens of the eye (Figure 3f). GFP expression in the lens of the eye is uniform as seen in figure 3g. Integration efficiency ranged from 37-41% (Table 1) and was confirmed by Southern blot analysis (Figure 2 lanes 3 and 4).
We show here both by expression analysis of GFP and by Southern blot analysis that genomic integration of plasmid can be obtained if the plasmid has an attB site and ϕC31 integrase is expressed. We have assayed 39 embryos by Southern blot analysis. Although we commonly found that a single integration event occurred, we have seen nine different sized bands containing the integrated plasmid on Southern blots with no consistent indication of insertion site preference. The Southern blot band sizes are not consistent with multiple repeats or extrachromosomal plasmid, but vary in size dependent upon the nearest common restriction site in the Xenopus genome. We have not yet been able to assign particular integration sites to the different patterns of tissue expression seen with uninsulated reporter plasmids, but further analysis of the sites of integration are underway. Although we do not know if the integrations using this technique will be transmitted in the germ line we have analyzed buccal cells derived from a juvenile frog grown from an embryo co-injected with integrase mRNA and pEGFPB2. We find that there is still an integrated copy of the reporter gene in the buccal cell DNA (data not shown), so we are cautiously optimistic.
The insulated GFP reporters render the expression of the integrated gene independent of chromosome position effects. In addition, we have shown that tissue specific gene expression can be achieved, as is demonstrated by the studies using crystallin lens promoter-GFP reporter plasmid. The plasmids successfully used in this study were between 5 and 6.2 Kbp. We are in the process of constructing larger plasmids to test the limits of the system. We have reason to be optimistic that plasmids as large as 41Kb pairs may be able to be used, as that is the size of the ϕC31 phage21.
The goal of this study was to determine if the ϕC31 integrase system could generate transgenic Xenopus laevis embryos at a high enough efficiency to be able to study the F0 generation of transgenes. This goal has been met. We feel that this technique has the potential to become a valuable tool for transgenic studies on Xenopus and in other systems.
Methods
DNA Constructs
Plasmids pET11phiC31 polyA, used to generate the integrase mRNA in vitro, and reporter plasmid pEGFPB2, which contains a CMV driven GFP along with an attB site, were kindly provided by Michele Calos (Stanford). Gary Felsenfeld (NCBI) kindly provided a plasmid encoding the duplicated 250 base-pair 5′ HS4 insulator. The 5′ HS4 double insulator core fragment was PCR amplified with either BmgBI or PciI ends using the primer pairs CCACGTCCGGGTACCGAGTTGGCGCG and CGACGTGTCGATGAATTCGAGTTGGC or CACATGTCCGGGTACCGAGTTGGCGCG and CACATGTCGATGAATTCGAGTTGGC. The 550 base-pair PCR fragments were cloned into TOPO 4 (Invitrogen) and sequenced to ensure insulator sequence fidelity. BmgBI -HS4 double insulator was ligated into the BmgBI site between the attB site and the 5′ end of the CMV-GFP reporter and the PciI -HS4 double insulator ligated into the PciI site 3′ of the CMV-GFP reporter.
To prepare the Xenopus minimal gamma-crystallin lens promoter 20 driving GFP reporter plasmid, the CMV promoter was removed from insulated CMV promoter driving GFP expression reporter plasmid by digesting with AseI and AgeI. The Xenopus gamma crystallin lens promoter was kindly provided by Paul Krieg (University of Arizona). A 551 base-pair region of the gamma crystallin lens promoter was PCR amplified with AseI and AgeI ends and inserted into the reporter plasmid.
Integrase mRNA synthesis
The plasmid pET11phiC3 1polyA was linearized with EcoRI. mRNA was synthesized in vitro using the T7 mMessage Machine Kit following the manufacture's instructions (Ambion). A total of 1 μg of digested DNA was used as a template for mRNA production.
Xenopus Injections
Xenopus laevis males and females were obtained either from Xenopus I or Nasco. Eggs were obtained from hormonally induced females and fertilized as previously described 22. Single-cell embryos were injected with either water, 5 pg of reporter plasmid, or 5 pg of reporter plasmid and 1ng, 500 pg, or 50 pg of ϕC31 integrase mRNA in a total volume of 10nl as previously described23. The developmental staging of stage 46 embryos was performed as described in Nieuwkoop and Faber24.
Embryo Analysis
Embryos were analyzed for GFP fluorescence through embryonic stage 46 with a fluorescent Zeiss dissecting microscope (Stemi SV 11) or for higher magnifications with a Zeiss Axioplan 2 microscope. Light passed through a GFP filter with an emission filter limit of 535 nanometers. Color brightfield and GFP embryo pictures were taken with a Diagnostic Instrument Spot Camera. Black and white images were taken with a Zeiss Axiocam. Images were oriented and processed with Adobe Photoshop 7.0.
Genomic DNA extraction and Southern blotting
DNA was extracted from stage 46 tadpoles according to the manufacturer's instructions (Qiagen DNeasy Tissue Kit). Genomic DNA was digested with HindIII (non-insulated experiments) or BamHI (insulated experiments) overnight at 37°C, separated on a 1% agarose gel, and transferred onto positively charged Hybond membrane (Amersham). For the non-insulated integrant DNA analysis, pEGFPB2 was linearized with HindIII and then labeled with P32- dCTP following the manufactures instructions of Rediprime II Random Prime Labeling System (Amersham). For the insulated integrant DNA analysis, a 790 base-pair GFP fragment from pEGFPB2 was isolated by digestion with AgeI and BamHI. The fragment was then labeled with P32- dCTP following the manufactures instructions of Rediprime II Random Prime Labeling System (Amersham). The filters were hybridized with the labeled probe and washed at 68°C to a stringency of 0.1XSSC, 0.1% SDS.
Acknowledgments
The authors would like to thank Drs. Michele Calos, Gary Felsenfeld and Paul Krieg for plasmids used in this study. We thank Heather Bartlett, John Dagle, Sandra Kolker, Craig Fett, Emma Hornick and and Ema Stokasimov for helpful discussions. This work was supported by funding to DLW from the NIH. BGA is a student in the MSTP training program at the Roy J. and Lucille A. Carver College of Medicine, University of Iowa.
References
- 1.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–14. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 3.Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–83. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- 4.Sparrow DB, Latinkic B, Mohun TJ. A simplified method of generating transgenic Xenopus. Nucleic Acids Research. 2000;28:E12. doi: 10.1093/nar/28.4.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorpe HM, Wilson SE, Smith MC. Control of directionality in the site-specific recombination system of the Streptomyces phage phiC31. Molecular Microbiology. 2000;38:232–41. doi: 10.1046/j.1365-2958.2000.02142.x. [DOI] [PubMed] [Google Scholar]
- 6.Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5505–10. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutz KA, Corneille S, Azhagiri AK, Svab Z, Maliga P. A novel approach to plastid transformation utilizes the phiC31 phage integrase. Plant Journal. 2004;37:906–13. doi: 10.1111/j.1365-313x.2004.02015.x. [DOI] [PubMed] [Google Scholar]
- 8.Chalberg TW, Genise HL, Vollrath D, Calos MP. phiC31 integrase confers genomic integration and long-term transgene expression in rat retina. Investigative Ophthalmology & Visual Science. 2005;46:2140–6. doi: 10.1167/iovs.04-1252. [DOI] [PubMed] [Google Scholar]
- 9.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Molecular & Cellular Biology. 2001;21:3926–34. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomason LC, Calendar R, Ow DW. Gene insertion and replacement in Schizosaccharomyces pombe mediated by the Streptomyces bacteriophage phiC31 site-specific recombination system. Molecular Genetics & Genomics: MGG. 2001;265:1031–8. doi: 10.1007/s004380100498. [DOI] [PubMed] [Google Scholar]
- 11.Olivares EC, et al. Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nature Biotechnology. 2002;20:1124–8. doi: 10.1038/nbt753. [DOI] [PubMed] [Google Scholar]
- 12.Held PK, et al. In vivo correction of murine hereditary tyrosinemia type I by phiC31 integrase-mediated gene delivery. Molecular Therapy: the Journal of the American Society of Gene Therapy. 2005;11:399–408. doi: 10.1016/j.ymthe.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nature Biotechnology. 2003;21:321–4. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- 14.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–82. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vize PD, Melton DA, Hemmati-Brivanlou A, Harland RM. Assays for gene function in developing Xenopus embryos. Methods in Cell Biology. 1991;36:367–87. doi: 10.1016/s0091-679x(08)60288-5. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Current Opinion in Cell Biology. 2003;15:259–65. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 17.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes & Development. 2002;16:271–88. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 18.Bell AC, Felsenfeld G. Stopped at the border: boundaries and insulators. Current Opinion in Genetics & Development. 1999;9:191–8. doi: 10.1016/S0959-437X(99)80029-X. [DOI] [PubMed] [Google Scholar]
- 19.Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes & Development. 1998;12:2852–62. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Offield MF, Hirsch N, Grainger RM. The development of Xenopus tropicalis transgenic lines and their use in studying lens developmental timing in living embryos. Development. 2000;127:1789–97. doi: 10.1242/dev.127.9.1789. [DOI] [PubMed] [Google Scholar]
- 21.Smith MC, Burns RN, Wilson SE, Gregory MA. The complete genome sequence of the Streptomyces temperate phage straight phiC31: evolutionary relationships to other viruses. Nucleic Acids Research. 1999;27:2145–55. doi: 10.1093/nar/27.10.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebagliati MR, Weeks DL, Harvey RP, Melton DA. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell. 1985;42:769–77. doi: 10.1016/0092-8674(85)90273-9. [DOI] [PubMed] [Google Scholar]
- 23.Dagle JM, Walder JA, Weeks DL. Targeted degradation of mRNA in Xenopus oocytes and embryos directed by modified oligonucleotides: studies of An2 and cyclin in embryogenesis. Nucleic Acids Research. 1990;18:4751–7. doi: 10.1093/nar/18.16.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin): a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. Garland Pub; New York: 1994. [Google Scholar]