Summary
p68 RNA helicase is a prototypical RNA helicase. Here we present evidence to show that, by interacting with Ca-calmodulin (CaM), p68 plays a role in cancer metastasis and cell migration. A peptide fragment that spans the IQ motif of p68 strongly inhibits cancer metastasis in two different animal models. The peptide interrupts p68 and CaM interaction and inhibits cell migration. Our results demonstrate that the p68-CaM interaction is essential for the formation of lamellipodia and filopodia in migrating cells. p68 interacts with microtubules in the presence of CaM. Our experiments show that interaction with microtubules stimulates p68 ATPase activity. Further, microtubule gliding assays demonstrate that p68, in the presence of CaM, can function as a microtubule motor. This motor activity may allow p68 to transport CaM to the leading edge of migrating cells.
Introduction
Migration is an essential characteristic of cells in numerous fundamental biological processes1–6, including cancer metastasis 7. Targeting cell migration is a very important strategy for metastasis intervention 8,9. Cell migration is accomplished by a series of highly organized cyclic processes. First, the cells polarize and extend protrusions towards the direction of stimuli. The polarization is led by lamellipodia and filopodia10. Accompanying the protrusion, an extensive rearrangement of cell adhesions to the extracellular matrix (ECM) stabilizes the protrusion and functions as an anchor for traction 10,11. Following protrusion, the cell adhesion at the rear side detaches from the ECM 12. It is believed that formation of the leading edge is driven by cytoskeleton polymerization 13,14 15. Many proteins play a role in regulating cytoskeletal rearrangement during migration 16. Another important feature in the cell migration is that many molecules that play critical role(s) in migration re-distribute to the migration leading edge 17,18. Transportation along microtubules by families of microtubule motors is the main mechanism by which proteins and organelles translocate toward the direction of migration 19,20 21,22.
Calmodulin (CaM) is a calcium trigger protein with four EF-hands. The protein activates a wide range of cellular targets to regulate multiple processes in response to Ca2+ signals 23. One important molecular mechanism that contributes to the capability of CaM in regulating many cellular processes is its rapid redistribution to subcellular compartments in response to various signals 24,25. CaM is a major player in connecting Ca2+ signaling to cell motility in many cell types 26,27. Migration signals trigger spatiotemporal redistribution of CaM to the leading edge of the migrating cell, which is essential for cell motility 28–30. Although, redistribution of CaM has long been recognized as a mechanism that regulates complex cellular Ca2+ signals, little is known about how redistribution of CaM is accomplished and its role in cancer metastasis.
Here we report that interaction of p68 RNA helicase with CaM is essential for cell migration. Disruption of p68-CaM interaction inhibits cell migration. Interruption of p68-CaM interaction also strongly inhibits cancer metastasis. Our experiments showed that p68, upon interacting with CaM, can act as a microtubule motor to transport CaM to the leading edge of migrating cells.
Results
A peptide fragment of p68 inhibits cancer metastasis
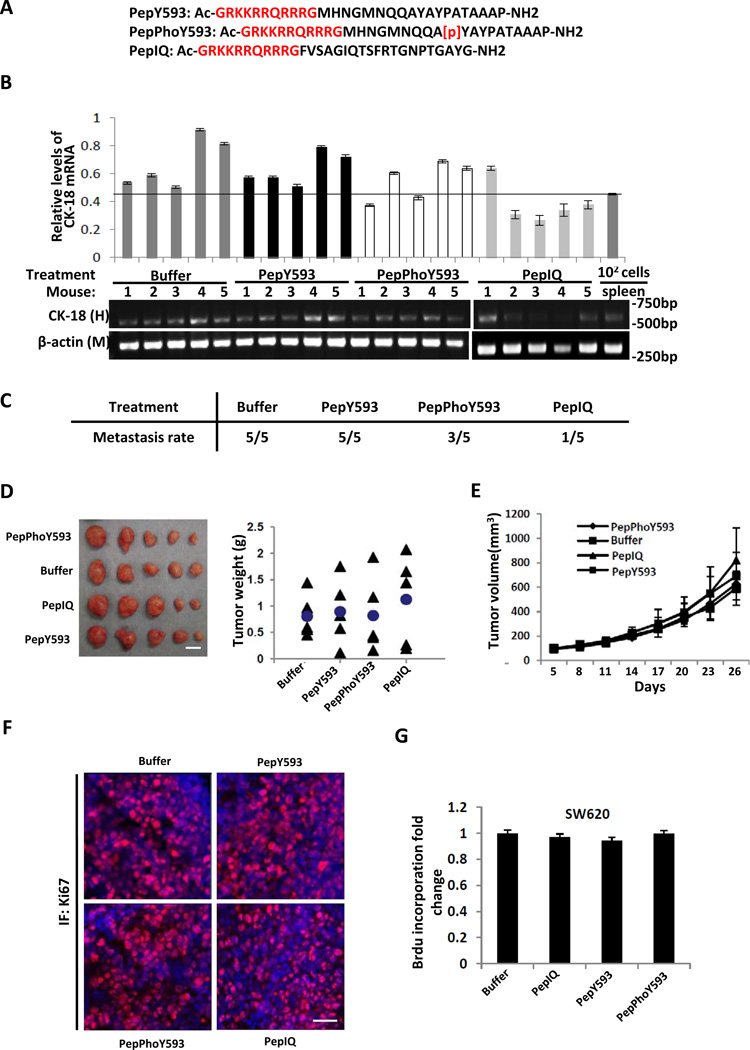
We previously reported that phosphorylation of p68 at Y593 mediated the effects of PDGF in promoting EMT by facilitating β-catenin nuclear localization 31. We asked whether a peptide that spans the nearby region of Y593 with the phosphorylation would be able to inhibit EMT, and could therefore potentially be used for metastasis intervention. To test this, three peptides were synthesized: two peptides span the region of aa 584 to 602 with/without Y593 phosphorylation (ref to as PepY593 and PepPhoY593 respectively), and a peptide spans the region of aa 549 – 568 (containing an IQ-like motif, therefore ref to as PepIQ). Both the PepIQ and PepY593 were used as control peptides. Three peptides were fused with the TAT cell permeable sequence at the N-terminus (Fig. 1A). The peptides were used to treat mouse xenografts of SW620 cells. We used SW620 because our previous studies revealed high p68 Y593 phosphorylation in these cells 32,33. Xenografts of SW620 metastasize to the lymph nodes, and their metastasis can be analyzed by examination of SW620 cells in the spleen 34. The PepPhoY593 peptide had small effects on SW620 tumor metastasis, while cancer metastasis was significantly reduced by the PepIQ peptide (Fig. 1 B & C). Tumor growth rates were not affected by treatment with any of the peptides as demonstrated by the anti-Ki-67 immunostaining (Fig. 1 D, E, and F). Consistent with this, the peptides had no effects on SW620 cell proliferation in vitro (Fig. 1G). We also examined whether the peptides would abolish PDGF stimulated EMT by analyzing the EMT markers E-cadherin and vimentin. In consistent with our previous report 31, the peptide PepPhoY593 treatment led to a decrease in vimentin and an increase in E-cadherin. However, the levels of E-cadherin and vimentin were not affected by the PepIQ and PepY593 (Supplementary Fig. S1A). The results suggested that, contrary to our original expectations, the PepPhoY593 had very small effects on cancer metastasis, while the PepIQ inhibited metastasis and the inhibitory effect was not due to inhibition of cell proliferation and EMT. The effect of the PepIQ on cancer metastasis was confirmed by an additional treatment using the PepIQ and a control peptide with a scrambled sequence (PepScram) (Supplementary Fig. S1 B, C, D). We further verified the effects of the IQ-like sequence on inhibiting cancer metastasis by expression of an eGFP fusion protein with the aa 549 – 568 sequence fused to the C-terminal of eGFP (ref to as GFP-IQ) in SW620 cells (Supplementary Fig. S2A). Expression of the GFP-IQ greatly reduced metastasis of the SW620 xenograft, while this reduction was not observed with expression of eGFP and vector alone (Supplementary Fig. S2 B, C, and D). The reduction of cancer metastasis was not due to changes in tumor growth upon expression of the fusion proteins (Supplementary Fig. S2E).
Figure 1. The effects of designed peptides on metastasis of xenograft of SW620 tumors.
(A) Sequences of three different peptides used in our study. (B) The mRNA levels of human cytokarotine-18 (CK-18) in the spleen extracts of tumor-bearing mice that were treated by different peptides (indicated) 1.5 mg/kg i.p. were analyzed by quantitative RT-PCR (Upper) or end-point RT-PCR (Bottom). The numbers represent individual mouse in each treatment group. The CK-18 mRNA levels were presented as relative levels of CK-18 mRNA by define the level of β-actin mRNA in the same mouse as 1. The mRNA levels of mouse β-actin in the spleen extracts are loading controls. The levels of the CK-18 mRNA in the extracts prepared from the harvested spleen (without tumor implantation) with direct injection of 100 SW620 cells were used as a reference (102 cells/spleen). (C) Metastatsis rates of the SW620 xenograft tumors in each treatment group. Tumor metastasis was judged by the CK-18 mRNA levels in the qRT-PCR using that of the 102 cells/spleen as the cut-off value. (D) Pictures (Left) and weight (Right) of harvested SW620 tumors after 28 days growth with 14 days treatment using the indicated peptides. The p values between “buffer” and “pepIQ” (p=0.46658) are calculated using pair-wise student t-test. (E) Tumor growth was monitored by measuring tumor volumes every three days and the tumor volumes were calculated by formula; Tumor volume = π/6 × (width) 2 × length. (F) Representative tissue sections were prepared from the harvested tumors and immune fluorescence stained with antibodies against Ki67 (IF: Ki67, Red). The blue is DAPI stain of cell nucleus. (G) Cell proliferation of SW620 cells that were treated by indicated peptides were measured using a commercial BrdU cell proliferation kit. The proliferation rates are presented as fold changes of BrdU incorporation of the same cultured cells before and after 48 hours culturing by defining the initial BrdU incorporation measurement as 1. The scale bars in D represent 1 cm and in F represent 100 µm.
PepIQ abrogates p68-CaM interaction and cell migration
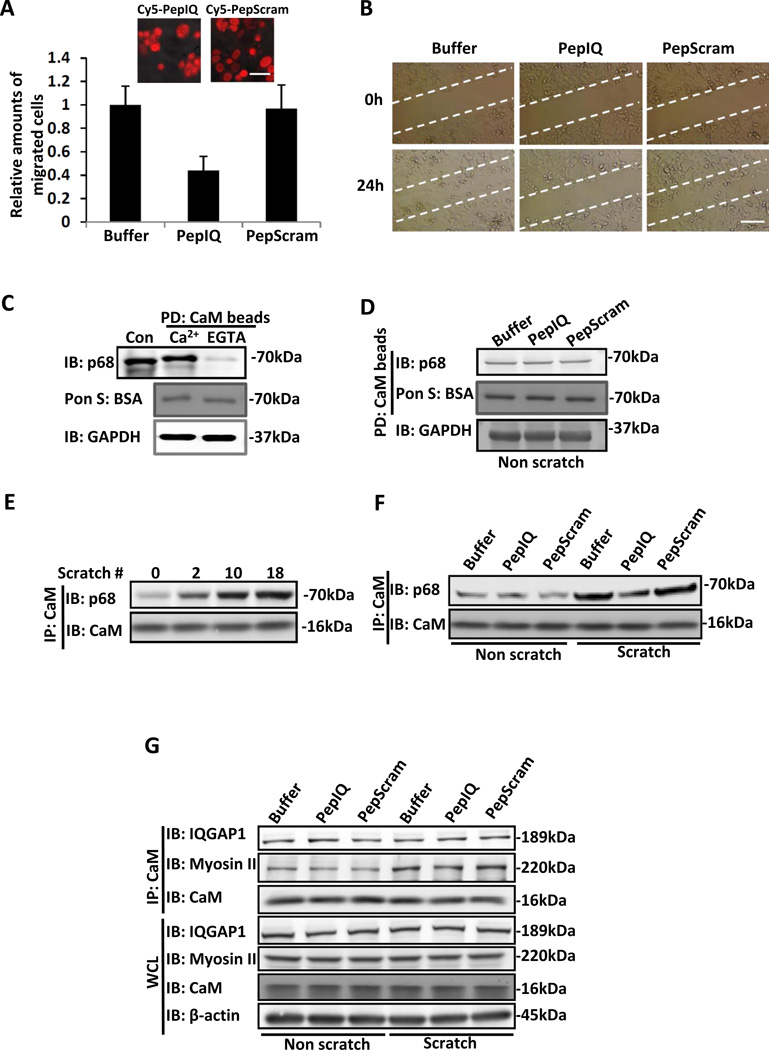
How does PepIQ substantially reduce cancer metastasis? Our preceding experiments showed that the peptide had no effects on expression of EMT markers, tumor growth or cell proliferation. It is well known that cell migration is vitally important for cancer metastasis 35. Thus, cell migration may be the target of the PepIQ peptide. We performed cell migration assays with the SW480 cell line, which is derived from the same cancer patient from whom the SW620 line was derived. SW480 cells have similar migration properties with lower proliferation rates compared to SW620. Clearly, cell migration was almost completely inhibited by PepIQ (Fig. 2 A&B). The observed cell migration effects were not due to effects of peptides on cell proliferation (Supplementary Fig. S2F). Similarly, cell migration was also dramatically inhibited by stable expression of the GFP-IQ in SW480 cells (Supplementary Fig. S2 G&H). We then questioned why PepIQ and expression of the GFP-IQ were so effective in inhibiting cell migration. It was demonstrated that p68 interacts with CaM in a number of in vitro binding studies and the IQ-like motif harbors a p68-CaM interaction site 36,37. Thus, it is possible that PepIQ and GFP-IQ competed with p68 to interact with CaM therefore inhibiting cell migration. Thus, we sought to test whether PepIQ interrupts the p68-CaM interaction and consequently inhibits cell migration. We first probed the p68-CaM interaction by co-precipitation with extracts of SW480 cells using CaM beads. Clearly, p68 was precipitated from the extracts by the CaM beads, and the interaction was Ca2+ dependent (Fig. 2C).
Figure 2. Cell migration and p68 - calmodulin interactions.
Cell migration of SW480 cells was analyzed by Boyden chamber assay (A) and monolayer scratch wound assay (B). Cells were treated by the indicated peptides (10 µg/ml). The upper two panels in (A) are representative confocal images of SW480 cells that were treated by the indicated Cy5.5-conjugated peptides, indicating peptide uptake. In (A), the number of cells migrating to the lower chamber is presented relative to the number of migrating cells that were treated by buffer. (C) Interaction of p68 with CaM in the presence of 2 mM CaCl2 (Ca2+) and 5 mM EGTA (EGTA) was probed by CaM pull-down (PD:CaM beads) followed by immunoblot using antibody against p68 (IB:p68). Cons are the control immunoblot of the cellular extracts using indicated antibody without CaM beads pull-down. (D) Interaction of p68 with CaM in extracts of SW480 cells was probed by CaM pull-down (PD:CaM beads) followed by immunoblot using antibody against p68 (IB:p68). The cells were treated by indicated peptides. Immunoblot of GAPDH (IB:GAPDH) is a loading control. (E) p68 and CaM interaction in extracts prepared from SW480 cells that were subjected to multiple scratch-wound treatment (number of scratches was indicated) analyzed by immunoprecipitation of CaM (IP:CaM). (F) p68 and CaM interaction in extracts prepared from SW480 cells that were subjected to 18 scratch wounds (Scratch) or no scratch wound (Non scratch), analyzed by immunoprecipitation of CaM (IP:CaM). Cells were treated by the indicated peptides. (G) CaM-IQGAP1 and CaM-Myosin II interactions in extracts prepared from SW480 cells that were subjected to 18 scratch wounds (Scratch) or no scratch wound (Non scratch), analyzed by immunoprecipitation of CaM (IP:CaM). The cells were treated by the indicated peptides. Immunoblots of IQGAP1 (IB:IQGAP1), Myosin (IB:Myosin II), and CaM (IB:CaM) in the whole cell lysate (WCL) indicate the cellular levels of these three proteins. Immunoblot of β-actin (IB:β-actin) in whole cell lysate is a loading control. The scale bars in A represent 20 µm and in B represent 100 µm.
We then probed whether PepIQ affected the p68-CaM interaction by the same CaM beads pull-down assay. Clearly, the peptide treatment did not lead to dramatic changes in the p68-CaM interaction (Fig. 2D). Since we observed that PepIQ greatly inhibited cell migration, we reasoned that the peptide might interfere with the p68-CaM interaction under conditions of migration stimulation, and the p68-CaM interaction itself could also be affected by migration induction. Thus, we examined the p68-CaM interaction in response to migration stimuli by co-immunoprecipitation with extracts of SW480 cells using an antibody against CaM. To induce migration, we used scratch-wounds on the culture plate. A strong increase in p68 and CaM co-immunoprecipitation was observed in the SW480 cells that were subjected to multiple scratch-wounds, and the extent of increase was dependent upon the number of scratches introduced (Fig. 2E). Similarly, there was a strong increase in p68 and CaM co-precipitation upon EGF treatment (Supplementary Fig. S3, A&B). Measurement of the changes in cell migration revealed a close correlation between the increase in the p68-CaM interaction and cell migration under EGF treatment (Supplementary Fig. S3C). The results suggest that cell migration strengthened the p68-CaM interaction. Since the IQ-like motif harbors the site for the Ca2+-dependent p68-CaM interaction 38, we asked whether the increase in the p68-CaM interaction induced by cell migration requires the IQ-like motif, and used a mutant that carries mutations IQ to MA (Ref to as IQ-M). HA-IQ-M was exogenously expressed in SW480 cells in which endogenous p68 was knocked down. HA-IQ-M co-precipitated with CaM. However, the scratch-wound dependent enhancement in p68-CaM interaction was not observed with the IQ-M mutant (Supplementary Fig. S3D). To test whether PepIQ abolished the enhancement of the p68-CaM interaction induced by cell migration, we probed the p68-CaM interaction in cell lysates of SW480 cells that were treated by the peptide. It was clear that the strong increase in the p68-CaM interaction upon multiple scratches was not observed with cells that were treated by the peptide (Fig. 2F). Similarly, the p68-CaM interaction was not dramatically strengthened upon migration stimuli in SW480 cells in which GFP-IQ was expressed. Co-precipitation of CaM and GFP-IQ was detected from extracts of the scratch-wound cells (Supplementary Fig. S3E), indicating that GFP-IQ competed with p68 for CaM interaction. Many proteins interact with CaM by their IQ motifs 39,40. Examples of the IQ motif containing proteins that interact with CaM are IQGAP1 and Myosin-II. It is well known that IQGAP1 and Myosin-II, and their interaction with CaM, are important for cell migration 41–43. To test whether PepIQ disrupted the interaction of IQGAP1 or Myosin-II with CaM, we carried out the co-immunoprecipitation with cell extracts of SW480 cells that were treated by PepIQ with/without multiple scratches, using the antibody against CaM. Immunoblots of the co-precipitates indicated that the IQGAP1-CaM and Myosin-CaM interactions were not affected by the PepIQ treatment regardless whether the cells were treated or not by the scratch-wound (Fig. 2G). The results indicate that PepIQ likely disrupts the p68 and CaM interaction specifically. The results also excluded a possibility that PepIQ sequestered all CaM in the cells therefore inhibiting cell migration. Thus, our experiments supported the notion that the cell migration strengthened the p68-CaM interaction, and that PepIQ disrupted the p68-CaM interaction induced by migration thus inhibiting cell migration.
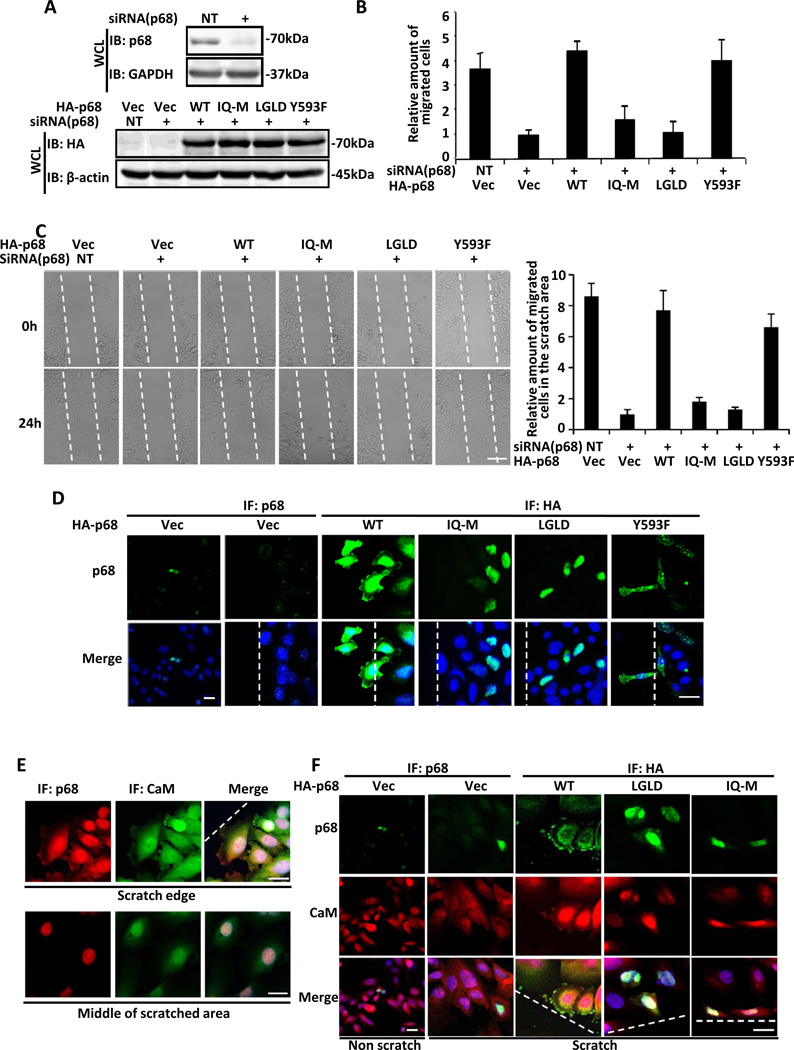
Based on the preceding experiments, we predicted that the p68-CaM interaction would be critical for cell migration. The hypothesis was tested by both Boyden chamber and scratch-wound assays. Endogenous p68 was knocked down in SW480 cells and HA-p68s, wt, IQ-M, LGLD (An ATPase activity deficiency mutant 44), or Y593F 31, was expressed in the cells (Fig. 3A). Boyden chamber assays showed that knockdown of p68 results in a dramatic reduction in cell migration, which could be fully recovered by expression of wt p68, but not by the IQ-M mutant. As a control, cell migration could also be rescued by expression of a mutant that carries a mutation at an irrelevant site (Y595F) (Fig. 3B). Interestingly, expression of an ATPase deficient mutant LGLD did not rescue cell migration (Fig. 3B), indicating that the ATPase activity of p68 is required for cell migration. We further confirmed the role of the p68-CaM interaction in cell migration by scratch-wound assays (Fig. 3C). Immunofluorecence staining using anti-HA antibody revealed that no HA-IQ-M and HA-LGLD expressing cells migrated to the wounds, while p68 wt expressing cells migrated to the wound areas normally (Fig. 3D). These experiments showed that the p68-CaM interaction is required for cell migration.
Figure 3. p68 and CaM interaction is essential for cell migration and for CaM localization.
(A) (Upper) Levels of p68 in the p68 knockdown SW480 cells were examined by immunoblot in whole cell lysates. (Bottom) Levels of exogenously expressed HA-p68s, wild type (WT) and indicated mutants, were analyzed by immunoblots of whole cell lysates using anti-HA antibody. Immunoblots of GAPDH and β-actin are loading controls. (B) Migration of SW480 cells in which endogenous p68 was knocked down by RNAi and HA-p68s, wild type (WT) and mutants (indicated), were exogenously expressed, was analyzed by Boyden chamber assays. The number of cells migrating to the lower chamber was measured relative to the number of migrating cells in which the empty vector was transfected. (C) (Left panel) Migration of SW480 cells in which endogenous p68 was knocked down by RNAi and HA-p68s, wild type (WT) and mutants (indicated), were exogenously expressed, revealed by phase contrast microscopy 0 h and 24 h after scratch-wounds were introduced into the cell culture plates. (Right panel) Quantification of the cells in a fixed area that migrated to the scratch area. (D) Representative images of SW480 cells in which endogenous p68 was knocked down by RNAi and HA-p68s, wild type (WT) and mutants were exogenously expressed. HA-p68s was immunostained using anti-HA antibody (green). Endogenous p68 was stained with the antibody p68-RGG (red). DAPI (blue). (E) Representative images of immunofluorescence staining of p68 (red) and CaM (green) in SW480 cells. The cells were treated by scratch-wounding. Upper panel shows cells that located to the scratch edges, bottom panel shows the middle of scratched areas. (F) Representative images of immunofluorescence staining of p68 (green) or exogenously expressed HA-p68s (green), wild type (WT) or mutants (indicated), and CaM (red) in SW480 cells. The endogenous p68 was knocked down by RNAi. The cells were treated by a scratch-wound (Scratch) or no scratch-wound (No scratch) in the culture plate. Error bars in (B) and (C) are standard deviations of three independent measurements. Vec is the cells transfected with the empty vector. NT represents the cells treated with non-target RNAi. The scale bars in C represent 100 µm and in D, E, and F represent 20 µm.
p68 and CaM interaction affect the localization of CaM and p68
In the immunostaining, we noted two interesting phenomena. (1) We observed a strong cytoplasmic p68 staining only in the migrating cells. When the cells reached their destination, p68 re-localized exclusively back to the cell nucleus (Fig. 3E). (2) There were very high levels of p68 staining in the cytoplasm and at the leading edge of the migrating cells and this staining pattern was not observed in the non-migrating cells (Fig. 3 D&E). Since p68 interacted with CaM in migration cells, we probed whether p68 and CaM would exhibit similar localization patterns. To this end, scratch-wounds were introduced to SW480 cells. The cells were then analyzed by immunostaining. p68 and CaM clearly co-localized, especially at the leading edge of the migration cells. In the non-migrating cells, p68 localized exclusively in the nucleus, while CaM could be detected almost everywhere in the cells. However, there were significant increases in CaM levels at the leading edges and the nucleus in the migrating cells compared to that in non-migrating cells (Fig. 3F).
We further examined the localizations of HA-p68 mutants LGLD and IQ-M and the effects of their expression on the cellular localization of CaM under scratch-wound conditions. Endogenous p68 was knocked down. The HA-p68s, wt, LGLD, or IQ-M was expressed in the cells. Scratch-wounds were subsequently introduced. Immunostaining using anti-HA antibody showed that wt HA-p68 localized to the leading edge of migrating cells. The ATPase deficient mutant LGLD, although detected in the nucleus and the cytoplasm, failed to accumulate at the membrane edge. Interestingly, the IQ-M almost exclusively localized in the cell nucleus (Fig. 3F). Immunostaining of CaM in the same HA-p68s expressing cells revealed that CaM also failed to accumulate at the membrane edge of the cells that expressed LGLD or IQ-M mutants under scratch-wound conditions (Fig. 3F). The results suggested that the p68-CaM interaction and the ATPase activity of p68 were required for the localization of CaM to the leading edge during cell migration.
p68 exhibits CaM-dependent microtubule motor activity
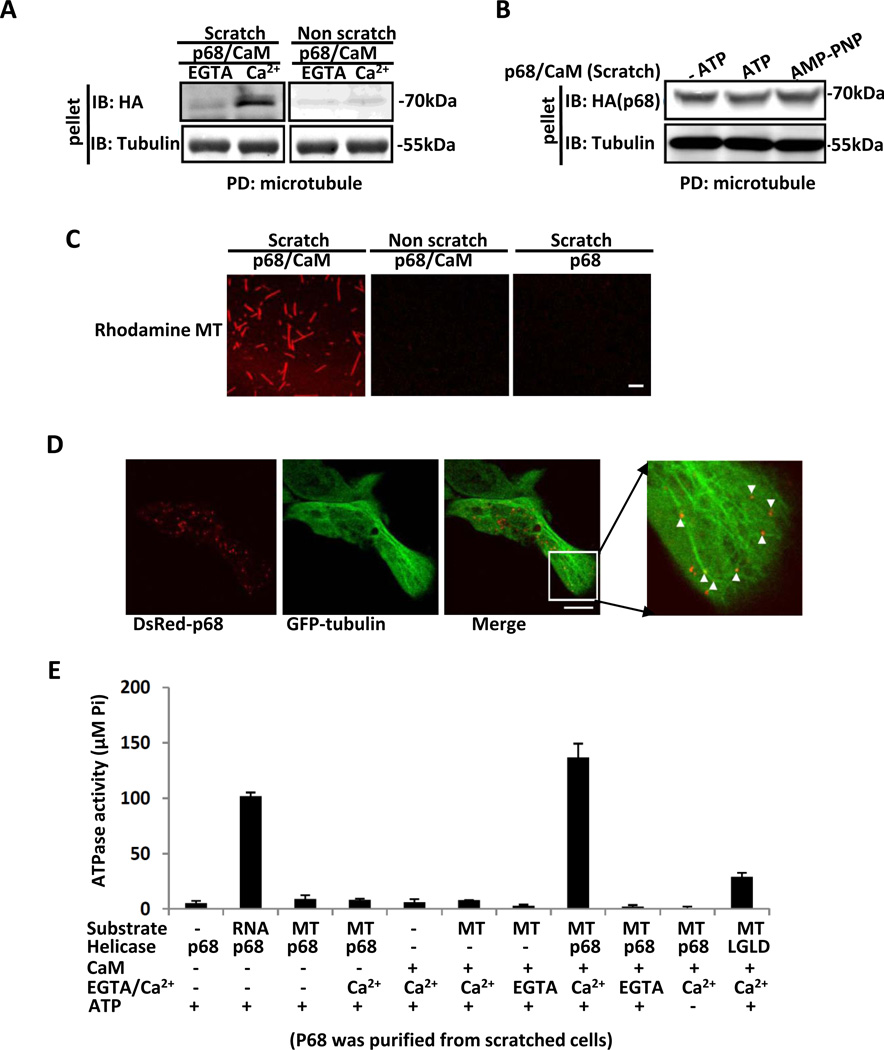
Redistribution of CaM to the leading edge areas is essential for cell migration 24,45. However, the molecular mechanism governing the localization of CaM to the leading edge is not well understood. It is documented that microtubule motors are responsible for transportation of various molecules to the leading edge of migrating cells. We questioned whether p68 could mediate CaM and microtubule motor interactions in the migrating cells. Thus, we probed by co-immunoprecipitation interactions between p68 and two well known microtubule motor molecules, kinesin and Dynein, in cell extracts prepared from SW480 cells that were treated by multiple scratch-wounds. No co-precipitations of either endogenous p68 or exogenous HA-p68 with the selected motor molecules were observed (Supplementary Fig. S4A). Since the ATPase activity of p68 was required for CaM to localize to the leading edge of the migration cells, we considered whether p68 itself could play a role in translocation of CaM to the leading edge in migrating cells by acting as an unconventional microtubule associated motor 17,18. To test this conjecture, we first analyzed whether p68 and/or CaM interact with microtubules. p68/CaM and microtubule interactions were first analyzed by microtubule co-precipitation assay. We carried out co-precipitation experiments with HA-p68 that was immunopurified from lysates of SW480 cells with/without multiple scratches (Supplementary Fig. S4B). The purified HA-p68 was incubated with microtubules. After precipitation of the microtubules by centrifugation, the presence of the HA-p68 in the microtubule precipitates was examined by immunoblot using an anti-HA antibody. HA-p68 that was purified from cells that were treated by scratch-wounds co-precipitated with microtubules in the presence of CaM. In contrast, HA-p68 immunopurified from SW480 cells that were not treated by scratch-wounds did not co-precipitate with microtubules even in the presence of CaM (Fig. 4A). The HA-p68 did not co-precipitate with unpolymerized α- and β- tubulin in the presence of Ca2+ and CaM (Supplementary Fig. S4C). Interestingly, co-precipitation of HA-p68 with microtubules in the presence of CaM was completely Ca2+ dependent, while ATP was dispensable as the HA-p68 co-precipitated with the microtubule in the presence and absence of ATP, and in the presence of non-hydrolysable ATP analogue AMP-PNP (Fig. 4B).
Figure 4. p68 and CaM interact with microtubules.
(A) & (B) Precipitation of HA-p68 that was immunopurified from cell lysates of multiple scratch wound treated (Scratch) or untreated (Non scratch) SW480 cells with in vitro assembled microtubule (pellet) in the presence of CaM (p68/CaM) was probed by immunoblot of the precipitated pellets using the anti-HA antibody (IB:HA). The precipitations were carried out in the presence of 0.5 mM CaCl2 (Ca2+) or 5 mM EGTA (EGTA) (A), or in the presence (ATP) and absence (−ATP) of ATP or in the presence of non-hydrolysable ATP analogue AMP-PNP (AMP-PNP) in addition to the presence of 0.5 mM CaCl2 (B). Immunoblot analyses of α-tubulin (IB:Tubulin) is the loading control indicating amounts of precipitated microtubule. (C) Representative fluorescence microscopy images reveal the bindings of Rhodamine labeled microtubule (Rhodamine MT) to the glass slides on which the HA-p68 that was immunopurified from cell lysates of multiple scratch wound treated (Scratch) or untreated (Non scratch) SW480 cells was attached (in the presence of CaM and Ca2+ [p68/CaM] and absence of CaM [p68]). (D) Representative fluorescence microscopy images of SW480 cells that expressed DsRed-p68 and eGFP-α-tubulin. The cells were treated by scratch-wound. The arrows indicated the DsRed-p68. (E) ATPase activities of HA-p68 or mutant (indicated) that was immunopurified from cell lysates of multiple scratch wound treated SW480 cells in the presence of various different substrates (indicated) were analyzed by measuring inorganic phosphate released from ATP hydrolysis. The ATPase activities were measured in the presence of 0.5 mM CaCl2 (Ca2+) or 5 mM EGTA (EGTA), and in the presence and absence of the recombinant CaM. In the ATPase assays, 4 mM ATP was always added unless specified (−ATP). The ATPase activity was expressed as µmole of inorganic phosphate hydrolyzed from ATP. MT is in vitro assembled microtubule. RNA is total RNA extracts from yeast. The scale bars in C represent 5 µm and in D represent 10 µm.
The interaction of p68-CaM with microtubules was further examined by binding of rhodamine-labelled microtubules to HA-p68 that was fixed on a glass slide. Binding of the microtubule to the glass slide was then visualized by fluorescence microscopy. The microtubules clearly bound to the HA-p68 purified from scratch-wound treated SW480 cells in the presence of CaM and Ca2+. The microtubules did not bind HA-p68 without addition of CaM (Fig. 4C). The microtubules also did not bind to HA-p68 that was purified from SW480 cells that were not treated by scratch-wounds (Fig. 4C). We further examined the co-localization of DsRed-p68 with microtubules labeled by eGFP. Under scratch-wound treatment, DsRed-p68 clearly co-localized with eGFP labeled microtubules (Fig. 4D). Thus, we could draw the conclusion that p68 interacts with microtubules and the interaction is dependent upon p68-CaM interaction.
If p68 could function as a microtubule motor-like molecule, we would expect that microtubules would stimulate ATPase activity of p68. Thus, the ATPase activity of the HA-p68 that was immunopurified from SW480 cells (Supplementary Fig. S4B) treated with multiple scratch-wounds was analyzed in the presence of various different possible substrates, including RNA and microtubules. As previously reported 46, HA-p68 exhibited strong RNA-dependent ATPase activity. Interestingly, the protein demonstrated an even stronger microtubule stimulated ATPase activity (Fig. 4E), and the microtubule-stimulated ATPase activity was Ca2+ dependent as addition of EGTA diminished it (Fig. 4E). The observed microtubule-stimulated ATPase activity came from p68 as the mutation that abolishes p68 ATPase activity (LGLD) also abolished the microtubule stimulated ATPase activity of the protein (Fig. 4E). In addition, this microtubule-dependent ATPase activity was not detected with the HA-p68 purified from the cells extracts without scratches (Supplementary Fig. S4D).
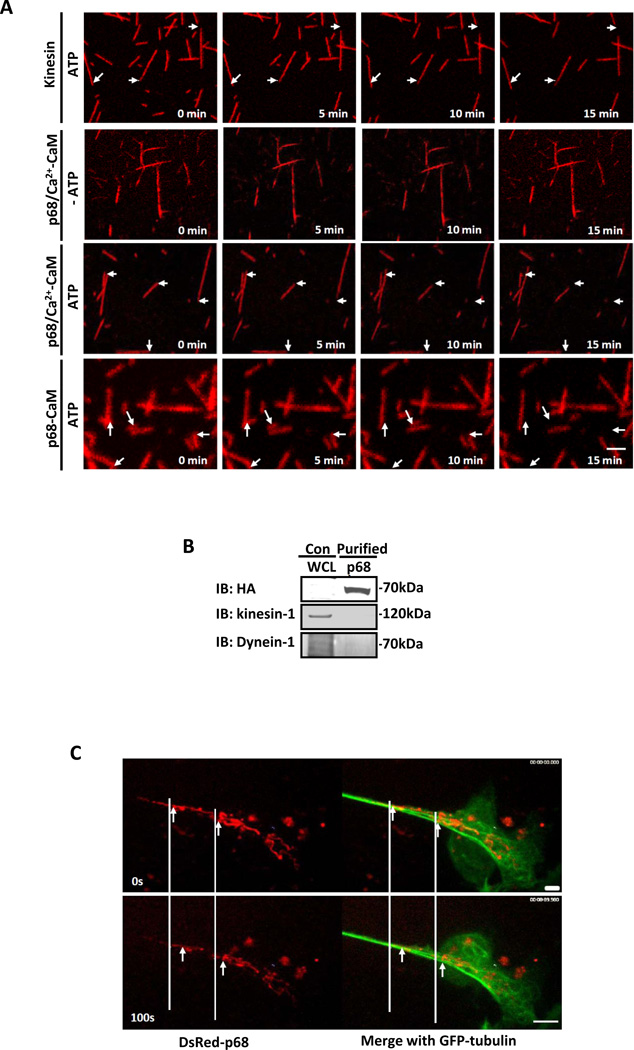
We next probed whether p68 indeed has microtubule motor activity. To this end, we employed microtubule gliding assays. HA-p68 was purified from SW480 cells with scratches and attached to a glass slide. Rhodamine-labelled microtubules were then added to the glass slide. Migration of the microtubule in the presence and absence of ATP was then recorded. First, migration of microtubules was clearly visualized with a positive control motor protein kinesin (Fig. 5A, Supplementary Movie 1). Migration of the microtubules on the glass slide with HA-p68 attached was also clearly evident in the presence of CaM and Ca2+. As a negative control, migration was not observed without addition of ATP (Fig. 5A, Supplementary Movie 2). Statistical analyses indicated that the HA-p68/CaM complex exhibits motor activity on microtubules very similar to that of a control motor kinesin (the number of gliding microtubles, mean gliding speed, and maximum gliding speed in a random selected group of 50 are similar, Table 1). Since p68 and HA-p68 did not co-immunoprecipitate with two very common microtubule associated motor proteins (Fig. 5B), it is less likely that the observed HA-p68 motor activity is due to co-purification of residual other cellular motor proteins. To further exclude the possibility that the motor activity was due to contamination during purification, we constructed a p68-CaM fusion protein (Fig. S5A). Based on computational modeling (Supplementary Fig. S6), the C-terminus of p68 is a flexible loop and is located close to the IQ-like motif. Thus, the fusion should bring the CaM in proximity to the IQ-like motif. Expression of the HA-p68-CaM fusion protein in SW480 cells led to greatly increased cell migration (Supplementary Fig S5 B&C) and the expressed fusion protein localized to the leading edge of migrating cells. In fact, expression of the fusion protein promoted formation of lamellipodia/filopodia without scratch-wounding (Supplementary Fig S5D). These observations indicated that the fusion protein functionally resembles the p68-CaM complex induced by migration signals. We then expressed and purified the fusion protein His-p68-CaM from E. coli. Gliding assays was performed with this bacterially expressed fusion protein. The bacterially expressed His-p68-CaM binds to microtubules (Supplementary Fig. S5E), possesses microtubule-dependent ATPase activity (Supplementary Fig. S5F), and has strong microtubule motor activity (Fig. 5A & table 1).
Figure 5. p68-CaM has microtubule motor activity.
(A) The movements of Rhodamine labeled microtubules on the glass slides on which the immunopurified HA-p68 (from SW480 cells) or bacterially expressed His-p68-CaM (p68-CaM) was attached in the presence (ATP) and absence (−ATP) and in the presence of 0.5 mM CaCl2 were recorded by the time lapse photography with confocal fluorescence microscopy (for video see on-line supplementary). The HA is a negative control with the HA peptide attached to the glass slides. (B) Existence of Kinesin-1 and Dynein-1 in the HA-p68 immunopurification was analyzed by immunoblots of the purified HA-p68 (IB:HA) using antibodies against kinesin-1 (IB:kinesin-1) and Dynein-1 (IB:Dynein-1). (C) Movements of DsRed-p68 along eGFP labeled microtubules in SW480 cells that were treated by scratch-wound were recorded by time lapse photography with confocal fluorescence microscopy (for video see on-line supplementary). In (A), arrows indicate the same positions of different time points. In (C), the arrows indicate the moved DsRed-p68. The scale bars in A and C represent 5 µm.
Table 1.
Motor activity of p68/CaM and p68-CaM fusion
| Kinesin (ATP) |
p68/Ca2+-CaM (−ATP) |
p68/Ca2+-CaM (ATP) |
His-p68-CaM (ATP) |
|
|---|---|---|---|---|
| % of gliding microtubules# (n=50) | 89.20±1.13 | 5.79±3.13 | 74.15±1.20 | 85.48±2.40 |
| Mean gliding speed* (µm/min) | 0.17±0.06 | 0.00±0.00 | 0.16±0.12 | 0.09±0.03 |
| Maximum gliding speed (µm/min) | 0.27±0.12 | 0.01±0.00 | 0.52±0.14 | 0.13±0.02 |
Only MTs with gliding speed > 0.025µM/min were counted.
Mean gliding speed only measure the MTs with the motility >0.05µM/min.
Finally, we examined whether p68 indeed migrates along microtubules in cells that are induced to migrate. p68 fused with DsRed (DsRed-p68) and eGFP labeled α-tubulin were co-expressed in SW480 cells. Cell migration was induced by scratch-wounds. Movement of the DsRed-p68 along the eGFP-labeled microtubule was recorded using confocal microscope with multi-photon excitation for imaging of living cells with time-lapse photography. The fluorescence images clearly indicated that DsRed-p68 migrated along the microtubule under cell migration induction. The movements were toward the migration leading edge (Fig. 5C, Supplementary Movie 3). The results suggest that by interacting with CaM, p68 RNA helicase can function as a microtubule motor.
PepIQ inhibits metastasis in a 4T1 orthotopic model
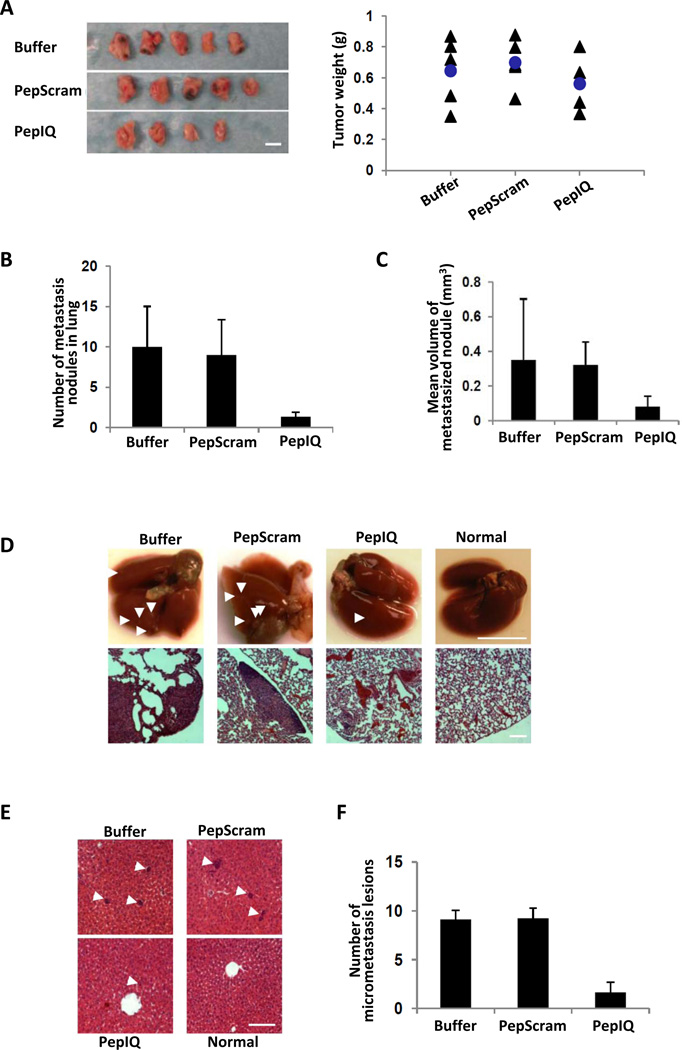
PepIQ was very effective in inhibiting cancer metastasis in a xenograft model using SW620 cells, suggesting a potential application for metastasis intervention using the peptide or its derivatives. Thus, we tested the effects of the peptide on inhibition of cancer metastasis in another cancer model. Mouse breast cancer 4T1 cells were implanted into the mammary glands of Balb/c mouse. Four days post tumor implantation, the mice were treated with buffer alone or with PepIQ or the control peptide (PepScram) at a 2 mg/kg daily dose for 14 days via i.p. injection. The mice were sacrificed 26 days after tumor implantation. The primary tumors and major metastasis organs (lungs and liver), were collected and examined. It was evident that the peptide treatments did not lead to any significant change in primary tumor growth (Fig. 6A, a tumor in one mouse in the PepIQ treated group did not grow). Strong metastasis to the lung with multiple metastasis sites was observed in the buffer and the PepScram treatment groups, while only 3 out of 4 mice displayed lung metastases in the PepIQ treated group (one mouse was completely metastasis free in the lungs). The numbers of visible metastatic nodules in the lungs of the PepIQ treated group were much less and the nodules were much smaller than those in the PepScram and buffer treated groups (Fig. 6 B, C, and D). No visible metastasis nodules were observed in livers of all mice. However, histological analyses revealed micrometastasis lesions in the livers (Fig. 6E). Quantification of the micrometastasis lesions in four randomly selected fields from three randomly selected tissue sections of liver from each mouse revealed that the average numbers of micrometastasis lesions were substantially less in the PepIQ treated group compared to those of buffer and PepScram treated groups (Fig. 6F). No micrometatstasis lesions were seen in the liver tissue sections from two out of four mice in the PepIQ treated group. The size of micrometastasis lesions in the PepIQ treated group was also substantially smaller than that of control groups (data not shown, and see Fig. 6E). To test whether PepIQ also affected the migration of 4T1 cells, we carried out cell migration assays. Both Boyden chamber and scratch-wound assays indicated that cell migration was dramatically reduced upon the PepIQ treatment (Supplementary Fig. S6 A&B).
Figure 6. PepIQ inhibits metatstasis of 4T1 tumors.
(A) Pictures (Left) and weight (Right) of harvested 4T1 tumors after 26 days growth with 14 days treatment using the indicated peptides. (B) & (C) The numbers (B) and sizes (C) of the metastasis nodules in the lung of 4T1 tumor bearing mice that were treated by different peptides (indicated) by physical inspection. (D) (Upper) representative lungs collected from 4T1 tumor bearing mice that were treated by different peptides (indicated). Arrows indicate the metastasis nodules. (Bottom) Representative images of H&E staining of tissue sections of lungs from 4T1 tumor bearing mice that were treated by different peptides (indicated). (E) Representative images of H&E staining of tissue sections of livers from 4T1 tumor bearing mice that were treated by different peptides (indicated). Arrows indicate micrometastasis lesions. (F) The numbers of the micrometastasis lesions in the tissue sections of liver of 4T1 tumor bearing mice that were treated by different peptides (indicated). The numbers of micrometastasis lesion were counted by randomly selected 4 fields from randomly selected three tissue sections of each liver. The p values (between “pepscram” and “pepIQ”, p=0.297525 in A, between “pepscram” and “pepIQ”, p=0.005257 in B, between “pepscram” and “pepIQ”, p=0.038065 in C, and between “pepscram” and “pepIQ”, ***p=1.71745E−08 in F) are calculated by the pair-wise student t-tests. The scale bars in A represent 1 cm, in D, upper panel 1 cm, lower panel 200 µm. in E represent 200 µm.
Discussion
We present evidence to demonstrate that strengthening the interaction of p68 with CaM is required for cell migration. Disrupting the p68-CaM interaction effectively inhibits cancer metastasis in two different animal models, suggesting a potential new approach for metastasis intervention. The actions of the PepIQ in inhibiting cancer metastasis provide a proof-of-principle that the strategy of disrupting the p68-CaM interaction can be a target for development of antimetastasis agents. Using the peptide may not be ideal pharmacologically. However, study with the peptide certainly provides important guidance for the further design of new molecules that can disrupt p68-CaM interaction and stop cancer metastasis.
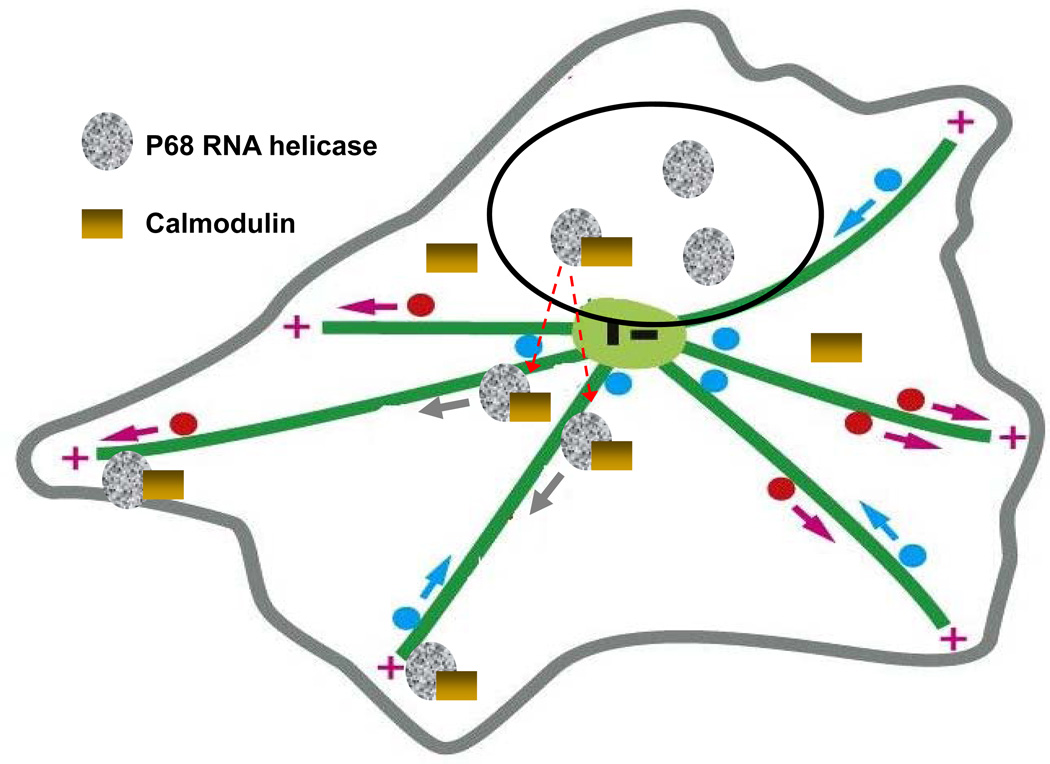
p68 was shown to interact with CaM by several independent in vitro analyses36–38. However, the actual interaction site(s) is not certain. The IQ region of p68 does not match a complete consensus sequence of IQ motif, rather an incomplete IQ-like motif characterized by (FILV)Qxxx(RK)xxxxx. The x represents any amino acid. Studies show that the IQ-like motif binds to CaM with various affinities 47,48. The biological function of p68-CaM interaction is not known. Our results suggested that the p68-CaM interaction is necessary for localization of CaM to the migration leading edge, which is an essential event during cell migration. p68, by interacting with CaM, exhibited a microtubule motor activity. Apparently, this motor activity allows CaM to be transported to the leading edge of migrating cells. Based on our current findings, our working model for the role of p68 in cell migration is illustrated in Figure 7. Interestingly, p68 stays at the leading edge of migrating cells until the cell reaches its migration destination, indicating possible additional function(s) of p68 at the migration leading edge during migration. One possibility is that p68 may also play a role in leading edge protrusion, e.g. actin filament polymerization.
Figure 7. Model of p68-calmodulin function in cell migration.
A hypothetical model that illustrates the functions of p68 and CaM interaction in cell migration, and the microtubule motor actions of p68 upon interacting with CaM
CaM regulates various cellular processes by targeting over a hundred proteins. A key feature for such functional diversity is temporal and spatial redistribution of the protein under stimulation by various different signaling pathways 24,25. A long standing question is how CaM is translocated to a specific subcellular compartments. We demonstrated an excellent example here, in which p68 interacts with CaM upon cell migration stimulation. This interaction allows p68 to interact with microtubules and to subsequently act as a microtubule motor, which consequently transports the bound CaM to the leading edge of migrating cells. An open question is whether a similar mechanism is also used to re-localize CaM in other cellular processes that require temporal and spatial redistribution of CaM, such as cytokinesis and mitosis.
It is traditionally believed that DEAD box RNA helicases use RNA as a substrate to stimulate ATPase activity. The energy is then used to displace duplex RNA or proteins that bind the RNA substrate. Along with our previously published observations 31, this is the second example showing that p68 RNA helicase can use protein as a substrate to stimulate its ATPase activity. It is conceivable that the energy derived from ATP hydrolysis can be used for translocation along the microtubule (motor like protein). However, a typical microtubule motor binds microtubules with two “motor head”. The movement along the microtubule is usually driven by cyclic hydrolysis of ATP at the two heads 49. Thus, an open question is how the p68 motor action on microtubules is achieved by one ATP hydrolysis site. One possibility is that the bound CaM may function as an anchor, while ATP hydrolysis in p68 powers the movement. This scenario would require that CaM interact with microtubules. Nevertheless, the interaction between CaM and microtubule is not observed. Alternatively, p68 RNA helicase may travel along the microtubule using a mechanism similar to that used by a DEAD box RNA helicase translocating along an RNA substrate 50.
Methods
Experimental materials
The HA peptide was synthesized by AnaSpec. Polyclonal (Pabp68) and monoclonal (P68-RGG) antibodies against p68 were the same as previously reported 32. Antibodies HA-tag, GAPDH, Lamin A/C, CaM, α-tubulin, β-tubulin, heavy chain of kinesin-1, Dynein intermediate 1, and β-actin were purchased from Cell Signaling, Abnova, SantaCruz, Abcam, and AnaSpec respectively. Microtubule and Rodamine labeled microtubule were purchased from Cytoskeleton. Cell line SW480/SW620 and 4T1 were purchased from ATCC and cultured by following the vendor’s instructions. HA-tagged p68 encoding DNA sequence was cloned into the pHM6 vector.
Calmodulin beads pull-down
Whole cell lysate from SW480 cells was prepared according to the procedure described in our previous report 31,32. The agarose beads conjugated with recombinant calmodulin (Abcam) were pre-washed with Ca2+ containing buffer (2 mM CaCl2, 50 mM NaCl in 50 mM Tris-HCl pH7.5) or EGTA containing buffer (5 mM EGTA, 50 NaCl in 50 mM Tris-HCl pH7.5). After washing, the beads were incubated with the whole cell lysates in the Ca2+ containing or EGTA containing buffer. The beads were extensively washed after incubation. Finally, 2 × SDS-loading buffer were added to the beads and the beads were heated to 85°C for 10 minutes. The proteins that were pulled down by the CaM beads were analyzed by 10% SDS-PAGE and subsequent immunoblot.
Boyden chamber assay
QCM™ 24-Well Fluorimetric Cell Migration Assay kit (ECM) was used to measure the migration of SW480 cells. The test cells were first treated under the different conditions (indicated in figure legends) in regular cell culture plates. The treated cells were re-suspended into optimum medium (without serum) and seeded into the inner chamber of the migration assay kit. The L-15 culture medium with 10% FBS was added to the outer chambers. After overnight incubation, medium in the inner chamber was removed and the cells attached to the outer bottom side were detached using the cell detachment buffer (included in the kit). The detached cells were then lysed using the cell lysis buffer (included in the kit). The amounts of the migrated cells were determined by measuring the fluorescence using λex=485nm and λem=535nm. The migration of the control cells with p68 knockdown was defined as 1.
Immunopurification of HA-tagged proteins from cell extracts
SW480 cells expressing HA-tagged proteins by transient transfection were cultured in medium L-15 with 10% FBS. The cells were subjected to different treatment (such as scratchwounds). The cells were collected and lysed in NETS buffer with addition of the protease inhibitors. To minimize the non-specific protein-protein interactions in the immunopurification, 600 mM (final concentration) 6-aminocaproic acid (6-AcA) along with 800 mM (final concentration) NaCl were added to the extracts and incubated for 2 hours at RT. A commercially available polyclonal anti-HA antibody was added to the prepared extracts (500 µl extracts/20 µl of Abs). The mixture was incubated overnight at 4 °C. After incubation, the protein G agarose beads (50 µl, pre-treated with BSA 3 mg/ml) were then added to the mixture and incubated for an additional 2 hours. The beads were washed in NETS buffer 6 times. The beads bound proteins were either directly used in some assays (such as ATPase assays) or the bound HA-tagged proteins were eluted using the HA peptide. The beads with bound proteins were suspended in elution buffer and packed in a small column. The HA peptide (Sigma) (500 µg/ml in elution buffer) was flowed through the column to elute the bead bound proteins. The fractions were collected and analyzed by SDS-PAGE and immunoblot to identify the fractions that contained HA-tagged protein. The eluted proteins were further dialyzed in the subsequent assay buffers (such as MT binding buffer) and concentrated by centrifugation.
ATPase assays
The ATPase activity of different proteins (specified in figures and legends) was assayed by an experimental procedure similar to that described previously with minor modifications 44,46. The reaction volume was 50 µl, containing 4 µg of MTs (or 2 µg of RNA), 4 mM ATP, and 0.5 mM Ca2+ or 5mM EGTA in the ATPase assay buffer (10mM Pipes pH 7.0, 1mM MgCl2, 25mM NaCl with taxol added). 10 µl of the purified HA tagged proteins (bound or un-bound to agarose beads) was added. The ATPase reactions were incubated for 30 minutes at 37°C. After incubation, 30 µl of the supernatant was added to 1ml of malachitegreen-molybdenum reagent. After 5 minutes incubation at room temperature, the absorbance of the reaction was then measured at 630nm (A630). The concentrations of released inorganic phosphate were determined by matching the A630nm in a standard curve of A630nm vs known phosphate concentrations.
Metastasis inhibition of xenograft tumors
All animal experiments were carried out in accordance with the guidelines of IACUC of Georgia State University. Nude mice or Balb/c mice (6 to 7 weeks old, Harlan Laboratory) were subcutaneously or orthotopically injected with 5 × 106 SW620 or 2 × 106 4T1 cells. Peptides at appropriate doses were i.p. injected to the tumor bearing mice at appropriate time intervals. Tumor formation and volumes were assessed every 3 days. Tumor volumes were measured by two perpendicular diameters of the tumors over a 4-week time course with the formula 4p/3 × (width/2)2 × (length/2). The tumors and organs were collected and weighed at the end of the experiments. Tissue sections were prepared from harvested tumors and organs. Tissue sections were analyzed by either H&E or immune fluorescence staining (IF). For metastasis analyses of SW620 tumors, tissue extracts were prepared from spleens, total RNAs were extracted from the tissue extracts. RT-PCRs were performed with the isolated RNAs. For metastasis of 4T-1 cells, lung and liver were inspected. Tissue sections were also prepared and analyzed by H&E or immune staining. Statistical analyses were done in comparison to the control group with a paired Student’s t test.
Scratch-wound treatments
A scratch or multiple scratches using pipette tips were introduced into a 6 well cell culture plate. The cells were further cultured under serum starvation for 30 minutes, and were subsequently cultured for an additional indicated time. The scratch-treated cells were either directly visualized by microscopes or the cells were fixed and subsequently examined by immunostaining. Alternatively, the cells were collected and lysized. The prepared cellular extracts were then used for other analyses.
Microtubule pull-down and gliding assay
Microtubule purchased was reconstituted in warm PM buffer (15 mM Pipes pH 7.0, 1 mM MgCl2) with the addition of taxol as instructed by the vendor. The MTs were ready to use after 15 minutes incubation at room temperature. The test protein(s) was incubated with the reconstituted MTs under various conditions. The MTs with the bound proteins were separated from the incubation solution by centrifugation at 100,000g for 30 minutes. Co-precipitates with the MTs were re-suspended in 50 µl of 2 × SDS PAGE loading buffer and heated to 85 °C for 10 minutes and subsequently analyzed by immunoblots.
The protein HA-p68-CaM, HA-p68, or His-p68-CaM (10 µl) (pre-treated by RNase cocktail, RNase A, T1, and V1) was loaded onto a glass slide. The slide was blocked with the blocking buffer. For the slide loaded with HA-p68, 10 µl of calmodulin with 0.5 mM of Ca2+ was added to the slide and was incubated for 10 minutes. After loading the slide was washed with the Ca2+ containing wash buffer to remove the unbound proteins. The prepared Rodamine labeled microtubules were then loaded onto the slide. The same Ca2+ containing wash buffer was used to remove unbound microtubules. The movement of microtubules was then recorded by the confocal microscope.
Supplementary Material
Acknowledgments
We thank Birgit Neuhaus, Yun Huang, and Jin Zhou for assistance in microscopic imaging. This manuscript is greatly improved by comments from Dan Adams and Michael Kirberger. This work is supported in part by research grants from National Institute of Health (CA118113) and Georgia Cancer Coalition to ZR Liu. X. Gao and H. Wang are supported by a MBD fellowship, GSU.
Footnotes
Contributions
ZR Liu conceptualized, planned, and coordinated the study. ZR Liu wrote the paper. HZ. Wang and X. Gao conducted most of experiments, data analyses, and participated in paper writing; Jenny Yang conducted the computation modeling of p68 structure, participated in CaM binding, image analyses, and paper writing. All authors discussed the results and commented on the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Vicente-Manzanares M, Webb DJ, Horwitz AR. Cell migration at a glance. J Cell Sci. 2005;118:4917–4919. doi: 10.1242/jcs.02662. [DOI] [PubMed] [Google Scholar]
- 2.Montell DJ. Morphogenetic cell movements: diversity from modular mechanical properties. Science. 2008;322:1502–1505. doi: 10.1126/science.1164073. [DOI] [PubMed] [Google Scholar]
- 3.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 4.Zoller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 5.Kurosaka S, Kashina A. Cell biology of embryonic migration. Birth Defects Res C Embryo Today. 2008;84:102–122. doi: 10.1002/bdrc.20125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bretscher MS. Recap on cell migration. Traffic. 2008;9:198–199. doi: 10.1111/j.1600-0854.2007.00676.x. [DOI] [PubMed] [Google Scholar]
- 7.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer TD, Ashby WJ, Lewis JD, Zijlstra A. Targeting tumor cell motility to prevent metastasis. Adv Drug Deliv Rev. 2011;63:568–581. doi: 10.1016/j.addr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai YC, Weissman AM. Dissecting the diverse functions of the metastasis suppressor CD82/KAI1. FEBS Lett. 2011;585:3166–3173. doi: 10.1016/j.febslet.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 11.Webb DJ, Parsons JT, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells -- over and over and over again. Nat Cell Biol. 2002;4:E97–E100. doi: 10.1038/ncb0402-e97. [DOI] [PubMed] [Google Scholar]
- 12.Horwitz R, Webb D. Cell migration. Curr Biol. 2003;13:R756–R759. doi: 10.1016/j.cub.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 14.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 15.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Roy C, Wrana JL. Signaling and endocytosis: a team effort for cell migration. Dev Cell. 2005;9:167–168. doi: 10.1016/j.devcel.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 18.de Curtis I. Cell migration: GAPs between membrane traffic and the cytoskeleton. EMBO reports. 2001;2:277–281. doi: 10.1093/embo-reports/kve072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 20.Lowery LA, Van Vactor D. The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10:765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 22.Vallee RB, Seale GE, Tsai JW. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 2009;19:347–355. doi: 10.1016/j.tcb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 24.Pezzella-D'Alessandro N, et al. Calmodulin distribution and the actomyosin cytoskeleton in Toxoplasma gondii. J Histochem Cytochem. 2001;49:445–454. doi: 10.1177/002215540104900404. [DOI] [PubMed] [Google Scholar]
- 25.Shariat-Madar Z, Goldsmith AM, Gnegy ME. Effect of continuous phorbol ester treatment on muscarinic receptor-mediated calmodulin redistribution in SK-N-SH neuroblastoma cells. J Neurochem. 1997;68:40–46. doi: 10.1046/j.1471-4159.1997.68010040.x. [DOI] [PubMed] [Google Scholar]
- 26.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 27.Hinrichsen RD. Calcium and calmodulin in the control of cellular behavior and motility. Biochim Biophys Acta. 1993;1155:277–293. doi: 10.1016/0304-419x(93)90010-a. [DOI] [PubMed] [Google Scholar]
- 28.Collins SR, Meyer T. Calcium flickers lighting the way in chemotaxis? Dev Cell. 2009;16:160–161. doi: 10.1016/j.devcel.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei C, et al. Calcium flickers steer cell migration. Nature. 2009;457:901–905. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans JH, Falke JJ. Ca2+ influx is an essential component of the positive-feedback loop that maintains leading-edge structure and activity in macrophages. Proc Natl Acad Sci U S A. 2007;104:16176–16181. doi: 10.1073/pnas.0707719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, Lin C, Liu ZR. P68 RNA Helicase Mediates PDGF-Induced Epithelial Mesenchymal Transition by Displacing Axin from beta-Catenin. Cell. 2006;127:139–155. doi: 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 32.Yang L, Lin C, Liu ZR. Phosphorylations of DEAD box p68 RNA helicase are associated with cancer development and cell proliferation. Mol Cancer Res. 2005;3:355–363. doi: 10.1158/1541-7786.MCR-05-0022. [DOI] [PubMed] [Google Scholar]
- 33.Carter CL, Lin C, Liu CY, Yang L, Liu ZR. Phosphorylated p68 RNA helicase activates Snail1 transcription by promoting HDAC1 dissociation from the Snail1 promoter. Oncogene. 2010;29:5427–5436. doi: 10.1038/onc.2010.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu K, McDuffie E, Abrams SI. Exposure of human primary colon carcinoma cells to anti-Fas interactions influences the emergence of pre-existing Fas-resistant metastatic subpopulations. J Immunol. 2003;171:4164–4174. doi: 10.4049/jimmunol.171.8.4164. [DOI] [PubMed] [Google Scholar]
- 35.Wels J, Kaplan RN, Rafii S, Lyden D. Migratory neighbors and distant invaders: tumor-associated niche cells. Genes Dev. 2008;22:559–574. doi: 10.1101/gad.1636908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen X, Valencia CA, Szostak JW, Dong B, Liu R. Scanning the human proteome for calmodulin-binding proteins. Proc Natl Acad Sci U S A. 2005;102:5969–5974. doi: 10.1073/pnas.0407928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang DJ, Guo M, Wang D. Proteomic and biochemical studies of calcium- and phosphorylation-dependent calmodulin complexes in Mammalian cells. J Proteome Res. 2007;6:3718–3728. doi: 10.1021/pr0703268. [DOI] [PubMed] [Google Scholar]
- 38.Buelt MK, Glidden BJ, Storm DR. Regulation of p68 RNA helicase by calmodulin and protein kinase C. J Biol Chem. 1994;269:29367–29370. [PubMed] [Google Scholar]
- 39.Bahler M, Rhoads A. Calmodulin signaling via the IQ motif. FEBS Lett. 2002;513:107–113. doi: 10.1016/s0014-5793(01)03239-2. [DOI] [PubMed] [Google Scholar]
- 40.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 41.White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583:1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tirnauer JS. A new cytoskeletal connection for APC: linked to actin through IQGAP. Dev Cell. 2004;7:778–780. doi: 10.1016/j.devcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 43.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin C, Yang L, Yang JJ, Huang Y, Liu ZR. ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol Cell Biol. 2005;25:7484–7493. doi: 10.1128/MCB.25.17.7484-7493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolsover SR. Calcium signalling in growth cone migration. Cell Calcium. 2005;37:395–402. doi: 10.1016/j.ceca.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Huang Y, Liu ZR. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J Biol Chem. 2002;277:12810–12815. doi: 10.1074/jbc.M200182200. [DOI] [PubMed] [Google Scholar]
- 47.Macdougall DA, Wachten S, Ciruela A, Sinz A, Cooper DM. Separate elements within a single IQ-like motif in adenylyl cyclase type 8 impart ca2+/calmodulin binding and autoinhibition. J Biol Chem. 2009;284:15573–15588. doi: 10.1074/jbc.M809585200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong Q, et al. Regulation of the transient receptor potential channel TRPM2 by the Ca2+ sensor calmodulin. J Biol Chem. 2006;281:9076–9085. doi: 10.1074/jbc.M510422200. [DOI] [PubMed] [Google Scholar]
- 49.Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Current opinion in cell biology. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fairman ME, et al. Protein displacement by DExH/D "RNA helicases" without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.