Abstract
Structural studies of the cystic fibrosis transmembrane conductance regulator (CFTR) are reviewed. Like many membrane proteins, full-length CFTR has proven to be difficult to express and purify, hence much of the structural data available is for the more tractable, independently expressed soluble domains. Therefore, this chapter covers structural data for individual CFTR domains in addition to the sparser data available for the full-length protein. To set the context for these studies, we will start by reviewing structural information on model proteins from the ATP-binding cassette (ABC) transporter superfamily, to which CFTR belongs.
Structural data for full-length CFTR are limited; however, data for individual CFTR domains and related ABC transporters are available. The structure of CFTR is likely to resemble that of the bacterial efflux transporter Sav1866 and the human multidrug efflux transporter P-glycoprotein.
INSIGHT FROM STRUCTURES OF MODEL ABC TRANSPORTERS
Domain Organization of ABC Transporters
The ABC transporter superfamily is characterized by a stereotyped ATP-binding cassette (ABC), alternatively called a nucleotide-binding domain (NBD), that is readily identified via simple sequence-homology searches (Higgins 1992; Linton and Higgins 1998; Dassa and Bouige 2001; Dean et al. 2001; Kerr 2002; Davidson and Chen 2004; Jones and George 2004; Dean 2005; Dean and Annilo 2005). This domain contains a series of signature sequences involved in binding ATP (described below) that enable an expert to identify most representatives based on visual inspection of the amino acid sequence (Hung et al. 1998; Karpowich et al. 2001; Smith et al. 2002). The first reported sequence of a mammalian ABC transporter was that of P-glycoprotein (Pgp, also called ABCB1) (Dean et al. 2001; Dean and Annilo 2005), a multidrug resistance protein overexpressed in a variety of advanced cancers. The publication of the Pgp sequence in 1986 represented a breakthrough in the application of molecular biology methods to understanding human disease (Fojo et al. 1985; Gerlach et al. 1986; Chen et al. 1986; Roninson et al. 1986). The identification of the human cystic fibrosis transmembrane conductance regulator (CFTR, also called ABCC7) (Dean et al. 2001; Dean and Annilo 2005), reported in 1989 (Riordan et al. 1989), represented another breakthrough marking the dawn of the era of genomic medicine. Although protein sequencing had led to the characterization of the molecular basis of other human diseases, the discovery of CFTR represented the first report of the DNA sequence of a human disease-causing gene. Most remarkably, the sequence of CFTR showed strong homology to that of Pgp as well as to a series of bacterial proteins involved in transmembrane active transport (Riordan et al. 1989; Higgins 1992; Dean et al. 2001). As sequence databases expanded, it became clear that these proteins belong to a ubiquitously distributed protein superfamily that came to be known as the ABC transporters (Higgins 1992; Saurin et al. 1999; Dean et al. 2001). Although some superfamily members are involved in DNA repair and protein translation (Kerr 2004), the vast majority are transmembrane proteins that function as ATP-dependent active transporters (Higgins 1992; Saurin et al. 1999; Dean et al. 2001; Davidson and Chen 2004; Jones and George 2004). Although the NBDs from ABC transporters share strongly conserved structures, the transmembrane domains (TMDs) are more diverse and come from a variety of sequence/structure families.
Although CFTR functions as an ATP-gated chloride channel (Rich et al 1990; Anderson et al. 1991a,b; Drumm et al. 1991) and is not believed to mediate active transport (Gadsby et al. 2006; Riordan 2008; Kim Chiaw et al. 2011), it shares the conserved domain architecture (Fig. 1A–D) characteristic of the ATP-dependent transporters that represent the vast majority of the ABC superfamily (Higgins 1992; Davidson and Chen 2004; Jones and George 2004; Hollenstein et al. 2007a; Cui and Davidson 2011; Zolnerciks et al. 2011; George and Jones 2012). This architecture involves a dimeric organization of TMDs (Fig. 1B,D) and tightly associated NBDs (Fig. 1B–D) that are encoded in anything from one to four polypeptide chains (Higgins 1992; Dean et al. 2001). In all ABC transporters, the TMDs and NBDs both interact with one another with approximate twofold symmetry (Fig. 1B,D), but the interacting domains can be either identical in sequence (i.e., making a homodimeric interaction) or homologous but divergent in sequence (i.e., making a heterodimeric interaction). Although bacterial ABC importers exhibit more diverse schemes of covalent organization (Higgins 1992; Linton and Higgins 1998; Saurin et al. 1999), human ABC transporters like bacterial ABC exporters have their TMDs and NBDs fused in a single polypeptide chain (Dean et al. 2001; Dean and Annilo 2005). In some human ABC transporters, including CFTR (Fig. 1A), both TMDs (TMD1 and TMD2) and both NBDs (NBD1 and NBD2) are fused in a single polypeptide chain at least 1100 amino acids in length, producing an effectively heterodimeric functional organization. The binding and hydrolysis of two ATP molecules at the interface between the homodimeric or heterodimeric NBDs drives functional conformational changes in ABC proteins (Fig. 1D) (Jones and George 1999; Smith et al. 2002; Davidson and Chen 2004; Vergani et al. 2005; Riordan 2008), as discussed in greater detail below. In the case of CFTR, the ATP-gated chloride channel resides in the interface between the heterodimeric TMDs (Anderson et al. 1991b; Mansoura et al. 1998; Linsdell et al. 2000, 2006; Liu et al. 2003, 2004; Alexander et al. 2009; Liu and Dawson 2011). The transported substrate binds at the equivalent location in the ATP-dependent transporters in the ABC superfamily (Davidson and Chen 2004; Hollenstein et al. 2007a; Cui and Davidson 2011; Kim Chiaw et al. 2011; Zolnerciks et al. 2011; George and Jones 2012).
Figure 1.
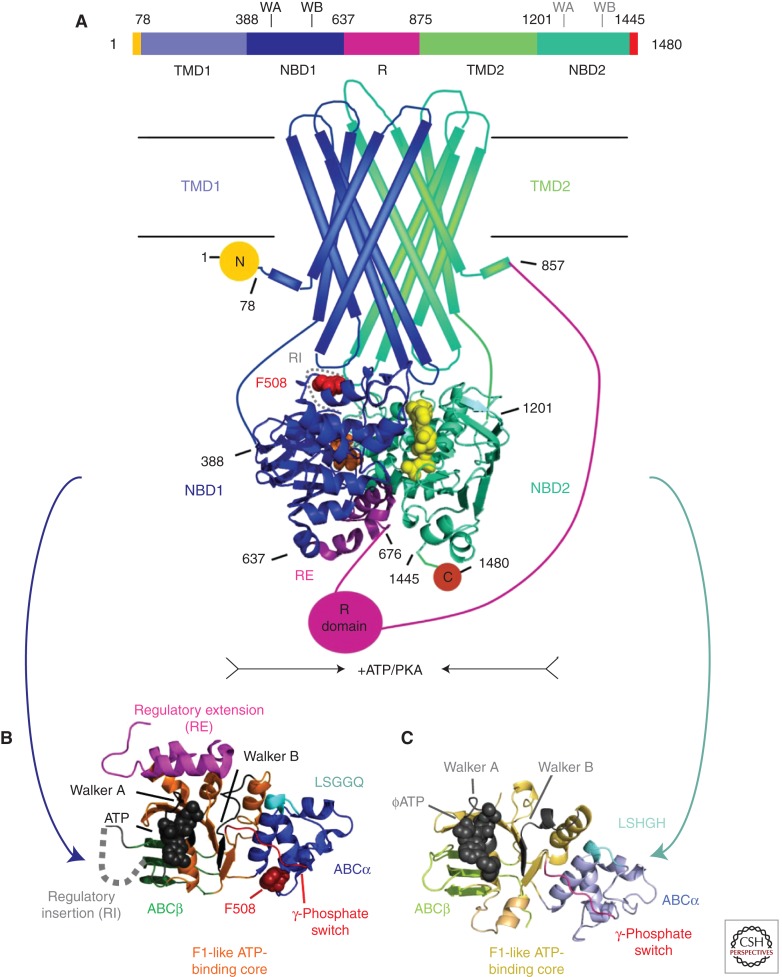
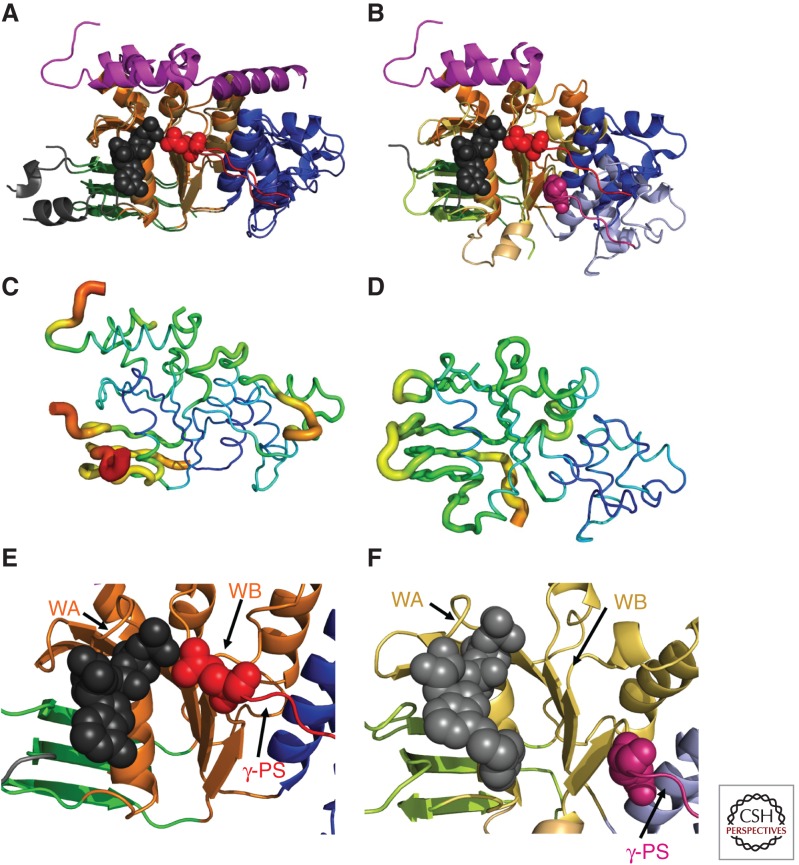
Domain organization of CFTR and simplified consensus gating model. (A) The three-dimensional domain organization of CFTR is schematized below a bar showing its linear sequence organization. NBD stands for nucleotide-binding domain, TMD for transmembrane domain, R for regulatory domain, RI for the regulatory insertion in NBD1, RE for the regulatory extension at the junction between NBD1 and the R domain, and N and C for the amino- and carboxy-terminal cytoplasmic tails, respectively. ATP is shown in orange (bound to the Walker motifs in NBD1) or yellow (bound to the Walker motifs in NBD2) space-filling representation. The NBDs are represented by ribbon diagrams of crystal structures of the isolated human domains (PDB IDs 2BBO for hNBD1 and 3GD7 for hNBD2), which are shown interacting in an ATP-sandwich heterodimer conformation modeled based on least-squares alignment to the crystal structure of the low-affinity homodimer of hNBD1-Δ(RI,RE) (PDB ID 2PZE). PKA stands for protein kinase A, which uses ATP to phosphorylate and thereby activate the CFTR channel; phosphorylation presumably occurs primarily on residues in the R domain. (B) Ribbon diagram showing the subdomain organization of hNBD1 (PDB ID 2BBO), with the F1-like ATP-binding core subdomain shown in orange, the ABCβ subdomain in dark green, the ABCα subdomain in blue, the γ-phosphate switch in light red, the LSGGQ signature sequence in light cyan, the RE in magenta, the RI in gray (depicted as a dotted line), and Mg-ATP in black space-filling representation. Residue F508 (dark red) and its Walker A and B motifs are labeled. (C) Ribbon diagram showing the subdomain organization of hNBD2 (PDB ID 3GD7), with the F1-like ATP-binding core subdomain shown in orange, the ABCβ subdomain in light green, the ABCα subdomain in gray-blue, the γ-phosphate switch in magenta, the LSHGH signature sequence in dark cyan, and Mg-ϕATP (phenyl-ATP) in black space-filling representation. Its Walker A and B motifs are labeled. (D) Schematic diagrams illustrating a simplified version of the accepted model for the mechanochemistry of CFTR channel gating. The upper row shows a top view looking down on the NBDs from the cytoplasm, whereas the lower row shows a side view of the transmembrane structure (with the phospholipid bilayer schematized as two parallel black lines). The coloring is equivalently to panel A except for ATP, which is shown here in red. The opening of the transmembrane chloride channel in CFTR is gated by mechanical reorientation of the NBDs upon ATP binding at their interface, which drives formation of an ATP-sandwich heterodimer structure. Phosphorylation of the R domain by PKA is believed to facilitate channel opening at least in part by disrupting binding interactions between polypeptide segments in the R domain and surfaces in the NBDs mediating tight interdomain structural interactions in the open conformation of the channel.
Some ABC transporters contain additional domains, most frequently at their amino terminus or immediately after their first NBD. CFTR has such a domain called the regulatory or R domain, which is ∼240 residues in length and inserted between NBD1 and TMD2. Its presence produces the following overall amino-terminal to carboxy-terminal domain organization in CFTR: TMD1, NBD1, R, TMD2, and finally NBD2 (Riordan et al. 1989; Gadsby et al. 2006; Riordan 2008) (Fig. 1A). CFTR additionally has an amino-terminal extension of ∼80 residues and a carboxy-terminal extension of ∼30 residues. These terminal sequences are likely disordered except when interacting with specific protein-binding partners. Similarly, the R domain is predominantly disordered (Ostedgaard et al. 1997; Baker et al. 2007; Kanelis et al. 2010; Lewis et al. 2010), at least in isolation from the other domains of CFTR, which has led to the proposal that it should be renamed the R region rather than the R domain. Low-resolution electron microscopy data on CFTR molecules with the R domain labeled with 1.8-nm immuno-gold spheres has identified a relatively well-defined location for some of its residues relative to the other domains in full-length CFTR (Zhang et al. 2009, 2010), as discussed further below. Furthermore, NMR data show some interaction of the isolated R domain with isolated CFTR nucleotide-binding domains when mixed together in vitro (Baker et al. 2007).
The R domain plays a central role in mediating activation of CFTR by protein kinase A (PKA) (Gregory et al. 1990; Zhu et al. 2002; Chappe et al. 2005; Csanády et al. 2005) and may function in other regulatory processes as well (Chappe et al. 2004). It has a series of phosphorylation sites and is believed to act as an integrator of PKA signaling and perhaps also signaling from some other physiological systems. Although full-length CFTR has minimal basal activity prior to phosphorylation by PKA, deletion of the R domain leads to 50% of maximal activity in the absence of phosphorylation (Chappe et al. 2005; Csanády et al. 2000). Therefore, the R domain acts primarily although perhaps not exclusively by inhibiting channel activation when it is in the unphosphorylated state. These inhibitory interactions probably involve binding to interdomain interfaces in CFTR and thereby blocking the changes in interdomain interactions required to open the chloride channel (Lewis et al. 2004; Baker et al. 2007). One specific site on CFTR believed to participate in such an inhibitory interaction is schematized in Figure 1D and described below.
Structural Organization of the NBDs from ABC Transporters
Several key features of the structural organization of the NBDs from ABC transporters were inferred from sequence analysis prior to the first experimental structure determination for any superfamily member (Hobson et al. 1984; Thomas et al. 1991; Mourez et al. 1997; Hung et al. 1998). Their sequences contain the canonical Walker A and Walker B motifs (Fig. 1A,C, and D) present in a variety of different ATPase superfamilies (Walker et al. 1982), including the superfamily I and II helicases (Hunt et al. 2002) and the AAA ATPases (Snider and Houry 2008). The residues from these motifs, which were first identified in the F1 ATPase (Walker et al. 1982), ligate the Mg-ATP substrate in a conserved manner in all F1-like ATPase superfamilies including the ABC transporters. The Walker A motif (GxxGxGKT, with x being any residue) ligates the α- and β-phosphates of ATP, while the aspartate residue at the end of the Walker B motif (ϕ4D, with ϕ being a hydrophobic residue) ligates its tightly bound Mg2+ cofactor. In ABC domains, the Walker B is followed immediately by a glutamate residue (i.e., ϕ4DE) that was recognized as being invariant in sequence alignments long before crystallographic studies showed that it is the catalytic base that activates a water molecule for hydrolysis of ATP (Hung et al. 1998; Smith et al. 2002; Moody et al. 2002). The conservation of the Walker A and B motifs in the NBDs from ABC transporters combined with secondary structure analyses led to the inference that the ATP-binding core of the domain would have the same fold as F1 ATPase and other F1-like ATPase superfamilies, an inference later verified by the crystal structures of isolated NBDs from bacterial ATP transporters (e.g., as shown for the NBDs from CFTR in Fig. 1B,C) (Hung et al. 1998; Karpowich et al. 2001).
These structures also verified the sequence-based inference that an α-helical subdomain unique to the ABC superfamily is inserted into topology of the F1-like ATP-binding core in the NBDs from ABC transporters (Fig. 1B,C) (Mourez et al. 1997). This “ABCα subdomain” (Karpowich et al. 2001) contains the so-called “signature sequence” of ABC transporters, a nearly invariant pentapeptide motif with sequence LSGGQ that precedes the Walker B motif in ABC domains by ∼20 amino acids (Higgins 1992; Jones and George 1999). Enzymological studies of model ABC transporters showed that this sequence, sometimes called the C motif, is essential for catalysis of ATP hydrolysis as well as substrate transport (as reviewed in Davidson and Chen 2004), leading to expectations that it would form part of the ATPase active site in ABC transporters (Jones and George 1999). In transporters with heterodimeric NBDs, mutation of one of the two LSGGQ motifs was shown to be sufficient to block activity (Davidson and Sharma 1997). This observation reinforced the inference, from Michaelis-Menten analyses showing twofold kinetic cooperativity (Davidson et al. 1996), that the functional organization of ABC transporters involves participation of the signature sequence in some kind of dimeric structural interaction (Jones and George 1999). Insightful analyses of bacterial ABC transporters suggested that the ABCα subdomain also makes important structural contacts to the TMDs (Mourez et al. 1997), another inference later verified by structural studies (Locher et al. 2002; Dawson and Locher 2006). These contacts are proximal to the site of the predominant disease-causing F508del mutation in CFTR (Fig. 1B), as discussed in detail below.
The first X-ray crystal structures of isolated NBDs from model bacterial ABC transporters verified most of these prior inferences concerning domain architecture but also provided some surprises (Hung et al. 1998; Karpowich et al. 2001). The core of the NBD indeed has an identical topology to that of the catalytic subunit of F1 ATPase, and it ligates the α/β-phosphates and Mg2+-cofactor of ATP with equivalent stereochemistry (Fig. 1B,C). As predicted, this F1-like ATP-binding core of the NBD is interrupted by the insertion of a bundle of three α-helices, the ABCα subdomain, between two of the six conserved β-strands in the parallel β-sheet forming the core of the domain (Karpowich et al. 2001).
Unexpectedly, the amino terminus of the NBD contains an antiparallel β-sheet, the ABCβ subdomain, formed in large part by residues preceding the F1-like ATP-binding core (Karpowich et al. 2001). Like the ABCα subdomain, the ABCβ subdomain is unique to the NBDs from ABC superfamily members and is not found in other F1-like ATPase superfamilies. The ABCβ subdomain contributes to forming the ATP-binding site in ABC transporters. A conserved aromatic residue at the carboxyl terminus of the first β-strand in the ABCβ subdomain makes a base-stacking interaction with the adenine of the ATP, and this interaction positions the entirety of the nucleotide in a solvent-exposed position on the surface of the domain, in a substantially different geometry from that observed in other F1-like ATPases (Hung et al. 1998; Karpowich et al. 2001).
The most surprising feature in the crystal structures of isolated NBDs from a series of model bacterial ABC transporters was their failure to form a consistent dimeric structure (Hung et al. 1998; Diederichs et al. 2000; Karpowich et al. 2001). In the protomer structures, the LSGGQ signature sequence is at least 15 Å away from the ATP-binding site (Fig. 1B,C). Given the well-established importance of this motif for catalytic activity (Davidson and Chen 2004), this structural observation suggested that there are likely to be conserved oligomeric interactions in functional ABC transporters that were not recapitulated in the early structures of isolated NBDs. One specific prediction that proved to be highly insightful was that the LSSGQ signature sequence in one protomer would interact with the ATP molecule bound to the Walker motifs in a second protomer in the functional dimeric conformation of ABC transporters (Jones and George 1999).
Another noteworthy feature in the crystal structures of isolated NBDs from ABC transporters was a wide variation in the alignment of the ABCα subdomain relative to the F1-like ATP-binding core of the domain (Diederichs et al. 2000; Karpowich et al. 2001; Yuan et al. 2001). Different crystallographic snapshots, even of the same NBD, showed up to 20° relative rotations of the ABCα subdomain. These rotations are clearly correlated with a change in the conformation of the protein loop connecting the F1-like core to the amino terminus of the ABCα subdomain. In ATP-bound structures, an invariant glutamine at the amino terminus of this loop contacts the Mg2+ counterion bridging the β- and γ- phosphates of the ATP molecule (Hung et al. 1998; Smith et al. 2002), and this interaction holds the ABCα subdomain in a consistent orientation in close proximity to the F1-like ATP-binding core of the domain. In contrast, in ADP-bound structures, in which the position of the Mg2+ cofactor shifts because of the absence of a γ-phosphate group, the ABCα subdomain is observed in a variety of different orientations, usually further away from the ATP-binding core (Diederichs et al. 2000; Karpowich et al. 2001; Yuan et al. 2001). Given the associated conformational change in the protein loop connecting the F1-like core subdomain to the ABCα subdomain, combined with the critical role played by the contact between the γ-phosphate group of ATP and the invariant glutamine at the amino terminus of this loop in controlling its conformation, this loop is alternatively called the γ-phosphate switch (Karpowich et al. 2001; Smith et al. 2002) or the Q-loop (Hopfner et al. 2000). The function of the γ-phosphate-dependent ABCα subdomain rotation is not understood in detail, but it has been proposed to function in facilitating release of the ADP product from the active site or in allosteric activation of ATPase activity in response to the binding of transport substrate (i.e., by modulating ATP-binding affinity in response to allosteric communication from the TMDs) (Karpowich et al. 2001; Yuan et al. 2001).
NBD Mechanochemistry
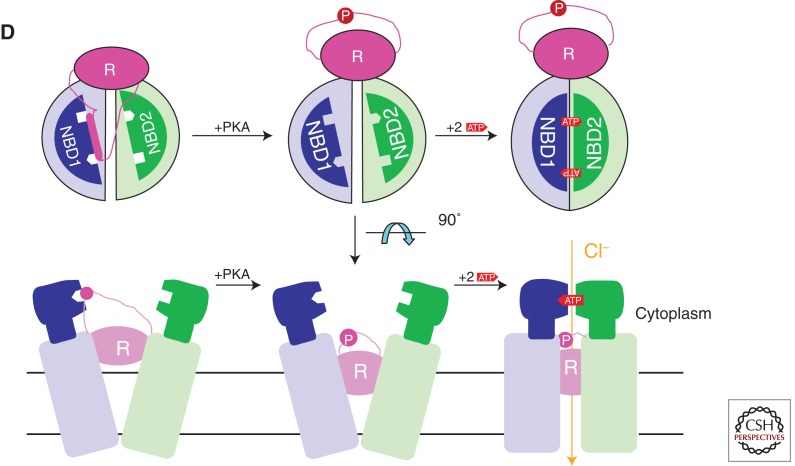
The foundation of the mechanochemistry of ABC transporters is the formation of an “ATP-sandwich dimer” by NBDs upon binding Mg-ATP (Jones and George 1999; Smith et al. 2002), as schematized in Figure 1D. This structure, which is sometimes called a “head-to-tail” NBD dimer, has two Mg-ATP molecules bound in the inter-NBD interface, with each molecule sandwiched between the Walker A/B motifs in one protomer and the LSGGQ signature sequence in the other protomer. The NBDs forming this structure can be either homodimeric (e.g., in most bacterial ABC transporters) or heterodimeric (as shown for CFTR in Fig. 1D). The binding of Mg-ATP to nucleotide-free NBDs drives their reorientation to form the ATP-sandwich dimer, and this ATP-driven reorientation provides a mechanical “power stroke” that is propagated to the attached TMDs to drive solute translocation in transporters (Smith et al. 2002; Oldham et al. 2011). A similar ATP-driven conformational rearrangement of the TMDs coupled to formation of the ATP-sandwich dimer by NBD1 and NBD2 is believed to mediate channel gating in CFTR (Fig. 1D) (Aleksandrov et al. 2002a,b, 2009; Vergani et al. 2005; Mense et al. 2006; Riordan 2008).
This widely accepted mechanochemical model was established primarily via studies of glutamate-to-glutamine (E-to-Q) mutations in the catalytic base that activates water for hydrolytic attack on ATP (Moody et al. 2002; Smith et al. 2002). As described above, in the NBDs from ABC transporters, this catalytic glutamate immediately follows the aspartate residue at the end of the Walker B motif, yielding a conserved hexapeptide sequence ϕ4DE (with ϕ being a hydrophobic residue). In a wide variety of ABC transporters, the E-to-Q mutation of the catalytic glutamate blocks the hydrolysis of ATP while preserving its high-affinity binding (Smith et al. 2002; Vergani et al. 2005; Oldham et al. 2007). The mechanochemistry of ABC transporters relies on the energy of ATP binding to drive functional conformational changes during formation of the ATP-sandwich dimer by the NBDs (as schematized in Fig. 1D). The energy of ATP binding to wild-type NBDs is very difficult to assess because of the transient lifetime of the prehydrolysis complex, preventing quantitative evaluation of the effect of the E-to-Q mutation on ATP-binding affinity. However, this mutation has enabled kinetic trapping and characterization of the functional ATP-bound conformation of a wide variety of ABC transporters (Smith et al. 2002; Janas et al. 2003; Vergani et al. 2005; Oldham et al. 2007; Zutz et al. 2010) and other ATPases in the ABC superfamily (Barthelme et al. 2011) (G Boel, PCM Smith, and JF Hunt, unpubl.). In some NBDs, similar thermodynamic, kinetic, and structural results are produced by introducing a histidine-to-alanine (H-to-A) mutation in an invariant active-site residue that hydrogen bonds (H-bonds) directly to the γ-phosphate of ATP (Zaitseva et al. 2005). In contrast, nonhydrolyzable analogs of ATP, all of which change the identity of one of the five atoms in its γ-phosphate group (i.e., the atoms distinguishing it from its hydrolysis product ADP), show variable and often poor efficacy in recapitulating the functional consequences of ATP binding (Hung et al. 1998; Moody et al. 2002; Smith et al. 2002; Horn et al. 2003). In this context, the binding of unmodified ATP to NBDs harboring the E-to-Q or H-to-A mutation in their active site is assumed to preserve affinity better than the binding of nonhydrolyzable analogs of ATP to wild-type NBDs.
A dramatic example of the difference between the two approaches to stabilizing the functional ATP-bound conformation of ABC proteins is provided by the early studies of isolated NBDs that led to the now standard model for ABC mechanochemistry. When expressed in the absence of their cognate TMDs, the NBDs from ABC transporters consistently purify as monomers with a minimal tendency to interact with one another. They remain monomeric in the presence of ADP or nonhydrolyzable analogs of ATP (Hung et al. 1998; Moody et al. 2002). In contrast, several NBDs harboring the E-to-Q mutation in their catalytic base, but not the corresponding wild-type NBDs, form hyperstable dimers on binding unmodified ATP (Moody et al. 2002; Smith et al. 2002). This phenomenon was used to determine the first crystal structure of an ATP-sandwich dimer of the NBDs from an ABC transporter, that of the MJ0796 protein from Methanococcus jannaschii (Smith et al. 2002). This model NBD comes from a homodimeric transporter likely involved in lipoprotein transport. Its crystal structure confirmed the earlier prediction that the LSGGQ signature sequence in one NBD directly ligates the ATP molecule bound to the Walker A/B motifs in another NBD in the functional dimeric state of ABC transporters (Jones and George 1999).
Crystallographic and thermodynamic studies on the E-to-Q mutant of MJ0796 suggest that electrostatic charge balance in the active site plays a critical role in modulating ABC mechanochemistry (Smith et al. 2002). The mutant protein, which removes a negative charge from the active site, binds sodium ATP (Na-ATP) rather than Mg-ATP in the ATP-sandwich dimer. Therefore, by compensating for the loss of a negative charge on the protein by the loss of a positive charge on the cofactor of the bound nucleotide, this hyperstable E-to-Q dimer complex maintains the same net electrostatic charge balance in the active site as the wild-type NBD bound to Mg-ATP. Electrostatic analyses of the interface of the ATP-sandwich dimer of MJ0796 showed that there should be substantial electrostatic repulsion between the two NBDs forming the complex (Smith et al. 2002). This observation suggests that a second electrostatically driven power stroke could push apart the NBDs in ABC transporters following ATP hydrolysis. However, the occurrence of such an electrostatic power stroke has yet to be clearly documented in functional studies of any superfamily member.
Extensive functional data on CFTR, summarized elsewhere in this book, suggests that, in the open state of the CFTR channel, NBD1 and NBD2 of CFTR form an ATP-sandwich complex stereochemically equivalent to that formed by the ATP-bound E-to-Q mutant of MJ0796 (as schematized in Fig. 1D) (Gadsby 2004; Gadsby et al. 2006; Mense et al. 2006; Riordan 2008; Aleksandrov et al. 2008, 2009; Hwang et al. 2009). One key observation supporting this model is that electrophysiological studies show that the E-to-Q mutation in NBD2 of CFTR (E1371Q) produces a very long-lived open state of the channel in the presence of ATP (Vergani et al. 2005; Gadsby et al. 2006; Hwang et al. 2009). However, the isolated domains have yet to be observed to form such a complex in vitro, as discussed below.
Structures of Homologous Transmembrane Proteins
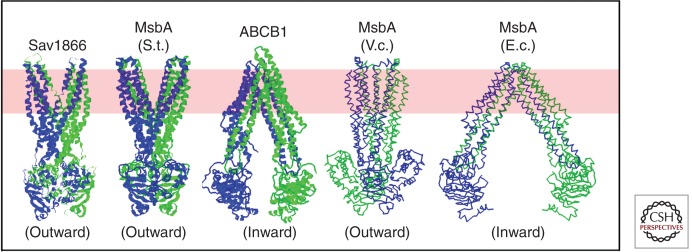
The structure of CFTR probably resembles the structures of the bacterial efflux transporters Sav1866 (Dawson and Locher 2006, 2007) and MsbA (Ward et al. 2007) and the mammalian multidrug resistance transporter Pgp (ABCB1) (Aller et al. 2009), which are illustrated in Figure 2. This class of ABC transporter has two TMDs, each containing six transmembrane α-helices. Four of these six α-helices have long cytoplasmic extensions that form two so-called intracellular loops (ICLs). Two of these loops, each comprising two of the cytoplasmic helical extensions and the short polypeptide segment connecting them, bind to one of the cytoplasmic NBDs, with one loop from each TMD crossing over to contact one loop from the other TMD. The resulting pair of loops coming from different TMDs binds in the groove on the surface of a single NBD (Dawson and Locher 2006, 2007).
Figure 2.
Published crystal structures of ABC transporters homologous to CFTR (Dawson and Locher 2006, 2007; Ward et al. 2007; Aller et al. 2009). Ribbon diagrams are shown for three structures in which sidechains were modeled (left), whereas backbone traces are shown for the two structures in which the limited resolution of the diffraction data prevented side-chain assignment (right) (Ward et al. 2007). Structures have been determined for the MsbA orthologs from Salmonella typhimurium (S.t.), Vibrio cholera (V.c.), and Escherichia coli (E.c). All structures shown are homodimers, with the exception of ABCB1 (also called P-glycoprotein or Pgp), which has two NBDs and two transmembrane domains fused in a single polypeptide chain. The two subunits in each homodimer are colored differently (i.e., blue vs. green), as are the amino-terminal and carboxy-terminal halves of ABCB1. The approximate location of the lipid bilayer is illustrated by a pink band. The SAV1866, MsbA (S.t.), and MsbA (V.c.) structures are in outward-facing conformations, whereas the ABCB1 and MsbA (E.c.) structures are in inward-facing conformations.
Examination of the structures of Sav1866, MsbA, and Pgp (Fig. 2) shows that the overall folds of the TMDs and NBDs are broadly similar. However, the existing structures of Pgp and some MsbA orthologs show a wide separation of the NBDs and an inward-facing (i.e., cytoplasm-facing) orientation of the two TMDs (Fig. 2) (Dawson and Locher 2007; Aller et al. 2009). In contrast, Sav1866 and other MsbA orthologs display closely associated NBDs and a more outward-facing configuration of the TMDs (Dawson and Locher 2006, 2007; Ward et al. 2007). Therefore, these structural studies suggest that major conformational changes can occur in the relative orientation of the four domains in this type of ABC protein. Similar, but smaller, conformational changes have also been observed in a separate group of bacterial ABC proteins, which carry out the import of substances into bacterial cells (Locher et al. 2002; Davidson and Chen 2004; Hollenstein et al. 2007b; Oldham et al. 2007; Pinkett et al. 2007; Kadaba et al. 2008; Khare et al. 2009). Large conformational changes were previously proposed to be part of the substrate translocation mechanism for ABC transporters (Rosenberg et al. 2001).
Inward versus Outward Facing Conformations of CFTR
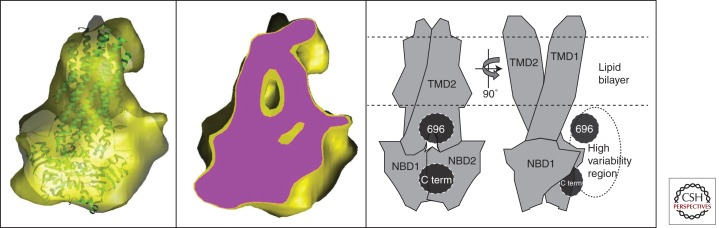
If CFTR’s molecular architecture can be understood in terms of the available structures for intact ABC transporters shown in Figure 2, then relatively coarse structural data (∼1/35 Å−1) should be reliable to detect the wide separation of the NBDs displayed by the Pgp conformation. The structural data published so far for full-length CFTR (Fig. 5), which exceeds this resolution, does not show a discrete separation of two globular domains (Awayn et al. 2005; Zhang et al. 2009, 2010), implying that CFTR more likely resembles the closed conformation observed in Sav1866. However, it is possible that CFTR undergoes major conformational rearrangements but that conditions have not yet been explored that favor the inward-facing conformation with separated NBDs.
Figure 5.
Cryo electron microscopy (EM) analysis of human CFTR. The yellow surface in the left panel shows structural data (Coulomb density map) derived from single-particle EM analysis of CFTR purified in the nonionic detergent β-dodecylmaltoside (β-DDM) but eluted from the final chromatography column in the charged detergent lyso-phosphatidylglycerol (l-PG) just above its critical micelle concentration (cmc) (Zhang et al. 2009). Similar images are obtained from samples eluted from the final column in β-DDM (Zhang et al. 2009). The crystal structure of Sav1866 (green ribbon trace) has been docked into the map. The center panel shows a slice through the same map (purple surface), revealing its internal features. The right panel shows two orthogonal views of a model for the domain organization of CFTR derived from the cryo-EM map shown in the other panels. The locations of the carboxyl terminus (C term) and the region around residue 696 in the R region were inferred from immuno-gold-labeling of poly-Histidine tags inserted into CFTR at these positions (Zhang et al. 2010). A high variability region revealed by cryo-EM (Zhang et al. 2009) is hypothesised to represent the R region.
CFTR Models Based on ABC Protein Structures
Several structural homology models for CFTR based on Sav1866 and Pgp have been published (Callebaut et al. 2004; Mornon et al. 2008, 2009; Serohijos et al. 2008). Although the two models based on Sav1866 are superficially very similar, especially in the NBD regions of the models (Callebaut et al. 2004; Serohijos et al. 2008), significant differences exist at the level of the individual residues, including alternative rotamers for side-chains and differing relative positions (phasing) of amino acids in the transmembrane regions. These discrepancies are not surprising, because the sequence homology between CFTR and Sav1866 or Pgp is low in the transmembrane regions, which limits the stereochemical accuracy of homology models. Inevitably, the beginning and end of transmembrane helices is rather uncertain, and a discrepancy of only two residues can result in a ∼180Å change in the orientation of a sidechain versus the long axis of the transmembrane α-helix.
Evolution of CFTR Ion Channel Function
Hypotheses for the evolution of the channel function of CFTR suggest that the ATP-bound, outward-facing transmembrane conformation observed in the model ABC transporters is likely to be similar to that adopted by CFTR when its channel is open, whereas the inward-facing transmembrane conformation is likely to be similar to that adopted by CFTR when its channel is closed (Vergani et al. 2003, 2005; Gadsby et al. 2006; Kos and Ford 2009). These hypotheses fit well with the observations that ATP is required to initiate CFTR channel opening both in patch-clamp measurements in vivo and in single-channel measurements from black lipid membranes in vitro (i.e., from such membranes fused with CFTR-containing vesicles) (Sheppard et al. 1993; Sheppard and Welsh 1999; Sheppard 2004; Vergani et al. 2003, 2005; Gadsby et al. 2006; Aleksandrov et al. 2007). Furthermore, CFTR channels show long quiescent periods approximating the expected rate of ADP release and ATP rebinding by NBD2 in CFTR (Sheppard et al. 1993; Sheppard and Welsh 1999; Vergani et al. 2003, 2005; Sheppard 2004; Gadsby et al. 2006). Similarly, the likely timescale of the large structural changes between the observed inward- and outward-facing conformations of the model ABC transporters seems to be consistent with the moderate ATPase velocity of these proteins, supporting the hypothesis that their large-scale conformational changes are coupled to ATP binding and hydrolysis (Davidson and Chen 2004; Dawson et al. 2007; Davidson and Maloney 2007; Oldham et al. 2007; McDevitt et al. 2008; Khare et al. 2009). On the other hand, electrophysiological recordings from CFTR can also show very rapid transitions from open to closed states and between subconductance states, on a timescale much faster than its rate of ATP hydrolysis (Cai et al. 2003; Da Paula et al. 2010). These observations suggest that small conformational shifts could produce transiently or partially closed states of the channel while the NBDs of CFTR remain in the ATP-bound state.
STRUCTURAL STUDIES OF THE NBDS FROM CFTR
Variations in ABC Domain Structure in NBD1 from CFTR
For many years, efforts to express and purify NBD1 from CFTR were impeded by ambiguities in its domain boundaries (Thomas et al. 1991; Qu et al. 1997). A breakthrough was made by the company Structural GenomiX Inc., in a project that was initiated and funded by the U.S. Cystic Fibrosis Foundation. This long-term project applied high-throughput robotic cloning and protein expression methods to evaluate many possible domain boundaries using CFTR orthologs from a variety of vertebrate species, leading eventually to the isolation and crystallization of soluble protein constructs containing NBD1 (Lewis et al. 2004). Insights developed in the course of determining crystal structures of NBD1 from murine CFTR (mNBD1) enabled subsequent determination of crystal structures of NBD1 from human CFTR (hNBD1—Figs. 1A–C, 3A,C,E, and 4) (Lewis et al. 2005, 2010). To date, nine crystal structures of mNBD1 and eight of hNBD1 have been deposited in the Protein Data Bank (PDB). (The mNBD1 structures spanning residues 389–673 or 389–655 are available under accession codes (IDs) 1Q3H, 1R0W, 1R0X, 1R0Y, 1R0Z, 1R10, 1XF9, 1XFA, and 3SI7, while the hNBD1 structures spanning residues 389–678 or 389–646 are available under IDs 1XMI, 1XMJ, 2BBO, 2BBS, 2BBT, 2PZE, 2PZF, and 2PZG.)
Figure 3.
Subdomain reorientation and backbone B-factors in hNBD1 and hNBD2 crystal structures. All panels show the hNBD1 (panels A, C, and E on the left and panel B on the right) and hNBD2 (panels B, D, and F on the right) domains in the same orientation after least-squares alignment of the β-strands in their ATP-binding cores (i.e., the ABCβ subdomain plus the F1-like core subdomain). The ribbon diagrams in panels A,B and E,F are colored equivalently to Fig. 1B,C (i.e., orange or yellow for the F1-like ATP-binding core subdomain, shades of blue for the ABCα subdomain, shades of green for the ABCβ subdomain, and red or magenta for the γ-phosphate switch or Q-loop). The bound ATP molecules (black or gray) and the invariant glutamines in the γ-phosphate switch (Q493 in hNBD1 in red and Q1291 in hNBD2 in magenta) are shown in space-filling representation. (A,B) Least-squares superpositions of two full-length F508-hNBD1 structures (panel A—PDB IDs 2BBO and 1 XMI) and of one of these with hNBD2 (panel B—PDB IDs 2BBO and 3GD7, respectively). The 7° rotation of the ABCα subdomain between the two hNBD1 structures shown here represents the largest displacement observed when comparing any two hNBD1 crystal structure, all of which have ATP bound in the active site. As discussed in the text, the 17° rotation and ∼10 Å displacement of the ABCα subdomain in hNBD2 relative to the ATP-bound conformation in hNBD1 likely prevents formation of a hydrolytically active ATP-sandwich heterodimer with hNBD1 in vitro, even though extensive evidence supports formation of such a complex coupled to opening of the CFTR channel in vivo (as schematized in Fig. 1D) (Gadsby 2004; Mense et al. 2006; Aleksandrov et al. 2008, 2009; Hwang et al. 2009). (C,D) Equivalent views of the hNBD1 (panel C) and hNBD2 (panel D) structures from panel B with the thickness and color of the backbone trace representing their mean backbone B-factors. B-factors quantify the variation or disorder in atomic position in a crystal structure, with larger B-factors indicating more disorder. The default encoding scheme in PyMOL was used to generate these images (i.e., 0.6–4.0 Å backbone thickness and a linear blue-to-green-to-red color ramp for B-factors running from 32–80 Å2 for hNBD1 and from 19–76 Å2 for hNBD2). (E,F) Close-up views of the ATP-binding sites in the crystal structures of F508-hNBD1-Δ(RI,RE) (panel E – PDB ID 2PZE) and hNBD2 (panel F – PDB ID 3GD7). Labels indicate the locations of the Walker A (WA) and Walker B (WB) motifs in the F1-like core subdomain and the γ-phosphate switch (γ-PS) linking the core subdomain to the amino terminus of the ABCα subdomain. Although hNBD1 is catalytically inactive, due at least in part to the substitution of a serine for the catalytic glutamate residue adjacent to the Walker B motif, its γ-PS adopts a catalytically active conformation in which its amino-terminal glutamine (Q493, red spheres) contacts the Mg2+ cofactor of ATP, as observed in ATP-bound structures of catalytically active NBDs from a wide variety of ABC transporters (Hung et al. 1998; Smith et al. 2002). In contrast, the γ-PS in hNBD2 adopts a different conformation resulting in an ∼17 Å displacement of the amide group of its amino-terminal glutamine (Q1291, magenta spheres) from the ATP molecule bound in the active site.
Even using domain boundaries equivalent to those in the crystal structures of mNBD1, the orthologous human domain could only be purified in sufficient yield and in monodisperse form after introducing a series of “solubilizing” point mutations (Lewis et al. 2005, 2010). Many sets of such mutations were evaluated using robotic mutagenesis and expression methods, and several sets yielded significant improvements in the apparent solubility of the domain during purification and concentration for crystallization. One such set, called the Teem set, comprised three point mutations (G550E/R553Q/R555K) that previously were shown to promote improved biogenesis of F508del CFTR in tissue cultures cells, presumably by improving the stability of the protein (Teem et al. 1993; DeCarvalho et al. 2002). (This mutation set is found in combination with the F409L, F429S, F433L, and H667R mutations in PDB IDs 1XMJ and 2BBO.) The other mutation sets that improved the yield of soluble hNBD1 involved substitution of surface-exposed residues in hNBD1 with more polar residues occurring at the same position in CFTR orthologs from other species (F429S/F494N/Q637R found in PDB ID 2BBS, F494N/Q637R found in PDB ID 2BBT, and F429S/H667R found in PDB ID 1XMI). Eventually, it was shown that the mutations that significantly improve the purification of hNBD1 all thermodynamically stabilize the native conformation of the domain compared to an aggregation-prone “molten globule” conformation (described further below) (Protasevich et al. 2010; Wang et al. 2010). Therefore, what had been perceived as a solubility problem, a technical problem related to requirements for purification in vitro, turned out to be a stability problem directly related to the molecular pathology caused by the F508del mutation (as explained below). Thus, the research program focused on crystallographic characterization of CFTR domains eventually provided vital insight not only into the atomic structure of CFTR but also into the molecular pathogenesis of CF.
There are no significant differences in the crystal structures of mNBD1 and hNBD1, irrespective of the presence or absence of stabilizing/solubilizing mutations in the human domain, even though their structures were determined in solvent environments with dramatic differences in salt concentration and in some cases pH (Lewis et al. 2010). Both structures show the canonical fold for the NBD from an ABC transporter, as described above. There are local conformational variations in NBD1 at the carboxyl terminus of the ABCβ subdomain and immediately preceding the LSGGQ signature sequence, in regions that show significant structural variations among different members of the ABC transporter superfamily (Zaitseva et al. 2005). However, in addition to these well-precedented structural variations, the crystal structures revealed two noteworthy structural variations specific to NBD1 from CFTR. The first of these, which was anticipated from sequence alignments with NBDs from ABC transporters, involves the substitution of two highly conserved residues in the ATPase active site with noncanonical amino acids (Lewis et al. 2004). The catalytic glutamate residue is substituted with a serine (S573), rendering the Walker B sequence ϕ4DS instead of ϕ4DE, and a catalytically vital histidine residue that contacts the γ-phosphate of ATP is also substituted with serine (S605). Although these changes do not produce a significant change in active site conformation relative to NBDs from model ABC transporters, they prevent ATP hydrolysis by NBD1 (Lewis et al. 2004; Thibodeau et al. 2005). This property allowed determination of both the murine (Lewis et al. 2004) and human (Lewis et al. 2005) structures bound to unmodified ATP with a physiological Mg2+ cofactor, which has not been possible with any catalytically active NBD. Enzymological studies of the domain in solution verified the lack of significant ATPase activity in purified preparations of NBD1 (Lewis et al. 2004; Thibodeau et al. 2005).
The second noteworthy structural variation revealed by the crystal structures of NBD1 from CFTR is the addition of two protein segments not found in NBDs from model ABC transporters (Lewis et al. 2004, 2010). One of these is an ∼35-residue insertion in the loop between the first two β-strands in domain (i.e., at the amino terminus of the ABCβ subdomain). NBD1 preserves the aromatic residue at the end of the first β-strand in the ABCβ subdomain (Lewis et al. 2005), which makes stacking interactions with the adenine of ATP exactly as observed in NBDs from model ABC transporters (Karpowich et al. 2001). However, the ∼5-residue tight turn that follows this conserved aromatic in most NBDs is replaced by a 35-residue loop that is largely disordered based on crystallographic analyses as well as hydrogen-deuterium exchange mass spectrometry (HDX-MS) studies conducted on the domain in solution (Lewis et al. 2010). This loop spanning approximately residues 402–436 in human CFTR has been called the regulatory insertion or RI (Figs. 1A, B and 3A,C). Its unexpected presence following the first ∼12 residues in NBD1 is largely responsible for the inability to accurately identify the amino terminus of the domain based on sequence analysis (Lewis et al. 2010).
A second protein segment that is not shared with NBDs from model ABC transporters is an ∼35-residue extension appended at the carboxyl terminus of NBD1 (Figs. 1A,B and 3C,E) (Lewis et al. 2004). This regulatory extension or RE segment spanning approximately residues 638–672 in human CFTR could be considered to comprise the amino terminus of the R domain. However, its inclusion improves the solubility of NBD1 protein constructs (Lewis et al. 2004, 2005), and it adopts a consistent folded conformation (Figs. 1B, and 3A,C) in most crystal structures of murine and human NBD1 constructs, including structures determined under widely varying conditions of ionic strength and pH (Lewis et al. 2010). HDX-MS studies show that the RE rapidly explores unfolded conformations, suggesting that it is likely to be predominantly unfolded in the isolated domain in solution (Lewis et al. 2010). However, elegant NMR dynamics studies support the inference from crystallographic studies that it also populates a folded state in which it interacts at specific sites on the surface of NBD1 (Baker et al. 2007; Kanelis et al. 2010).
Eventually, a construct of hNBD1 was designed with the RI (residues 405–436) and the RE (residues after Gly646) both deleted (i.e., yielding a construct spanning residues 389–646 in human CFTR with a 32-residue internal deletion) (Atwell et al. 2010). Although removal of the RE has little influence on the stability of the domain, removal of the RI strongly stabilizes the domain in vitro (Protasevich et al. 2010) and significantly improves biogenesis of F508del-CFTR in tissue culture cells (Aleksandrov et al. 2010). This effect is presumably attributable to favorable energetic interaction of the RI with the molten globule conformation of NBD1, which reduces the free energy difference between the native state and the unfolded state (Protasevich et al. 2010; Wang et al. 2010). The hNBD1 construct without either the RI or RE, called NBD1-Δ(RI,RE), has been an important tool for biochemical and biophysical studies of CFTR because of its improved stability compare to the full-length domain (Aleksandrov et al. 2010; Protasevich et al. 2010; Wang et al. 2010). A crystal structure has been determined for this NBD1-Δ(RI,RE) construct in the presence of Mg-ATP (Atwell et al. 2010), which shows it forming a homodimeric ATP-sandwich complex with two ATP molecules bound between the Walker motif in one protomer and the LSGGQ signature sequence in the other protomer. This structure is similar in conformation and oligomeric organization to the physiological ATP-sandwich complexes formed by the NBDs from model homodimeric bacterial ABC transporters. However, the NBD1-Δ(RI,RE) construct, like all other NBD1 constructs characterized to date, does not homodimerize in solution in the presence of ATP (Atwell et al. 2010). Thus, the crystal structure of NBD1-Δ(RI,RE) has trapped a low affinity and likely nonphysiological homodimer that nonetheless provides a potentially useful stereochemical model for the physiological ATP-sandwich heterodimer formed with NBD2 in CFTR.
A Structural Paradigm for CFTR Regulation by the R Domain
The crystal structures of NBD1 constructs including the RI and RE segments have provided a model for the structural basis of CFTR activation by covalent modification of the R domain (Lewis et al. 2004, 2010). In different crystal structures, both segments show variable conformations (Lewis et al. 2010), consistent with biophysical assays showing that these segments are dynamic in solution (Baker et al. 2007; Lewis et al. 2010). In some structures, each segment is observed to adopt a conformation interacting with the NBD2-binding surface of NBD1, suggesting that the R domain could inhibit channel activation by physically blocking formation of the ATP-sandwich complex by NBD1 and NBD2 (Fig. 1D). Although it is unclear whether the RI functions in this manner physiologically, substantial evidence exists supporting participation of the RE in such a regulatory mechanism. Most crystal structures of NBD1 show a long segment of the RE forming an α-helix docked at the same site on the NBD2-binding surface of NBD1, which would clearly block formation of any ATP-sandwich complex by NBD1 (Lewis et al. 2004). This conformation of the RE is observed in all crystal structures of both the murine and human domains determined at neutral pH, including structures from crystals grown at drastically different ionic strengths ranging from low millimolar to several molar (Lewis et al. 2010).
NMR studies show that the RE is largely disordered in solution but populates some partially folded and likely α-helical conformational states (Baker et al. 2007). Chemical-shift perturbation studies show that several regions of NBD1, including some observed to bind to the RE in crystal structures, interact with unphosphorylated RE, and these interactions are weakened by phosphorylation of the RE by PKA. These studies suggest that multiple segments of the RE interact with multiple regions of NBD1 to enable integration of diverse signaling pathways mediating covalent modifications of different segments of the R domain (Csanády et al. 2006; Baker et al. 2007; Kanelis et al. 2010). One such interaction is likely to involve the crystallographically observed docking of the RE on the NBD2-binding surface of NBD1 (Lewis et al. 2004, 2010), which will block formation of the physiological ATP-sandwich heterodimer with NBD2 until this interaction is somehow disrupted by phosphorylation of the RE or an allosterically coupled site elsewhere in the R domain (Fig. 1D).
Structural Consequences of the F508del Mutation in NBD1
The deletion of the phenylalanine residue at position 508 in NBD1 of CFTR is the predominant CF-causing mutation in the human population (Cutting 1993, 2005). This F508del mutation accounts for at least one mutant allele in ∼90% of CF patients, making it a critical priority to understand the molecular defects that it causes and to develop methods to remediate them. Extensive efforts to stabilize NBD1 for structural studies, as described above, eventually led to successful purification and crystallization of the domain harboring the destabilizing F508del mutation (Lewis et al. 2005). The crystal structure of the mutant protein shows conformational perturbations only in the immediate vicinity of the mutation (Fig. 4A) (Lewis et al. 2005, 2010), and hydrogen–deuterium exchange studies show that even perturbations in backbone dynamics are limited to modest enhancements at sites immediately adjacent to F508 in the primary sequence (Lewis et al. 2010). Extensive in vitro biophysical studies of NBD1 (Protasevich et al. 2010; Wang et al. 2010), correlated with a wide variety of in vivo studies (DeCarvalho et al 2002; Pissarra et al. 2008; Aleksandrov et al. 2010, 2012), have established that thermodynamic destabilization of NBD1 by the F508del mutation plays a central role in the molecular pathogenesis of CF, as outlined in the next section. However, the local structural perturbations caused by the F508del mutation in the folded conformation of NBD1 are likely to contribute at least indirectly to pathogenesis by disrupting structural interactions between NBD1 and the TMDs of CFTR (Lewis et al. 2005, 2010; Serohijos et al. 2008; He et al. 2010; Loo et al. 2010; Thibodeau et al. 2010). Recently published work suggests that developing effective drugs to correct the molecular defects caused by the F508del mutation in CFTR may require correction of these disrupted interdomain interactions (Mendoza et al. 2012; Rabeh et al. 2012).
Figure 4.
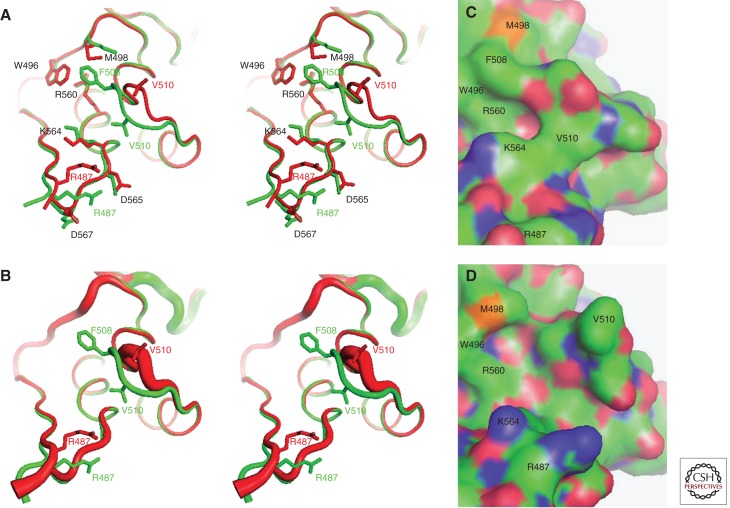
Structural consequences of the F508del mutation in hNBD1. Structures of hNBD1 with and without the F508del mutation were aligned based on least-squares superposition of the three α-helices comprising the core of the ABCα subdomain. (A) Stereopair showing the interface between the F1-like core subdomain and the ABCα subdomain in F508 (green, PDB ID 2BBO) and F508del (red, PDB ID 1XMJ) hNBD1 structures. Selected sidechains are shown in ball-and-stick representation. The site of attachment of hNBD1 to the transmembrane domains of CFTR is proximal to the viewer in this orientation, and the γ-PS is at the upper left (i.e., the backbone segment including residues W496 and M498). (B) Stereopair showing the same view of the same hNBD1 structures using the same color scheme, but with the thickness of the backbone traces encoding their mean backbone B-factors (using the default scheme in PyMOL, with 0.6–4.0 Å radius representing 33–77 Å2 and 12–69 Å2 for the F508 and F508del structures, respectively). (C,D) Surface representations of the same structures showing the alterations in local surface topography caused by the F508del mutation. These two panels show the same view of the molecular surfaces of F508 (panel C) and F508del (panel D) hNBD1, with carbon atoms colored green, oxygen atoms colored red, nitrogen atoms colored blue, and sulfur atoms colored orange. Note that the structures shown here contain seven point mutations included in hNBD1 constructs because of their beneficial influence on yield during purification—F409L, F429S, F433L, G550E, R553Q, R555K, and H667R. Extensive analyses led to the conclusion that these mutations do not significantly perturb the native ground-state conformation of hNBD1 (Lewis et al. 2010) but that they do stabilize it thermodynamically compared to an aggregation-prone molten globule intermediate that accounts for both the instability of F508del-CFTR in vivo and the progressive loss of hNBD1 constructs in vitro during purification (Protasevich et al. 2010; Wang et al. 2010). (The images shown here are from Lewis et al. 2010; adapted, with permission, from the authors.)
The F508 residue packs onto the surface of the ABCα subdomain in NBD1 in a broad but shallow groove at the interface between this subdomain and the F1-like ATP-binding core (Fig. 4A) (Lewis et al. 2004, 2010), as described briefly above. Crystal structures of full-length ABC transporters show that this region of the NBD is the principal site of packing interactions with the cytoplasmic loops of the TMDs (Fig. 1A) (Locher et al. 2002; Dawson and Locher 2006; Serohijos et al. 2008; Aller et al. 2009). The conservation of equivalent interdomain interactions in CFTR is supported by the crosslinking of pairs of cysteines replacing residues on either side of the predicted interface in an otherwise cysteine-less CFTR construct (Serohijos et al. 2008). Moreover, the functional significance of this interface is supported by electrophysiological experiments showing reversible arrest of single-channel gating on addition of a bifunctional sulfhydryl crosslinker to such a CFTR construct with cysteines replacing residues F508 in NBD1 and F1068 in TMD2 (Serohijos et al. 2008).
These important interdomain structural interactions between NBD1 and the TMDs of CFTR are likely to be significantly perturbed by the deletion of F508, even though the structural consequences of this mutation are relatively modest and localized within NBD1 (Lewis et al. 2004, 2010). The F508del mutation changes the conformation of the adjacent loop that connects the first and second α-helices in the ABCα subdomain, effectively shortening it (Fig. 4A,B) (Lewis et al. 2010). The glycine residue at position 509, which is invariant in CFTR orthologs and conserved at this site in many other ABC transporters, participates in a “capping motif” that H-bonds to the carboxyl terminus of the α-helix containing F508. These H-bonding interactions are altered by the deletion of F508, which causes a significant change in the backbone conformation of the loop spanning residues 509–511 in NBD1. This change flips the sidechain of the valine residue at position 510 from being partially buried on the surface of the ABCα domain to projecting outward (Fig. 4) into the likely interface between NBD1 and the TMDs of CFTR (Fig. 1A). The removal of the sidechain of residue F508 combined with the conformational change in the adjacent loop produces a substantial change in the topography and qualitative chemical characteristics of the surface of NBD1 at its likely site of interaction with the TMDs of CFTR (Fig. 4C,D) (Serohijos et al. 2008; Lewis et al. 2010).
These structural variations suggest that the F508del mutation is likely to perturb the interaction between NBD1 and the TMDs of CFTR (Lewis et al. 2004, 2010; Serohijos et al. 2008). Although the inability to date to purify the TMDs of CFTR in a native conformational state has prevented direct biophysical evaluation of this hypothesis, its validity is supported by the results from mutagenesis experiments conducted by several different laboratories (He et al. 2010; Loo et al. 2010; Thibodeau et al. 2010; Mendoza et al. 2012; Rabeh et al. 2012). These experiments provide evidence that the defective trafficking of F508del-CFTR in tissue culture cells can be suppressed by strengthening the interaction between the perturbed surface on NBD1 and the cognate region of the TMD. A second-site R1070W mutation in the proximal region of the TMDs produces significant suppression of the defect (Thibodeau et al. 2010), presumably by increasing hydrophobic interactions with the altered conformation of residue V510 in F508del-NBD1. Moreover, restoration of the trafficking of F508del-NBD1 by the V510D suppressor mutation, which introduces a negative charge into a generally apolar region of the interdomain interface (Fig. 4C,D), is strongly attenuated by introducing the R1070A or R1070D mutations that remove a complementary positive charge from the proximal surface of the TMD (Loo et al. 2010). In addition to providing insight into the molecular pathologies caused by the F508del mutation, these observations provide additional evidence that homology models based on model ABC transporters accurately predict the structural interactions between NBD1 and the TMDs of CFTR (Serohijos et al. 2008; Alexander et al. 2009; Mornon et al. 2009; Dalton et al. 2012).
Two recent papers both concluded that strengthening the interface between NBD1 and the TMDs in F508del-CFTR would be required for effective pharmacological correction of its trafficking defect (Mendoza et al. 2012; Rabeh et al. 2012). However, another recent paper directly challenges this critical pharmacological inference (Aleksandrov et al. 2012). The two papers supporting this inference both present systematic evaluations of different combinations of suppressor mutations in correcting the trafficking defect in F508del-CFTR in tissue culture cells. In these studies, none of the suppressor mutation sets confined to NBD1 restored maturation to wild-type levels, but full restoration or correction of the F508del defect was observed when combining suppressor mutations in NBD1 with mutations likely to strengthen the interdomain interface between NBD1 and the TMDs in F508del-CFTR (Mendoza et al. 2012; Rabeh et al. 2012). In contrast, the third recent paper showed essentially wild-type levels of maturation and stability in F508del-CFTR containing second-site suppressor mutations exclusively in NBD1 (i.e., the I539T mutation plus four proline substitutions found in chicken CFTR, which is naturally more thermostable than human CFTR) (Aleksandrov et al. 2012). Based on this latter finding, small molecules that bind tightly to properly folded NBD1, and thereby significantly stabilize its native conformation, could be sufficient to correct the molecular pathologies caused by the F508del mutation and cure the disease in patients having this mutation. However, the conflicting interpretation offered in the other two recent papers implies that effective pharmacological correction would additionally require stabilization of the interface between NBD1 and the TMDs in F508del-CFTR. Given the critical implications of this controversy for CF drug development, future cell biological and pharmacological investigations should focus on establishing the operative constraints in a system that accurately reflects the dynamics of CFTR maturation and degradation in human lungs.
Perturbation of the NBD1 Folding Pathway by F508del
The folding pathway of NBD1 plays an important role in the molecular pathology caused by the F508del mutation. Recent publications describe this pathway in detail (Protasevich et al. 2010; Wang et al. 2010), so only its central features will be summarized here. The native conformation of NBD1 (Figs. 1 and 3) exists in equilibrium with a relatively low energy “molten globule” conformation. The molten globule, first described by Privalov and coworkers (Griko and Privalov 1994; Griko et al. 1994), is a dynamic species that retains a substantial amount of native-like 2Å structure but does not form stable tertiary structure (Arai and Kuwajima 2000; Englander 2000; Redfield 2004; Sampson et al. 2011). Its thermodynamic hallmark, which has been clearly observed in hNBD1, is a very low enthalpy of unfolding for a species with a substantial amount of regular secondary structure. Increasing temperature decreases the stability of the native conformation of NBD1 compared to the molten globule, while the binding of ATP has the opposite effect and substantially increases the stability of the native conformation compared to the molten globule (Protasevich et al. 2010; Wang et al. 2010).
Extensive thermodynamic analyses show that the F508del mutation destabilizes the native conformation of NBD1 compared to the molten globule, making the latter species the dominant conformation of the isolated domain at 37°C even in the presence of physiological concentrations of ATP (Protasevich et al. 2010). In contrast, only a small proportion of the population of the wild-type domain adopts the molten globule conformation under these conditions. The molten globule of NBD1 aggregates aggressively and irreversibly in vitro (Wang et al. 2010). A series of second-site mutations in NBD1 have parallel effects in rescuing the trafficking defect in CFTR in vivo (DeCarvalho et al. 2002; Pissarra et al. 2008; Aleksandrov et al. 2010) and inhibiting molten globule formation by isolated NBD1 in vitro (G550E/R553Q/R555K, F494N/Q637R, or V510D) (Protasevich et al. 2010; Wang et al. 2010). These results suggest that the influence of F508del in promoting formation of the aggregation-prone molten globule conformation is likely to be the principal factor underlying the molecular pathologies caused by the F508del mutation in vivo.
In this context, small molecules that bind to NBD1 and stabilize it in the native conformational state should be able to remediate the trafficking defect caused by the F508del mutation in CFTR. The efficacy of this approach is supported by observations on two related compounds that display significant corrector activity in tissue culture cells and that also appear to stabilize purified NBD1 against thermal denaturation in vitro (Sampson et al. 2011). Broader and deeper investigations are merited of this pharmacological approach focusing on thermodynamic correction of the folding defect in F508del-NBD1. It will be important to identify the location of compound binding sites in natively folded F508del-NBD1 and also to evaluate whether stabilization of NBD1 must be accompanied by stabilization of the NBD1-TMD interface to produce effective correction of the molecular defect caused by the F508del mutation, a claim supported by some studies cited in the last section (Mendoza et al. 2012; Rabeh et al. 2012) but questioned by others (He et al. 2010; Aleksandrov et al. 2012).
The Existing Crystal Structure of NBD2 Shows an Inactive Conformation
A crystal structure of NBD2 from human CFTR (hNBD2) bound to unhydrolyzed phenyl-ATP (ϕATP) has been solved and deposited into the Protein Data Bank by Structural GenomiX Inc., the same company that produced the crystal structures of NBD1 described above. They used equivalent high-throughput robotic cloning and expression methods in combination with more aggressive and ambitious protein-engineering methods to generate a crystal structure of an hNBD2 construct. This construct spans residues 1193–1427 in human CFTR and has the regulatory domain from the E. coli MalK protein fused at its carboxyl terminus (PDB ID 3GD7; X Zhao, S Atwell, and S Emtage, unpubl.). This domain, which is found at the carboxyl terminus of the NBD from the E. coli maltose transporter (Diederichs et al. 2000; Oldham et al. 2007), was observed to mediate extensive crystal-packing contacts in earlier crystal structures containing the domain (Chen et al. 2003). These observations suggested that fusion of this domain at an equivalent position in hNBD2 might help promote its crystallization. Therefore, a homology model was constructed for hNBD2 and used to engineer a fusion joint likely to preserve the rigid orientation of the regulatory domain of MalK relative to the F1-like ATP-binding subdomain of the NBD. The design strategy involved superimposing the carboxy-terminal α-helices from the homology model of hNBD2 and the experimentally determined crystal structure of MalK to create a precise in-frame fusion construct. This fusion construct, like all other constructs of hNBD2 characterized to date, suffers from poor stability and solubility during purification in vitro. Improvements in the yield of soluble protein were obtained by introducing the hydrolytically inactivating H1402A mutation in ATPase active site of hNBD2 plus a series of “solubilizing” mutations on its surface (Q1280E/Y1307N/W1310H/Q1411D) (X Zhao, S Atwell, JF Hunt, et al., unpubl.). Two additional “surface entropy reduction” mutations (E1308A/ Q1309A) were introduced into the construct to promote crystallization. The H1402A mutation as well as at least some of the “solubilizing” mutations seem likely to stabilize the native conformation of the domain thermodynamically, as observed for the solubilizing surface mutations that improved the stability and yield of preparations of hNBD1 (as described above). However, because of the limited stability of the domain in the absence of these mutations, it has not been possible to verify this inference experimentally.
Nonetheless, indirect evidence supports the hypothesis that the H1402A mutation stabilizes hNBD2. Based on the ATP-bound crystal structures of model NBDs from bacterial transporters, histidine 1402 in hNBD2 contacts the γ-phosphate of ATP in the prehydrolysis complex, while glutamate 1371 is the catalytic base that actives water for hydrolytic attack on the γ-phosphate of ATP (Hung et al. 1998; Smith et al. 2002; Zaitseva et al. 2005). Electrophysiological studies of the H1402A and E1371Q mutations in intact human CFTR support these inferences concerning catalytic geometry by showing that the H1402A (Kloch et al. 2010) and E1371Q (Vergani et al. 2005) mutations both greatly increase the lifetime of the open state of the CFTR chloride channel, presumably because they block ATP hydrolysis and stabilize ATP-sandwich heterodimer formed by hNBD1 and hNBD2. This interpretation is consistent with the effects of equivalent mutations in a wide variety of model NBDs and ABC transporters (Smith et al. 2002; Janas et al. 2003; Vergani et al. 2005; Oldham et al. 2007; Zutz et al. 2010), as explained above. However, despite their similar influence on the gating properties of intact CFTR, the H1402A and E1371Q mutations have dramatically different effects on the stability and yield of hNBD2 during purification, with the first greatly improving yield compared to the second (X Zhao, S Atwell, JF Hunt, et al., unpubl.). In silico energetic calculations suggest that the H1402A mutation stabilizes isolated hNBD2 bound to Mg-ATP, while the E1371Q mutation destabilizes it (P Kumar, C Wang, JF Hunt, et al., unpubl.). Future experimental studies will be required to evaluate this computational inference that the influence of these mutations on the yield of purified hNBD2 reflects their differential effects on the thermodynamic stability of the native conformation of the domain.
Although the crystal structure of the extensively engineered hNBD2 construct shows a canonical fold, without any elaborations like the RI in hNBD1, it adopts a surprising and likely inactive conformation. This structure (PDB id 3GD7) shows the ABCα subdomain pulled away from the F1-like ATP-binding core in hNBD2 by ∼10 Å compared to its position in hNBD1 (Fig. 3B), even though unhydrolyzed Mg-ϕATP is observed in the active site. (In this crystal structure, the phenyl group covalently attached to the adenine ring in ϕATP mediates intermolecular packing contacts that seem likely to stabilize the lattice, presumably explaining why the structure could be obtained only with ϕATP; X Zhao, S Atwell, and S Emtage, unpubl.) Furthermore, the γ-phosphate switch (or Q-loop) in hNBD2 adopts a different conformation that moves the terminal atoms in the sidechain of the invariant glutamine at its amino terminus out of the active site, to a position 17 Å away from the position of the equivalent residue in hNBD1 (i.e., comparing Gln1291 in hNBD2 in Fig. 3F to Gln493 in hNBD1 in Fig. 3E). Although qualitatively similar displacements of the ABCα subdomain and alternative conformations of the γ-phosphate switch have been observed in nucleotide-free and Mg-ADP-bound structures of a variety of NBDs from model ABC transporters (Diederichs et al. 2000; Karpowich et al. 2001; Yuan et al. 2001), as described above, a conformation of this kind has not previously been observed in any structure with unhydrolyzed ATP or a nonhydrolyzable analog of ATP bound in the active site. The observed conformation of the ABCα subdomain and γ-phosphate switch are likely to be incompatible with binding to hNBD1 and formation of the functional ATP-sandwich heterodimer complex believed to gate opening of the CFTR channel (as described above and schematized in Fig. 1D).
All previously determined NBD structures in which a γ-phosphate group or an analog of this group are present in the active site show the ABCα domain aligned in close apposition to the F1-like core subdomain (Hung et al. 1998; Smith et al. 2002; Chen et al. 2003; Oldham et al. 2007; Atwell et al. 2010), with the conserved glutamine residue at the amino terminus of the γ-phosphate switch contacting the metal cofactor bound to the nucleotide (e.g., as shown for Gln493 in hNBD1 in Fig. 3E). Among the 17 different crystal structure of NBD1 currently available in the PDB, there is at most a 7° rotation of the ABCα subdomain producing minor displacements of its putative NBD2-interacting surface (Fig. 3A), even though these structures come from two different species and were determined in some cases in solutions with drastically different pHs and ionic strengths (Lewis et al. 2010). In contrast, the ∼10 Å displacement of the ABCα subdomain observed in all four copies of hNBD2 present in the asymmetric unit of its crystal structure moves the domain diagonally away from the likely location of its interface with hNBD1 (i.e., based on the geometry of the ATP-sandwich dimer complexes observed in structures of NBDs from model ABC transporters as well as the structure of the low-affinity hNBD1 homodimer described above) (Atwell et al. 2010). The ABCα subdomain in hNBD2 appears to move primarily as a rigid body, based on both comparison of hNBD2 to hNBD1 and analysis of the smaller 1–2 Å variations in its location in the four different molecules of hNBD2 present in its crystal structure (data not shown). However, the α-helix following the Walker B motif, which makes extensive packing interactions with the ABCα subdomain, shows more complex variations in conformation involving smaller displacements in the same direction as the ABCα subdomain, which are coupled to differential bending of its helical axis (Fig. 3F and additional data not shown). These conformational changes in the α-helix following the Walker B and in the ABCα subdomain seem very likely to block functional interaction of hNBD2 with hNBD1.
Indeed, a wide variety of experiments conducted on the crystallized hNBD2 construct have failed to detect interaction with purified hNBD1 in solution (X Zhao, C Wang, S Atwell, et al., unpubl.). These experiments suggest that, even in solution, this extensively engineered hNBD2 construct adopts an inactive conformation. Therefore, the unusual conformation observed in its crystal structure might accurately reflect its inherent conformational tendencies. It is unclear whether wild-type hNBD2 would adopt a similarly inactive conformation in isolation from the other domains with which it interacts in CFTR. However, none of the mutations present in the crystallized construct provide an obvious explanation for its adoption of an inactive conformation with an ATP analog bound in the active site. Electrophysiological (Vergani et al. 2005; Kloch et al. 2010) and biochemical (Mense et al. 2006; Aleksandrov et al. 2009) experiments on CFTR strongly support formation of a canonical ATP-sandwich heterodimer by hNBD1 and hNBD2 in the open state of the CFTR channel (as discussed above and schematized in Fig. 1A) (Gadsby et al. 2006; Riordan 2008; Hwang et al. 2009), and the electron crystallography studies reviewed below provide additional circumstantial support for this model (Rosenberg et al. 2004, 2011; Ford et al. 2011). In this context, future studies will be required to determine whether hNBD2 has evolved to adopt an inactive conformation in the absence of proper interdomain packing interactions in intact CFTR or whether the sequence variations present in the crystallized hNBD2 construct are responsible for its adoption of an inactive conformation.
STRUCTURAL DATA FOR FULL-LENGTH CFTR
Crystallization of Full-Length CFTR
A method for two-dimensional (2D) crystallization of full-length CFTR has been published by Rosenberg and coworkers (2004). The method uses detergent (dodecylmaltoside) solubilized CFTR that was produced in stably-transfected baby hamster kidney (BHK) cells. CFTR obtained in this way has been relatively well characterized, and it appears to be fully glycosylated but mostly dephosphorylated (Zhang et al. 2009). This lack of phosphorylation could imply that CFTR is kept inactive in the BHK cells (which may be needed for cell growth when they express significant levels of this ion channel). However, it also seems possible that CFTR is rapidly dephosphorylated by cellular phosphatases at an early stage in the purification procedure. Dephosphorylation of CFTR may be important for 2D crystallization, as it probably removes some heterogeneity in the properties of the purified protein (i.e., in surface charge characteristics). There is some evidence that PKA-catalyzed phosphorylation of purified CFTR yields protein that crystallizes poorly (Awayn 2008). Glycosylation of CFTR will also yield heterogeneity in the physicochemical properties of the protein, but interestingly, this form of heterogeneity appears to be compatible with crystallization (Rosenberg et al. 2004; Zhang et al. 2009), perhaps because 2D crystals can accommodate the extracellular sugar polymers outside the planar lattice. The only method published so far for the crystallization of CFTR involves epitaxial (surface) crystallization in which the protein is slowly brought out of solution by concentration of solutes within the crystallization droplet. This methodology is essentially an extension of the sitting-drop vapor-diffusion method for generating three-dimensional (3D) crystals of biomolecules (Scarborough 1994; Auer et al. 1998, 1999; Rosenberg et al. 2001), but it is conducted at a 10- to 100-fold lower protein concentration. The method can also yield thin, multilayered 3D crystals at the carbon–water interface as well as 2D crystals (Scarborough 1994; Auer et al. 1998, 1999; Rosenberg et al. 2001), but the low concentration of protein ensures that most of the generated crystals are thin enough for electron crystallography (i.e., <50 nm) (Ford and Holzenburg 2008).
Structural data derived from 2D crystals of full-length CFTR using electron crystallography methods were published in 2004 (Rosenberg et al. 2004). The crystals were grown in the presence of the nonhydrolyzable ATP analog AMP-PNP (Rosenberg et al. 2004), and two separate crystal forms were detected. The crystals were stained with uranyl acetate to provide contrast and studied in the high vacuum of a conventional transmission electron microscope. The resolution of the data generated by these studies was ∼20 Å, which is typical for such specimens because visualization of molecular details is limited by the granular nature of the uranyl acetate stain and by dehydration of the specimen (Harris 1999; Ford and Holzenburg 2008). Furthermore, stain penetration tends to be uneven, which causes some parts of the specimen to be better preserved or contrasted than others (Harris 1999; Ford and Holzenburg 2008). Despite these drawbacks, the structural data from these electron crystallographic analyses showed that CFTR is in a monomeric state in the 2D crystals, and that its conformation bears strong similarity to that of Pgp (ABCB1), as previously predicted (Rosenberg et al. 2003).
Crystallization conditions have recently been identified that yield larger, better ordered, and more reproducible 2D arrays of full-length CFTR, and higher resolution electron crystallography data have been obtained for these 2D crystals in an unstained and frozen-hydrated state. Such crystals, which only grow in the absence of nucleotide in the crystallization reaction, have the protein packed in two crystalline layers stacked on top of each other.
Single-Particle Structural Studies of Full-Length CFTR
Structural data for purified CFTR has also been obtained for noncrystalline specimens using various microscopy techniques. An early atomic force microscopy (AFM) study of CFTR-expressing Xenopus oocytes observed donut-shaped objects within the plasma membrane that were associated with gold-labeled secondary antibodies bound to primary anti-CFTR antibodies (Schillers et al. 2004). However, the large dimensions (∼60 nm diameter) of these objects in the plasma membrane were difficult to reconcile with the expected molecular structure of CFTR. For example, the central pore of the donut-shaped objects was larger (∼20 nm diameter) than the dimensions of any ABC transporter characterized to date at atomic resolution. AFM of antibody-labeled CFTR has also been applied to try to assess its expression level in erythrocyte membranes (Schillers 2008). Despite the above-mentioned reservations concerning the AFM studies, they potentially represent structural data for CFTR resident in its native environment in the plasma membrane. If the large objects observed by AFM are indeed antibody-labeled CFTR, then our understanding of the behavior of CFTR in the plasma membrane will need to be revised. The objects detected in these studies would imply that several CFTR molecules encircle a large central cavity, and that CFTR expression in plasma membranes is at much higher levels than previously thought (Haggie et al. 2006). Further structural studies on CFTR localized to the plasma membrane will needed to evaluate the significance of these AFM observations.
Electron microscopy (EM) has also been employed to study the structure of purified full-length CFTR solubilized in the nonionic detergent β-dodecylmaltoside (β-DDM) or in the charged detergent lyso-phosphatidyl glycerol (l-PG). Dimeric complexes in β-DDM were reported in an early study conducted using single-particle averaging of purified CFTR embedded in a heavy-atom stain (Awayn et al. 2005). A subsequent study of purified CFTR particles in β-DDM also reported dimeric complexes, although these complexes were unexpectedly hollow in the center (Mio et al. 2008), perhaps because of lack of stain penetration or other problems associated with negative staining (Harris 1999; Ford and Holzenburg 2008). Later, EM structural data were obtained for single particles of purified CFTR in the unstained and frozen-hydrated state solubilized in either β-DDM or l-PG (Fig. 5), which provide similar images (Zhang et al. 2009). These data, in which stain artifacts are eliminated, confirmed the dimeric nature of the purified CFTR preparation but provided no evidence for a hollow cavity in the center of the dimer. Instead, they showed that the 3D structure was consistent with two side-by-side ABC transporters with a similar structure to Sav1866. Labeling of these CFTR particles using 1.8-nm diameter immuno-gold particles verified many conclusions from the earlier studies (Zhang et al. 2010), including the potential location of the R domain in the complex.
Regions Unique to CFTR
The interpretation of the available structural data for full-length CFTR has relied on the substantially higher resolution structures available for the homologous ABC exporters Sav1866, MsbA, and Pgp (Dawson and Locher 2006, 2007; Ward et al. 2007; Aller et al. 2009). This set of proteins has been assumed to represent a structural archetype for most eukaryotic ABC transporters. The Sav1866 structure has been used most frequently for homology modeling of such structures, including that of CFTR. Recently, these homology models have begun to be tested experimentally in mutagenesis studies (Stenham et al. 2003; Zolnerciks et al. 2007; Serohijos et al. 2008; Alexander et al. 2009; Mornon et al. 2009; He et al. 2010). However, Sav1866 is a homodimeric protein, with each protomer consisting of a single TMD fused to a single NBD. Hence, Sav1866 lacks the R domain that covalently connects TMD1/NBD1 at the amino terminus of CFTR to TMD2/NBD2 at its carboxyl terminus. Despite the extensive evidence discussed above that the R domain is substantially disordered (Ostedgaard et al. 1997; Baker et al. 2007; Kanelis et al. 2010; Lewis et al. 2010), EM imaging of CFTR labeled with immuno-gold suggests that part of the R region may be spatially localized in the full-length protein, at a site spanning the interface between NBD1 and NBD2 and proximal to the cytoplasmic surface of the plasma membrane (Fig. 5) (Zhang et al. 2009, 2010).
When activated, CFTR must contain a hydrophilic channel that is selective for negatively charged ions (anions), especially chloride. This channel must run continuously through its TMDs and be open to both sides of the membrane for significant periods of time when CFTR is activated by PKA and ATP. Comparison of the available high-resolution structures for ABC exporters shows that they are all sealed to either the inside or the outside surface of the membrane. A partially sealed conformation of this kind is presumably a requirement of the translocation mechanism, because the transported substrate should be exposed alternatively to one side of the membrane or the other, but not simultaneously to both of them. Hence, at least when the anion channel in CFTR is open, its detailed transmembrane structure must be significantly different from that of Sav1866 and other ABC exporters. Hypotheses have been advanced to explain how the anion-selective channel in CFTR could have evolved and how it may be gated so that anion flux can be controlled by the cell (Jordan et al. 2008; Alexander et al. 2009).
CFTR’s Channel and Gate
Examination of the outward-facing configuration of ABC exporters, as exemplified by the structure of Sav1866, suggests an appealing model to explain the existence of a continuous transmembrane channel in CFTR. The 12 transmembrane α-helices in the functional Sav1866 dimer form a cone-shape molecular surface surrounding a central water-filled cavity that is connected via a wide (∼10 Å diameter) mouth to the extracellular milieu (as shown on the left side of Fig. 2) (Dawson and Locher 2006). This cavity is sealed on the cytoplasmic side of the membrane by a group of amino acid residues in the ICLs of Sav1866. A significant conformational change, presumably coupled to reorientation or separation of its NBDs, would be needed to open this interface in Sav1866 to the cytoplasm. Notably, such a configuration of the cytoplasmic ICLs is found in the structure of the Pgp dimer (Aller et al. 2009), which is in an inward-facing conformation. However, the central cavity in this structure is blocked on the extracellular side of the membrane (center of Fig. 2). Intriguingly, the sealed region on the extracellular side of the transmembrane domains in this structure is relatively short. Hence, conceptually, it is easier to envision an inward-facing, Pgp-like conformation as being the active form of CFTR and residues on the extracellular side of its transmembrane domains as forming the gate of its channel. However, the existing mutagenesis and electrophysiological studies have been interpreted to imply that an outward-facing, Sav1866-like conformation is more likely to represent the active form of CFTR and that residues on the cytoplasmic side of the central portal are likely to form the gate of its channel. Nonetheless, experimental evidence shows that some ion selectivity is conferred by residues near the extracellular surface of CFTR (Jordan et al. 2008; Alexander et al. 2009). (The electrophysiological properties of the CFTR channel and predictions about its structure from homology models are covered in greater depth in a separate article in this work.) Further structural and functional studies will be required to reconcile these various inferences about the structure of the CFTR channel and understand the mechanism by which it is gated.
The molecular graphics images in Figs. 1, 3, and 4 in this article were prepared using PyMOL (www.pymol.org). The atomic models and 3D maps in Figs. 2 and 5 were displayed using Chimera (Pettersen et al. 2004) (www.cgl.ucsf.edu/chimera). Semi-automated fitting of the Sav1866 structure (PDB ID 2HYD) was performed by placing the models within the map by hand and then refining using the Fit-in-Map routine with optimization for correlation between the experimental CFTR maps and a simulated map based on the fitted models, which was arbitrarily restricted to a resolution corresponding to the reported resolution of the experimental map (Zhang et al. 2009). Both positive and negative regions of the map were included in the correlation calculation.
ACKNOWLEDGMENTS
We thank the Cystic Fibrosis Foundation (U.S.) for financial support via grants FORD08XX0 and HUNT09XX0.
Footnotes
Editors: John R. Riordan, Richard C. Boucher, and Paul M. Quinton
Additional Perspectives on Cystic Fibrosis available at www.perspectivesinmedicine.org
REFERENCES
- Aleksandrov AA, Aleksandrov L, Riordan JR 2002a. Nucleoside triphosphate pentose ring impact on CFTR gating and hydrolysis. FEBS Lett 518: 183–188 [DOI] [PubMed] [Google Scholar]
- Aleksandrov L, Aleksandrov AA, Chang XB, Riordan JR 2002b. The first nucleotide binding domain of cystic fibrosis transmembrane conductance regulator is a site of stable nucleotide interaction, whereas the second is a site of rapid turnover. J Biol Chem 277: 15419–15425 [DOI] [PubMed] [Google Scholar]
- Aleksandrov AA, Aleksandrov LA, Riordan JR 2007. CFTR (ABCC7) is a hydrolyzable-ligand-gated channel. Pflugers Arch 453: 693–702 [DOI] [PubMed] [Google Scholar]
- Aleksandrov L, Aleksandrov A, Riordan JR 2008. Mg2+-dependent ATP occlusion at the first nucleotide-binding domain (NBD1) of CFTR does not require the second (NBD2). Biochem J 416: 129–136 [DOI] [PubMed] [Google Scholar]
- Aleksandrov AA, Cui L, Riordan JR 2009. Relationship between nucleotide binding and ion channel gating in cystic fibrosis transmembrane conductance regulator. J Physiol 587: 2875–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksandrov AA, Kota P, Aleksandrov LA, He L, Jensen T, Cui L, Gentzsch M, Dokholyan NV, Riordan JR 2010. Regulatory insertion removal restores maturation, stability and function of δF508 CFTR. J Mol Biol 401: 194–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksandrov AA, Kota P, Cui L, Jensen T, Alekseev AE, Reyes S, He L, Gentzsch M, Aleksandrov LA, Dokholyan NV, et al. 2012. Allosteric modulation balances thermodynamic stability and restores function of δF508 CFTR. J Mol Biol 419: 41–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C, Ivetac A, Liu X, Norimatsu Y, Serrano JR, Landstrom A, Sansom M, Dawson DC 2009. Cystic fibrosis transmembrane conductance regulator: Using differential reactivity toward channel-permeant and channel-impermeant thiol-reactive probes to test a molecular model for the pore. Biochemistry 48: 10078–10088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, et al. 2009. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323: 1718–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MP, Berger HA, Rich DP, Gregory RJ, Smith AE, Welsh MJ 1991a. Nucleoside triphosphates are required to open the CFTR chloride channel. Cell 67: 775–784 [DOI] [PubMed] [Google Scholar]
- Anderson MP, Gregory RJ, Thompson S, Souza DW, Paul S, Mulligan RC, Smith AE, Welsh MJ 1991b. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science 253: 202–205 [DOI] [PubMed] [Google Scholar]
- Arai M, Kuwajima K 2000. Role of the molten globule state in protein folding. Adv Protein Chem 53: 209–282 [DOI] [PubMed] [Google Scholar]
- Atwell S, Brouillette CG, Conners K, Emtage S, Gheyi T, Guggino WB, Hendle J, Hunt JF, Lewis HA, Lu F, et al. 2010. Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant. Protein Eng Des Sel 23: 375–384 [DOI] [PubMed] [Google Scholar]
- Auer M, Scarborough GA, Kuhlbrandt W 1998. Three-dimensional map of the plasma membrane H+-ATPase in the open conformation. Nature 392: 840–843 [DOI] [PubMed] [Google Scholar]
- Auer M, Scarborough GA, Kuhlbrandt W 1999. Surface crystallisation of the plasma membrane H+-ATPase on a carbon support film for electron crystallography. J Mol Biol 287: 961–968 [DOI] [PubMed] [Google Scholar]
- Awayn NH 2008. Structural analyses of the human cystic fibrosis transmembrane conductance regulator (CFTR) protein using electron microscopy, PhD thesis, University of Manchester, Manchester, UK [Google Scholar]
- Awayn NH, Rosenberg MF, Kamis AB, Aleksandrov LA, Riordan JR, Ford RC 2005. Crystallographic and single-particle analyses of native- and nucleotide-bound forms of the cystic fibrosis transmembrane conductance regulator (CFTR) protein. Biochem Soc Trans 33: 996–999 [DOI] [PubMed] [Google Scholar]
- Baker JM, Hudson RP, Kanelis V, Choy WY, Thibodeau PH, Thomas PJ, Forman-Kay JD 2007. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol 14: 738–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampé R 2011. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci 108: 3228–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Scott-Ward TS, Sheppard DN 2003. Voltage-dependent gating of the cystic fibrosis transmembrane conductance regulator Cl- channel. J Gen Physiol 122: 605–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Eudes R, Mornon JP, Lehn P 2004. Nucleotide-binding domains of human cystic fibrosis transmembrane conductance regulator: Detailed sequence analysis and three-dimensional modeling of the heterodimer. Cell Molec Life Sci 61: 230–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappe V, Hinkson DA, Howell LD, Evagelidis A, Liao J, Chang XB, Riordan JR, Hanrahan JW 2004. Stimulatory and inhibitory protein kinase C consensus sequences regulate the cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci 101: 390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappe V, Irvine T, Liao J, Evagelidis A, Hanrahan JW 2005. Phosphorylation of CFTR by PKA promotes binding of the regulatory domain. EMBO J 24: 2730–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Chin JE, Ueda K, Clark DP, Pastan I, Gottesman MM, Roninson IB 1986. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell 47: 381–389 [DOI] [PubMed] [Google Scholar]
- Chen J, Lu G, Lin J, Davidson AL, Quiocho FA 2003. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell 12: 651–661 [DOI] [PubMed] [Google Scholar]
- Csanády L, Chan KW, Seto-Young D, Kopsco DC, Nairn AC, Gadsby DC 2000. Severed channels probe regulation of gating of cystic fibrosis transmembrane conductance regulator by its cytoplasmic domains. J Gen Physiol 116: 477–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanády L, Seto-Young D, Chan KW, Cenciarelli C, Angel BB, Qin J, McLachlin DT, Krutchinsky AN, Chait BT, Nairn AC, et al. 2005. Preferential phosphorylation of R-domain Serine 768 dampens activation of CFTR channels by PKA. J Gen Physiol 125: 171–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanády L, Nairn AC, Gadsby DC 2006. Thermodynamics of CFTR channel gating: A spreading conformational change initiates an irreversible gating cycle. J Gen Physiol 128: 523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Davidson AL 2011. ABC solute importers in bacteria. Essays Biochem 50: 85–99 [DOI] [PubMed] [Google Scholar]
- Cutting GR 1993. Spectrum of mutations in cystic fibrosis. J Bioenerg Biomembr 25: 7–10 [DOI] [PubMed] [Google Scholar]
- Cutting GR 2005. Modifier genetics: Cystic fibrosis. Annu Rev Genomics Hum Genet 6: 237–260 [DOI] [PubMed] [Google Scholar]
- Dalton J, Kalid O, Schushan M, Ben-Tal N, Villa-Freixa J 2012. New model of CFTR proposes active channel-like conformation. J Chem Inf Model 10.1021/ci2005884 [DOI] [PubMed] [Google Scholar]
- Da Paula AC, Sousa M, Xu Z, Dawson ES, Boyd AC, Sheppard DN, Amaral MD 2010. Folding and rescue of a cystic fibrosis transmembrane conductance regulator trafficking mutant identified using human-murine chimeric proteins. J Biol Chem 285: 27033–27044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassa E, Bouige P 2001. The ABC of ABCS: A phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol 152: 211–229 [DOI] [PubMed] [Google Scholar]
- Davidson AL, Chen J 2004. ATP-binding cassette transporters in bacteria. Annu Rev Biochem 73: 241–268 [DOI] [PubMed] [Google Scholar]
- Davidson AL, Maloney PC 2007. ABC transporters: How small machines do a big job. Trends Microbiol 15: 448–455 [DOI] [PubMed] [Google Scholar]
- Davidson AL, Sharma S 1997. Mutation of a single MalK subunit severely impairs maltose transport activity in Escherichia coli. J Bacteriol 179: 5458–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AL, Laghaeian SS, Mannering DE 1996. The maltose transport system of Escherichia coli displays positive cooperativity in ATP hydrolysis. J Biol Chem 271: 4858–4863 [PubMed] [Google Scholar]
- Dawson RJ, Locher KP 2006. Structure of a bacterial multidrug ABC transporter. Nature 443: 180–185 [DOI] [PubMed] [Google Scholar]
- Dawson RJ, Locher KP 2007. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett 581: 935–938 [DOI] [PubMed] [Google Scholar]
- Dawson RJ, Hollenstein K, Locher KP 2007. Uptake or extrusion: Crystal structures of full ABC transporters suggest a common mechanism. Mol Microbiol 65: 250–257 [DOI] [PubMed] [Google Scholar]
- Dean M 2005. The genetics of ATP-binding cassette transporters. Methods Enzymol 400: 409–429 [DOI] [PubMed] [Google Scholar]
- Dean M, Annilo T 2005. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet 6: 123–142 [DOI] [PubMed] [Google Scholar]
- Dean M, Rzhetsky A, Allikmets R 2001. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res 11: 1156–1166 [DOI] [PubMed] [Google Scholar]
- DeCarvalho AC, Gansheroff LJ, Teem JL 2002. Mutations in the nucleotide binding domain 1 signature motif region rescue processing and functional defects of cystic fibrosis transmembrane conductance regulator δ f508. J Biol Chem 277: 35896–35905 [DOI] [PubMed] [Google Scholar]
- Diederichs K, Diez J, Greller G, Müller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W 2000. Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J 19: 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drumm ML, Wilkinson DJ, Smit LS, Worrell RT, Strong TV, Frizzell RA, Dawson DC, Collins FS 1991. Chloride conductance expressed by δ F508 and other mutant CFTRs in Xenopus oocytes. Science 254: 1797–1799 [DOI] [PubMed] [Google Scholar]
- Englander SW 2000. Protein folding intermediates and pathways studied by hydrogen exchange. Annu Rev Biophys Biomol Struct 29: 213–238 [DOI] [PubMed] [Google Scholar]
- Fojo AT, Whang-Peng J, Gottesman MM, Pastan I 1985. Amplification of DNA sequences in human multidrug-resistant KB carcinoma cells. Proc Natl Acad Sci 82: 7661–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford RC, Holzenburg A 2008. Electron crystallography of biomolecules: Mysterious membranes and missing cones. Trends Biochem Sci 33: 38–43 [DOI] [PubMed] [Google Scholar]
- Ford RC, Birtley J, Rosenberg MF, Zhang L 2011. CFTR three-dimensional structure. Methods Mol Biol 741: 329–346 [DOI] [PubMed] [Google Scholar]
- Gadsby DC 2004. Ion transport: Spot the difference. Nature 427: 795–797 [DOI] [PubMed] [Google Scholar]
- Gadsby DC, Vergani P, Csanady L 2006. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature 440: 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George AM, Jones PM 2012. Perspectives on the structure-function of ABC transporters: The switch and constant contact models. Prog Biophys Mol Biol 109: 95–107 [DOI] [PubMed] [Google Scholar]
- Gerlach JH, Endicott JA, Juranka PF, Henderson G, Sarangi F, Deuchars KL, Ling V 1986. Homology between P-glycoprotein and a bacterial haemolysin transport protein suggests a model for multidrug resistance. Nature 324: 485–489 [DOI] [PubMed] [Google Scholar]
- Gregory RJ, Cheng SH, Rich DP, Marshall J, Paul S, Hehir K, Ostedgaard L, Klinger KW, Welsh MJ, Smith AE 1990. Expression and characterization of the cystic fibrosis transmembrane conductance regulator. Nature 347: 382–386 [DOI] [PubMed] [Google Scholar]
- Griko YV, Privalov PL 1994. Thermodynamic puzzle of apomyoglobin unfolding. J Mol Biol 235: 1318–1325 [DOI] [PubMed] [Google Scholar]
- Griko YV, Gittis A, Lattman EE, Privalov PL 1994. Residual structure in a staphylococcal nuclease fragment. Is it a molten globule and is its unfolding a first-order phase transition? J Mol Biol 243: 93–99 [DOI] [PubMed] [Google Scholar]
- Haggie PM, Kim JK, Lukacs GL, Verkman AS 2006. Tracking of quantum dot-labeled CFTR shows near immobilization by C-terminal PDZ interactions. Mol Biol Cell 17: 4937–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JR 1999. Negative staining of thinly spread biological particulates. Methods Mol Biol 117: 13–30 [DOI] [PubMed] [Google Scholar]
- He L, Aleksandrov LA, Cui L, Jensen TJ, Nesbitt KL, Riordan JR 2010. Restoration of domain folding and interdomain assembly by second-site suppressors of the δF508 mutation in CFTR. FASEB J 24: 3103–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CF 1992. ABC transporters: From microorganisms to man. Annu Rev Cell Bio 8: 67–113 [DOI] [PubMed] [Google Scholar]
- Hobson AC, Weatherwax R, Ames GF 1984. ATP-binding sites in the membrane components of histidine permease, a periplasmic transport system. Proc Natl Acad Sci 81: 7333–7337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein K, Dawson RJ, Locher KP 2007a. Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol 17: 412–418 [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Frei DC, Locher KP 2007b. Structure of an ABC transporter in complex with its binding protein. Nature 446: 213–216 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101: 789–800 [DOI] [PubMed] [Google Scholar]
- Horn C, Bremer E, Schmitt L 2003. Nucleotide dependent monomer/dimer equilibrium of OpuAA, the nucleotide-binding protein of the osmotically regulated ABC transporter OpuA from Bacillus subtilis. J Mol Biol 334: 403–419 [DOI] [PubMed] [Google Scholar]
- Hung LW, Wang IX, Nikaido K, Liu PQ, Ames GF, Kim SH 1998. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature 396: 703–707 [DOI] [PubMed] [Google Scholar]
- Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J 2002. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 297: 2018–2026 [DOI] [PubMed] [Google Scholar]
- Hwang TC, Sheppard DN 2009. Gating of the CFTR Cl- channel by ATP-driven nucleotide-binding domain dimerisation. J Physiol 587: 2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas E, Hofacker M, Chen M, Gompf S, van der Does C, Tampé R 2003. The ATP hydrolysis cycle of the nucleotide-binding domain of the mitochondrial atp-binding cassette transporter Mdl1p. J Biol Chem 278: 26862–26869 [DOI] [PubMed] [Google Scholar]
- Jones PM, George AM 1999. Subunit interactions in ABC transporters: Towards a functional architecture. FEMS Microbiol Lett 179: 187–202 [DOI] [PubMed] [Google Scholar]
- Jones PM, George AM 2004. The ABC transporter structure and mechanism: Perspectives on recent research. Cell Mol Life Sci 61: 682–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Kota KC, Cui G, Thompson CH, McCarty NA 2008. Evolutionary and functional divergence between the cystic fibrosis transmembrane conductance regulator and related ATP-binding cassette transporters. Proc Natl Acad Sci 105: 18865–18870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC 2008. The high-affinity E. coli methionine ABC transporter: Structure and allosteric regulation. Science 321: 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanelis V, Hudson RP, Thibodeau PH, Thomas PJ, Forman-Kay JD 2010. NMR evidence for differential phosphorylation-dependent interactions in WT and δF508 CFTR. EMBO J 29: 263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowich N, Martsinkevich O, Millen L, Yuan YR, Dai PL, MacVey K, Thomas PJ, Hunt JF 2001. Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter. Structure 9: 571–586 [DOI] [PubMed] [Google Scholar]
- Kerr ID 2002. Structure and association of ATP-binding cassette transporter nucleotide-binding domains. Biochim Biophys Acta 1561: 47–64 [DOI] [PubMed] [Google Scholar]
- Kerr ID 2004. Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem Biophys Res Commun 315: 166–173 [DOI] [PubMed] [Google Scholar]
- Khare D, Oldham ML, Orelle C, Davidson AL, Chen J 2009. Alternating access in maltose transporter mediated by rigid-body rotations. Mol Cell 33: 528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Chiaw P, Eckford PD, Bear CE 2011. Insights into the mechanisms underlying CFTR channel activity, the molecular basis for cystic fibrosis and strategies for therapy. Essays Biochem 50: 233–248 [DOI] [PubMed] [Google Scholar]
- Kloch M, Milewski M, Nurowska E, Dworakowska B, Cutting GR, Dolowy K 2010. The H-loop in the second nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator is required for efficient chloride channel closing. Cell Physiol Biochem 25: 169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos V, Ford RC 2009. The ATP-binding cassette family: A structural perspective. Cell Mol Life Sci 66: 3111–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA, et al. 2004. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J 23: 282–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Zhao X, Wang C, Sauder JM, Rooney I, Noland BW, Lorimer D, Kearins MC, Conners K, Condon B, et al. 2005. Impact of the δF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J Biol Chem 280: 1346–1353 [DOI] [PubMed] [Google Scholar]
- Lewis HA, Wang C, Zhao X, Hamuro Y, Conners K, Kearins MC, Lu F, Sauder JM, Molnar KS, Coales SJ, et al. 2010. Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry. J Mol Biol 396: 406–430 [DOI] [PubMed] [Google Scholar]
- Linsdell P 2006. Mechanism of chloride permeation in the cystic fibrosis transmembrane conductance regulator chloride channel. Exp Physiol 91: 123–129 [DOI] [PubMed] [Google Scholar]
- Linsdell P, Evagelidis A, Hanrahan JW 2000. Molecular determinants of anion selectivity in the cystic fibrosis transmembrane conductance regulator chloride channel pore. Biophys J 78: 2973–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton KJ, Higgins CF 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol Microbiol 28: 5–13 [DOI] [PubMed] [Google Scholar]
- Liu X, Dawson DC 2011. Cystic fibrosis transmembrane conductance regulator: Temperature-dependent cysteine reactivity suggests different stable conformers of the conduction pathway. Biochemistry 50: 10311–10317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Smith SS, Dawson DC 2003. CFTR: What’s it like inside the pore? J Exp Zoolog A Comp Exp Biol 300: 69–75 [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang ZR, Fuller MD, Billingsley J, McCarty NA, Dawson DC 2004. CFTR: A cysteine at position 338 in TM6 senses a positive electrostatic potential in the pore. Biophys J 87: 3826–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locher KP, Lee AT, Rees DC 2002. The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science 296: 1091–1098 [DOI] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM 2010. The V510D suppressor mutation stabilizes δF508-CFTR at the cell surface. Biochemistry 10.1021/bi100807h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoura MK, Smith SS, Choi AD, Richards NW, Strong TV, Drumm ML, Collins FS, Dawson DC 1998. Cystic fibrosis transmembrane conductance regulator (CFTR) anion binding as a probe of the pore. Biophys J 74: 1320–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt CA, Shintre CA, Grossmann JG, Pollock NL, Prince SM, Callaghan R, Ford RC 2008. Structural insights into P-glycoprotein (ABCB1) by small angle X-ray scattering and electron crystallography. FEBS Lett 582: 2950–2956 [DOI] [PubMed] [Google Scholar]
- Mendoza JL, Schmidt A, Li Q, Nuvaga E, Barrett T, Bridges RJ, Feranchak AP, Brautigam CA, Thomas PJ 2012. Requirements for efficient correction of δF508 CFTR revealed by analyses of evolved sequences. Cell 148: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mense M, Vergani P, White DM, Altberg G, Nairn AC, Gadsby DC 2006. In vivo phosphorylation of CFTR promotes formation of a nucleotide-binding domain heterodimer. EMBO J 25: 4728–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio K, Ogura T, Mio M, Shimizu H, Hwang TC, Sato C, Sohma Y 2008. Three-dimensional reconstruction of human cystic fibrosis transmembrane conductance regulator chloride channel revealed an ellipsoidal structure with orifices beneath the putative transmembrane domain. J Biol Chem 283: 30300–30310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody JE, Millen L, Binns D, Hunt JF, Thomas PJ 2002. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J Biol Chem 277: 21111–21114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mornon JP, Lehn P, Callebaut I 2008. Atomic model of human cystic fibrosis transmembrane conductance regulator: Membrane-spanning domains and coupling interfaces. Cell Mol Life Sci 65: 2594–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mornon JP, Lehn P, Callebaut I 2009. Molecular models of the open and closed states of the whole human CFTR protein. Cell Mol Life Sci 66: 3469–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourez M, Hofnung M, Dassa E 1997. Subunit interactions in ABC transporters: A conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. EMBO J 16: 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham ML, Chen J 2011. Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science 332: 1202–1205 [DOI] [PubMed] [Google Scholar]
- Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J 2007. Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450: 515–521 [DOI] [PubMed] [Google Scholar]
- Ostedgaard LS, Rich DP, DeBerg LG, Welsh MJ 1997. Association of domains within the cystic fibrosis transmembrane conductance regulator. Biochemistry 36: 1287–1294 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC 2007. An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science 315: 373–377 [DOI] [PubMed] [Google Scholar]
- Pissarra LS, Farinha CM, Xu Z, Schmidt A, Thibodeau PH, Cai Z, Thomas PJ, Sheppard DN, Amaral MD 2008. Solubilizing mutations used to crystallize one CFTR domain attenuate the trafficking and channel defects caused by the major cystic fibrosis mutation. Chem Biol 15: 62–69 [DOI] [PubMed] [Google Scholar]
- Protasevich I, Yang Z, Wang C, Atwell S, Zhao X, Emtage S, Wetmore D, Hunt JF, Brouillette CG 2010. Thermal unfolding studies show the disease causing F508 deletion mutation in cystic fibrosis transmembrane conductance regulator (CFTR) thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci 19: 1917–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu BH, Strickland EH, Thomas PJ 1997. Localization and suppression of a kinetic defect in cystic fibrosis transmembrane conductance regulator folding. J Biol Chem 272: 15739–15744 [DOI] [PubMed] [Google Scholar]
- Rabeh WM, Bossard F, Xu H, Okiyoneda T, Bagdany M, Mulvihill CM, Du K, di Bernardo S, Liu Y, Konermann L, et al. 2012. Correction of both NBD1 energetics and domain interface is required to restore δF508 CFTR folding and function. Cell 148: 150–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield C 2004. Using nuclear magnetic resonance spectroscopy to study molten globule states of proteins. Methods 34: 121–132 [DOI] [PubMed] [Google Scholar]
- Rich DP, Anderson MP, Gregory RJ, Cheng SH, Paul S, Jefferson DM, McCann JD, Klinger KW, Smith AE, Welsh MJ 1990. Expression of cystic fibrosis transmembrane conductance regulator corrects defective chloride channel regulation in cystic fibrosis airway epithelial cells. Nature 347: 358–363 [DOI] [PubMed] [Google Scholar]
- Riordan JR 2008. CFTR function and prospects for therapy. Annu Rev Biochem 77: 701–726 [DOI] [PubMed] [Google Scholar]
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL 1989. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 245: 1066–1073 [DOI] [PubMed] [Google Scholar]
- Roninson IB, Chin JE, Choi KG, Gros P, Housman DE, Fojo A, Shen DW, Gottesman MM, Pastan I 1986. Isolation of human mdr DNA sequences amplified in multidrug-resistant KB carcinoma cells. Proc Natl Acad Sci 83: 4538–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MF, Velarde G, Ford RC, Martin C, Berridge G, Kerr ID, Callaghan R, Schmidlin A, Wooding C, Linton KJ, et al. 2001. Repacking of the transmembrane domains of P-glycoprotein during the transport ATPase cycle. EMBO J 20: 5615–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MF, Kamis AB, Callaghan R, Higgins CF, Ford RC 2003. Three-dimensional structures of the mammalian multidrug resistance P-glycoprotein demonstrate major conformational changes in the transmembrane domains upon nucleotide binding. J Biol Chem 278: 8294–8299 [DOI] [PubMed] [Google Scholar]
- Rosenberg MF, Kamis AB, Aleksandrov LA, Ford RC, Riordan JR 2004. Purification and crystallization of the cystic fibrosis transmembrane conductance regulator (CFTR). J Biol Chem 279: 39051–39057 [DOI] [PubMed] [Google Scholar]
- Rosenberg MF, O’Ryan LP, Hughes G, Zhao Z, Aleksandrov LA, Riordan JR, Ford RC 2011. The cystic fibrosis transmembrane conductance regulator (CFTR): 3D structure and localisation of a channel gate. J Biol Chem 286: 42647–42654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson HM, Robert R, Liao J, Matthes E, Carlile GW, Hanrahan JW, Thomas DY 2011. Identification of a NBD1-binding pharmacological chaperone that corrects the trafficking defect of F508del-CFTR. Chem Biol 18: 231–242 [DOI] [PubMed] [Google Scholar]
- Saurin W, Hofnung M, Dassa E 1999. Getting in or out: Early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J Mol Evol 48: 22–41 [DOI] [PubMed] [Google Scholar]
- Scarborough GA 1994. Large single-crystals of the neurospora crassa plasma membrane H+-Atpase: An approach to the crystallization of integral membrane-proteins. Acta Crystallogr D Biol Crystallogr 50: 643–649 [DOI] [PubMed] [Google Scholar]
- Schillers H 2008. Imaging CFTR in its native environment. Pflugers Arch 456: 163–177 [DOI] [PubMed] [Google Scholar]
- Schillers H, Shahin V, Albermann L, Schafer C, Oberleithner H 2004. Imaging CFTR: A tail to tail dimer with a central pore. Cell Physiol Biochem 14: 1–10 [DOI] [PubMed] [Google Scholar]
- Serohijos AW, Hegedus T, Aleksandrov AA, He L, Cui L, Dokholyan NV, Riordan JR 2008. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci 105: 3256–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard DN 2004. CFTR channel pharmacology: Novel pore blockers identified by high-throughput screening. J Gen Physiol 124: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard DN, Welsh MJ 1999. Structure and function of the CFTR chloride channel. Physiol Rev 79: S23–S45 [DOI] [PubMed] [Google Scholar]
- Sheppard DN, Rich DP, Ostedgaard LS, Gregory RJ, Smith AE, Welsh MJ 1993. Mutations in CFTR associated with mild-disease-form Cl- channels with altered pore properties. Nature 362: 160–164 [DOI] [PubMed] [Google Scholar]
- Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF 2002. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell 10: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider J, Houry WA 2008. AAA+ proteins: Diversity in function, similarity in structure. Biochem Soc Trans 36: 72–77 [DOI] [PubMed] [Google Scholar]
- Stenham DR, Campbell JD, Sansom MS, Higgins CF, Kerr ID, Linton KJ 2003. An atomic detail model for the human ATP binding cassette transporter P-glycoprotein derived from disulfide cross-linking and homology modeling. FASEB J 17: 2287–2289 [DOI] [PubMed] [Google Scholar]
- Teem JL, Berger HA, Ostedgaard LS, Rich DP, Tsui LC, Welsh MJ 1993. Identification of revertants for the cystic fibrosis δ F508 mutation using STE6-CFTR chimeras in yeast. Cell 73: 335–346 [DOI] [PubMed] [Google Scholar]
- Thibodeau PH, Brautigam CA, Machius M, Thomas PJ 2005. Side chain and backbone contributions of Phe508 to CFTR folding. Nat Struct Mol Biol 12: 10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau PH, Richardson JM, Wang W, Millen L, Watson J, Mendoza JL, Du K, Fischman S, Senderowitz H, Lukacs GL, et al. 2010. The cystic fibrosis-causing mutation δF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis. J Biol Chem 285: 35825–35835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PJ, Shenbagamurthi P, Ysern X, Pedersen PL 1991. Cystic fibrosis transmembrane conductance regulator: Nucleotide binding to a synthetic peptide. Science 251: 555–557 [DOI] [PubMed] [Google Scholar]
- Vergani P, Nairn AC, Gadsby DC 2003. On the mechanism of MgATP-dependent gating of CFTR Cl- channels. J Gen Physiol 121: 17–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergani P, Lockless SW, Nairn AC, Gadsby DC 2005. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature 433: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1: 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Protasevich I, Yang Z, Seehausen D, Skalak T, Zhao X, Atwell S, Spencer Emtage J, Wetmore DR, Brouillette CG, et al. 2010. Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis. Protein Sci 19: 1932–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A, Reyes CL, Yu J, Roth CB, Chang G 2007. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci 104: 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Blecker S, Martsinkevich O, Millen L, Thomas PJ, Hunt JF 2001. The crystal structure of the MJ0796 ATP-binding cassette. Implications for the structural consequences of ATP hydrolysis in the active site of an ABC transporter. J Biol Chem 276: 32313–32321 [DOI] [PubMed] [Google Scholar]
- Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L 2005. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J 24: 1901–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Aleksandrov LA, Zhao Z, Birtley JR, Riordan JR, Ford RC 2009. Architecture of the cystic fibrosis transmembrane conductance regulator protein and structural changes associated with phosphorylation and nucleotide binding. J Struct Biol 167: 242–251 [DOI] [PubMed] [Google Scholar]
- Zhang L, Aleksandrov LA, Riordan JR, Ford RC 2010. Domain location within the cystic fibrosis transmembrane conductance regulator protein investigated by electron microscopy and gold labelling. Biochim Biophys Acta 1808: 399–404 [DOI] [PubMed] [Google Scholar]
- Zhu T, Hinkson DA, Dahan D, Evagelidis A, Hanrahan JW 2002. CFTR regulation by phosphorylation. Methods Mol Med 70: 99–109 [DOI] [PubMed] [Google Scholar]
- Zolnerciks JK, Wooding C, Linton KJ 2007. Evidence for a Sav1866-like architecture for the human multidrug transporter P-glycoprotein. FASEB J 21: 3937–3948 [DOI] [PubMed] [Google Scholar]
- Zolnerciks JK, Andress EJ, Nicolaou M, Linton KJ 2011. Structure of ABC transporters. Essays Biochem 50: 43–61 [DOI] [PubMed] [Google Scholar]
- Zutz A, Hoffmann J, Hellmich UA, Glaubitz C, Ludwig B, Brutschy B, Tampé R 2010. Asymmetric ATP hydrolysis cycle of the heterodimeric multidrug ABC transport complex TmrAB from Thermus thermophilus. J Biol Chem 286: 7104–7115 [DOI] [PMC free article] [PubMed] [Google Scholar]