Abstract
Adult mammalian cardiac stem cells express the LIM-homeodomain transcription factor Islet1 (Isl1). They are considered remnants of Isl1-positive embryonic cardiac progenitor cells. During amniote heart development, Isl1-positive progenitor cells give rise mainly to the outflow tract, the right ventricle, and parts of the atria. This led to the hypothesis that the development of the right ventricle of the amniote heart depends on the recruitment of additional cells to the primary heart tube. The region from which these additional, Isl1-positive cells originate is called second heart field, as opposed to the first heart field whose cells form the primary heart tube. Here, we review the available data about Isl1 in different species, demonstrating that Isl1 is an important component of the core transcription factor network driving early cardiogenesis in animals of the two clades, deuterostomes, and protostomes. The data support the view of a single cardiac progenitor cell population that includes Isl1-expressing cells and which differentiates into the various cardiac lineages during embryonic development in vertebrates but not in other phyla of the animal kingdom.
Electronic supplementary material
The online version of this article (doi:10.1007/s00427-012-0400-1) contains supplementary material, which is available to authorized users.
Keywords: Islet1, Heart development, Cardiac progenitor cell, Cardiac stem cell
Evolution of the heart
As their size increased, animals have developed pumping organs to ensure proper distribution of oxygen and nutrients throughout the body. In principle, there are two types of circulatory systems: Blood can either flow in a closed vascular network (e.g., in annelids and chordates), or it can be released into the body cavity, such as the hemolymph in arthropods. The morphology of the different circulatory pumps ranges from simple, pulsating (peristaltic) vessels, and tubular hearts, to the more complex multi-chambered hearts (Xavier-Neto et al. 2010). The existence of a contractile organ in both bilaterian clades together with molecular phylogenetic analyses suggests that the common ancestor of protostomes and deuterostomes may have had a simple pumping organ. In this review, we summarize the characteristics of pumping organs of selected developmental and evolutionary biology model organisms focusing on those organisms in which an Isl1-positive cell population has been identified. For more in depth data regarding the evolution of the four-chambered heart, we refer the reader to a recent review by Xavier-Neto et al. (2010).
The closed circulatory system has evolved in coelomates in which the body cavity is lined by a layer of epithelial cells, the mesothelium. In annelids, for example, parts of the mesothelium form the contractile vessels, and no specialized cell types, such as endothelial cells and cardiomyocytes have been identified (Jamieson 1992; Hartenstein and Mandal 2006). In fact, the mesothelium lines the blood vessels in most invertebrate coelomates and, through its ability to contract, causes blood circulation (Hartenstein and Mandal 2006). The dorsal vessel of Drosophila melanogaster consists of contractile myoepithelial cells and is similar to the contractile blood vessels of annelids. It was suggested to consider the dorsal vessel of the fly as a “differentiated mesothelium” (Hartenstein and Mandal 2006). This is interesting because there is a strong relationship between the invertebrate mesothelium and the vertebrate vascular system, the latter being derived from the splanchnopleura, the inner layer of the vertebrate mesothelium. This renders the Drosophila heart more similar to a vertebrate blood vessel. The heart of molluscs, which also belong to the protostomes, exceeds the simple morphology seen in most protostomes. The mollusc heart generally has two chambers; however, cephalopods have the most complex heart among molluscs consisting of two atria and one ventricle (Budelmann et al. 1997). Studying the molecular mechanisms of cardiogenesis in molluscs would provide valuable insights regarding the degree of conservation of the molecular network between protostomes and deuterostomes that leads to a multi-chambered heart. However, whether the subdivisions of the mollusc heart are homologous to those of vertebrates remains an interesting open issue.
In contrast to the contractile organs present in protostomes, the morphology of the heart in deuterostomes became more complex the higher the demands on oxygen distribution became. Tunicates have a V-shaped contracting vessel, and thus, the pumping organ roughly resembles the heart morphology of the protostomes. The lamprey (agnatha) heart consists of four consecutive compartments: the sinus venosus, the atrium, the ventricle, and the conus arteriosus (Kokubo et al. 2010). In teleosts, the two cardiac chambers, ventricle and atrium, are distinguished by the expression of chamber-specific genes, such as specific myosin heavy chains (Scott and Yelon 2010). The transition from aquatic life to terrestrial life of tetrapods required the development of a more sophisticated and more effective circulatory system, including the appearance of a pulmonary circulatory system. Amphibians serve as a good example for animals whose life cycle is partly aquatic and terrestrial. The heart of Xenopus laevis consists of two atria and a single, partially septated ventricle which pumps blood into both the pulmonary and the systemic circuit. Amniotes are characterized by the transition to the fully separated heart with four chambers as found in crocodiles, avians, and mammals (Koshiba-Takeuchi et al. 2009). These more complex, chambered hearts are composed of differently differentiated cell types. Contractile cardiomyocytes have replaced contractile mesothelial cells, and endocardial cells line the inner surface of the heart. The blood vessels are composed of endothelial and smooth muscle cells. Thus, in vertebrates, a variety of specialized cell types has replaced the “cardiac” mesothelium that forms the pumping organs in annelids and arthropods.
Figure 1 provides a simplified overview of the evolutionary relationships of the animals discussed in this review and the composition of their hearts. These contracting organs of different architecture may have evolved by parallelism or may represent homologous entities that share a common evolutionary origin. The finding of a core set of ortholog transcription factors active during the embryonic development of the hearts in different species suggests that these contracting organs of different architecture share a common evolutionary origin rather than having evolved by parallelism.
Fig. 1.
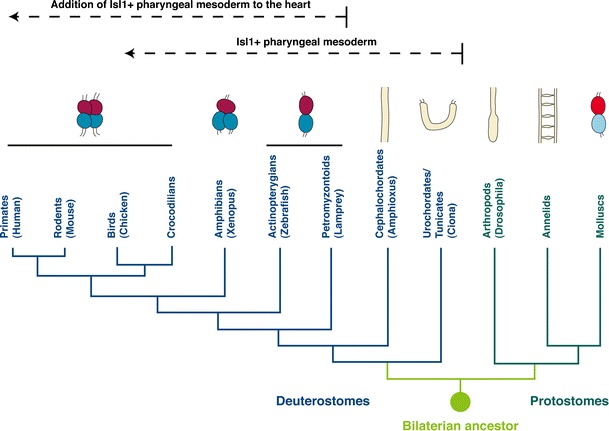
The evolutionary relationship of the organisms discussed in this review and a schematic presentation of the gross morphology of their pumping organs. Ventricular chambers are depicted in red; atrial chambers are shown in blue. The atrium and ventricle in molluscs are colored in light blue and light red since it is not clear whether they are indeed homologous to their vertebrate counterparts. The dashed arrows indicate the appearance of Isl1-positive pharyngeal mesodermal cells in deuterostomes that contribute to the head musculature and, in craniata, to the heart. The phylogenetic tree is based on Simoes-Costa et al. (2005)
A core set of cardiac transcription factors
A wealth of data from different model organisms has allowed us to identify a conserved core set of transcription factors that is essential for the specification of cardiomyocytes. The bHLH transcription factor Mesp1 is considered to be the earliest marker gene in the specification process of cardiac progenitor cells in vertebrate embryos. The closest Mesp1 ortholog in Drosophila, sage, seems to be exclusively expressed in the salivary glands and is therefore unlikely to play a role in fly cardiogenesis (Moore et al. 2000). However, it is tempting to speculate whether sage holds a function in salivary gland migration. If so, it would indicate a partial functional overlap of Sage and Mesp1, since Mesp1 is required for proper cardiac mesoderm migration in mouse embryos (Saga et al. 1999). However, the important role of Mesp1 in cardiovascular progenitor specification is a characteristic of heart development in deuterostomes.
Additional transcription factors required for cardiogenesis in protostomes and deuterostomes include members of the Nk family of homeobox transcription factors with Nkx2.5 being the factor with the most prominent role in cardiogenesis. Orthologs of Nkx2.5 with a crucial role in cardiogenesis are present in the annelid Platynereis (PduNK4), in Ciona (Ci-Nkx) and in Drosophila (Tinman) (Saudemont et al. 2008; Wada et al. 2003; Bodmer 1993). Regarding the GATA family of transcription factors, there are three members in vertebrates, GATA4, 5, and 6 that have distinct as well as partially redundant functions in cardiogenesis whereas there is only one GATA factor present in Ciona (Ci-GATAa) and in Drosophila (Pannier) that functions in cardiac development (Sirbu and Pandur 2009; Gajewski et al. 2001; Davidson 2007). Additionally, members of the T-Box family such as Tbx1, 2, 3, 4, 5, 18, and 20 in vertebrates, and Dorsocross1–3 (Doc) (which are related to the vertebrate Tbx6 subfamily) in Drosophila are required to ensure proper cardiogenesis (Stennard and Harvey 2005; Reim et al. 2003).
Orthologs of the LIM-homeodomain transcription factor Isl1 have been described in humans (Bu et al. 2009), mice (Cai et al. 2003), chicken (Yuan and Schoenwolf 2000), Xenopus (Brade et al. 2007; Gessert and Kühl 2009), zebrafish (Hami et al. 2011), lamprey (Kokubo et al. 2010), Amphioxus (Jackman et al. 2000), Ciona (Stolfi et al. 2010), and Drosophila (Mann et al. 2009; Tao et al. 2007). The evolutionary relationships of Isl1 in the different organisms are depicted in Fig. 2. The phylogenetic tree (Fig. 2a) and the accompanying comparison of amino acid sequences of previously characterized Isl1 proteins (Fig. 2b) reveal that invertebrate Isl1 is highly divergent from vertebrate Isl1. While this finding may not come as a surprise, it is interesting that Isl1 proteins among invertebrates are equally highly divergent. However, an alignment of the amino acid sequences reveals the remarkable high conservation of amino acids in the functional domains, that is, the two LIM domains and the homeodomain (Electronic supplementary material Fig. 1). Isl1 was demonstrated to play a role in heart development in all organisms listed above with the exception of Amphioxus and annelids where a possible cardiogenic role of Isl1 has not been studied so far. The presence of Isl1-positive adult stem cells in mice and humans has initiated research into a possible function of Isl1 in cardiac repair and regeneration (Moretti et al. 2007). It was demonstrated that these Isl1-positive cardiac stem cells have the capacity to differentiate into functional cardiomyocytes in vitro. Moreover, during cardiac regeneration in urodeles, Isl1 expression is reactivated emphasizing the importance of this transcription factor for cardiomyocyte formation (Witman et al. 2011). Here, we provide an overview of the expression and function of Isl1 during cardiogenesis in different species.
Fig. 2.
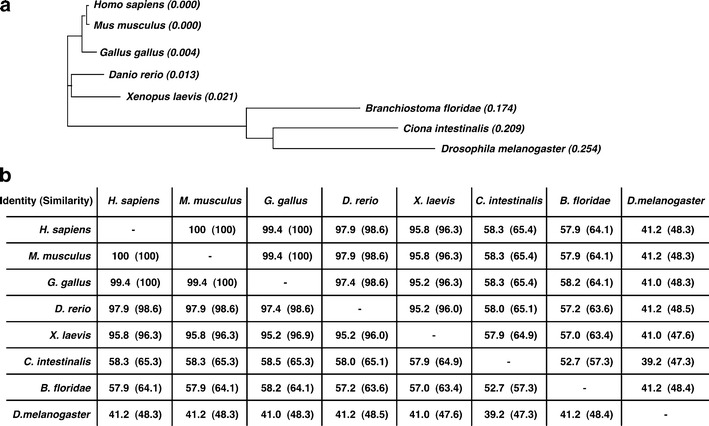
Illustration of the evolutionary relationship of Isl1 genes in different species. a The phylogenetic tree was generated using Clustal W (http://www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny/) (Chenna et al. 2003). Numbers indicate the genetic distance between the nodes of the tree. Sequences used were: NP_002193.2 (Homo sapiens), NP_067434.3 (M. musculus), NP_571037.1 (D. rerio), NP_990745.1 (G. gallus), NP_001104188.1 (X. laevis), AF226616_1 (Branchiostoma floridae), NP_001027767.1 (C. intestinalis), NP_476775.1 (D. melanogaster). b Comparison of Isl1 protein sequences. The percentages depict the degree of identical and similar (in parentheses) amino acids
Expression and function of Isl1 during heart development across species
Mus musculus
Cardiovascular cells originate from Brachyury-positive cells in the mesoderm of mouse embryos which start expressing Mesp1 at early precardiac/primitive streak stages (Solloway and Harvey 2003). Mesp1-positive cells are heterogeneous in fate, as they are able to give rise to all cardiac cells, skeletal muscles of the head and neck, and paraxial mesoderm derivatives (Saga et al. 1999). As soon as these cells leave the primitive streak and populate the anterior and lateral plate mesoderm to form the cardiac crescent, they turn off Mesp1 and become irreversibly committed to a cardiac fate by expressing Nkx2.5. Further differential expression of Tbx5 and Isl1 subdivides the cardiac crescent into two distinct domains, the first heart field (FHF) and the second heart field (SHF). As the cells of the FHF give rise to the left ventricle and parts of the right ventricle and atrium and as the cells of the SHF give rise to the right ventricle, outflow tract, and parts of both atria, the term lineage is more expedient and also used in this context. Expression of Tbx5 and Isl1 also marks an important restriction in the differentiation potency of cardiac progenitors, since these cells differentiate mainly to cardiomyocyte and smooth muscle vascular cells and, to a lesser extent, to endothelial cells (Watanabe and Buckingham 2010). Irreversible genetic marking of progenitor cells using the Isl1–Cre;Rosa26 fsLz system showed that Isl1 + progenitors contribute to cardiomyocyte, smooth muscle, and endothelial cells in the right ventricle, outflow tract, portions of the atria, and the inner curvature of the left ventricle (Cai et al. 2003; Sun et al. 2007). These studies are in accordance with the phenotype of Isl1 -/- mouse embryos, which die at midgestation due to a severely altered heart characterized by a single ventricular chamber with left ventricle identity, absence of outflow tract and of the right ventricle, and a severe reduction of atria (Cai et al. 2003).
The SHF is situated medial, slightly caudal, and dorsal to the first heart field, in the so-called pharyngeal mesoderm (Fig. 3a). During heart looping stages, Isl1-positive cells from the pharyngeal mesoderm are continuously added to the poles of the embryonic heart (Waldo et al. 2001; Mjaatvedt et al. 2001). Of note, the pharyngeal cardiogenic mesoderm has not only cardiogenic but also skeletal myogenic potential and strongly contributes to the development of the head muscles derived from the first and second branchial arches (with a lesser extent to the masseter, pterygoid, and temporalis). This finding has led not only to the description of the so-called cardio-craniofacial field of multipotent cardio-skeletal progenitors but also to a better understanding of the intricate relationship between pharyngeal muscles and cardiac development during evolution (Tirosh-Finkel et al. 2006; Lescroart et al. 2010; Tzahor and Evans 2011; Nathan et al. 2008).
Fig. 3.
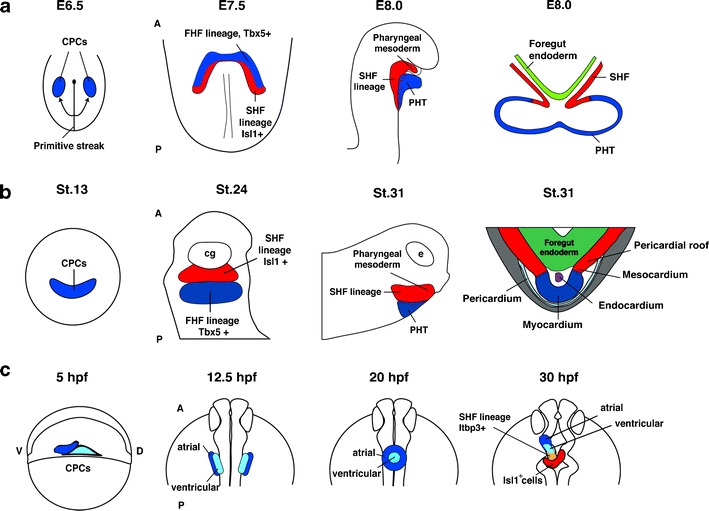
A simplified schematic presentation of heart development in different species. a Mouse. Developmental stages are indicated in embryonic days (E). The cartoons show a dorsal view of a flattened embryo (E6.5), an anterior view (E7.5), and a lateral view (E8.0). Additionally, a schematic illustration of a transverse section at E8.0 is provided. The SHF is characterized by the presence of Isl1-positive cells and is indicated in red. b X. laevis. Developmental stages are according to Nieuwkoop and Faber (1975). The cartoons show an anterior view (st.13), a ventral view (st.24), and a lateral view (st.31). Additionally, a schematic illustration of a transverse section through the heart at st.31 is provided. At st.24, cells of the SHF (red) express isl1 whereas cells of the FHF (blue) are positive for tbx5. c D. rerio. Developmental stages are indicated in hours post-fertilization (hpf). The cartoons show a lateral view for stage 5 hpf and dorsal views for stages 12.5, 20, and 30 hpf with the anterior side up. The localization of early cardiac progenitor cells at 40 % epiboly, as well as the distinction between an atrial and a ventricular fate was analyzed by fate-mapping experiments. An Isl1-positive cell population that contributes to the arterial pole is located posterior and dorsal to the ventricle and is indicated in red. It only partially contributes to the heart. The zebrafish SHF is characterized by the expression of ltbp3. Of note, the arrangement of the atrium and ventricle along the anterior–posterior axis is inverted at later stages (not shown here). As a result, the atrium will come to lie posterior to the ventricle. CPC: common cardiac progenitor cell population, A: anterior, D: dorsal, P: posterior, V: ventral, FHF: first heart field; SHF: second heart field, PHT: primary heart tube. Partly adopted from Gessert and Kühl (2009)
Transcription of Isl1 is shut off as soon as the progenitor cells enter the forming embryonic heart per se, suggesting that Isl1 is required for continuous proliferation, expansion, and migration of cardiovascular precursors. However, Isl1 + cells persist in the hearts of fetal and young (up to 10 months old) adult rodents, in quiescent cells with either cardiomyocyte or peripheral nervous ganglia characteristics (Genead et al. 2010; Laugwitz et al. 2005; Khattar et al. 2011).
Interestingly, a population of Isl1-positive adult cardiac stem cells can be identified in the postnatal heart of mice, rats, and humans (Laugwitz et al. 2005, 2008). These cells can develop into functional cardiomyocytes in vitro as demonstrated by marker gene expression, intracellular calcium release, and their ability to initiate action potentials (Laugwitz et al. 2005). A possible involvement of these cells in the recently described replacement of adult cardiomyocytes in vertebrates awaits further investigation (for review, see Kühl and Kühl 2011). In general, the Isl1-positive adult cardiac stem cells are detected in regions of the postnatal heart that originated from the SHF and thus are considered to be remnants of the aforementioned Isl1-positive cells present during the development of the heart.
Gallus gallus
In chicken, the cardiac progenitors migrate from the cranial primitive streak (Hamburger Hamilton stage 3, HH3) rostrally to the lateral plate mesoderm (HH6) where they irreversibly differentiate towards a cardiac identity by expressing Nkx2.5 and GATA4 (Garcia-Martinez and Schoenwolf 1993; Schultheiss et al. 1995). The two lateral plate cardiogenic areas come together and fuse at the midline at stage HH8–9 to form the heart tube around HH10 (Abu-Issa and Kirby (2007) and references therein). The newly formed heart tube is basically non-proliferating, and its growth requires continuous addition of cells to the anterior and posterior poles from the second heart field situated dorsally in the pharyngeal mesoderm (Waldo et al. 2001; Mjaatvedt et al. 2001; Tzahor and Evans 2011). Note that the first description of the second heart field in chicken used the term anterior heart field and only described a subpopulation of the SHF contributing to the arterial pole of the heart. The contribution of cells from the second heart field to the heart primordium is exhausted around HH18 (Waldo et al. 2001) and correlates with cells expressing Isl1 (van den Berg and Moorman 2009). Recent experiments have demonstrated that the chicken pharyngeal mesoderm also contributes to the pharyngeal muscles (Tirosh-Finkel et al. 2006; Nathan et al. 2008). Similar to Xenopus embryos (see below), the expression of chicken Isl1 is peculiar since it initially (HH4) overlaps with the entire cardiogenic area (Yuan and Schoenwolf 2000; Brade et al. 2007). The function of Isl1 during chicken heart development has not been studied so far. For example, it would be interesting to see whether Isl1 also plays a crucial role in the formation of the right ventricle of the four-chambered chicken heart as it does in the mouse.
X. laevis
X. laevis has a three-chambered heart consisting of one partially septated ventricle and two atria. The expression and function of isl1 during Xenopus cardiac development has been analyzed in two previous studies (Brade et al. 2007; Gessert and Kühl 2009). Isl1 expression is first detected at the end of gastrulation (Nieuwkoop Faber stage 13) in the anterior ventral mesoderm of Xenopus embryos by whole-mount in situ hybridization (Gessert and Kühl 2009; Brade et al. 2007). At this stage, isl1 is coexpressed with other cardiac transcription factors such as nkx2.5, gata6, or tbx1, although expression data on a single cell level are not yet available. To analyze the fate of these isl1-positive cells in Xenopus, fate-mapping experiments were performed using DiI as a lineage tracer. To this end, a small group of cells in the anterior ventral region of stage 13 Xenopus embryos was labeled by the injection of DiI, and the fate of these cells was followed during further development (Gessert and Kühl 2009). Cells of this region contribute to all parts of the tadpole Xenopus heart when the different components of the heart, the outflow tract, the ventricle, the atrium, and the inflow tract can be distinguished. Of note, the atrium is not yet septated at this stage. Both sets of data, the expression pattern and fate mapping results, strongly indicate that, in Xenopus, isl1-expressing cells are part of a common cardiac progenitor cell (CPC) population.
In situ hybridization studies further demonstrated that, starting at early tailbud stages, the CPC population separates into an isl1-positive anterior domain and a tbx5-positive posterior domain (Fig. 3b; Gessert and Kühl 2009). These data could also be supported by single-cell RT-PCR experiments performed at stage 24 in which cardiac cells could be identified being either positive for isl1 and nkx2.5 or tbx5 and nkx2.5 (Gessert and Kühl 2009). Additionally, isl1 expression could be detected in the pharyngeal mesoderm and in the mesenchyme of the pharyngeal pouches, similar to isl1 expression in the mouse (Brade et al. 2007). Consistent with this expression pattern, labeling of the anterior isl1-positive cells at stage 24 showed that these cells contribute to the aortic arch arteries, the aortic sac, the distal and proximal outflow tract (OFT), and also to the jaw muscles (Lee and Saint-Jeannet 2011; Gessert and Kühl 2009). This suggests the existence of cells with SHF lineage properties in Xenopus comparable to the SHF lineage described in chicken and mouse (Fig. 3b). Labeling of cells located more posterior to the isl1-positive cell population, which express nkx2.5 and tbx5 resulted in DiI-positive cells in the ventricle, the atrium, and the inflow tract, which may be called a FHF lineage. Thus, the separation into FHF and SHF lineages in Xenopus is indicated by the differential expression of isl1 and tbx5 which resembles the situation in the mouse.
When the heart tube starts to form, isl1 expression within the heart tube is restricted to the dorsolateral regions, the mesocardium, and pericardial roof. Isl1 transcripts are absent from the differentiating myocardium that starts to express the cardiomyocyte differentiation marker troponin Ic (Gessert and Kühl 2009; Brade et al. 2007). This isl1 expression pattern led to the hypothesis that the isl1-positive cells are added to the growing heart tube when it starts to undergo looping to establish the different compartments of the heart as shown in mouse and chicken. However, detailed experimental evidence for this hypothesis is still missing.
To interfere with isl1 function during Xenopus cardiac development, an isl1 antisense morpholino oligonucleotide (MO) was unilaterally injected into the dorsal marginal zone region of early cleavage-stage embryos (Brade et al. 2007). MO-induced knock-down of isl1 results in severe heart abnormalities during development. Dissected hearts revealed a significant size reduction of isl1 MO-injected hearts compared with untreated hearts. In some embryos, the heart tube failed to undergo looping. Molecular analyses showed that expression of core members of the cardiac transcriptional network, e.g., gata 4, gata 6, nkx2.5, and tbx20, are downregulated after isl1 MO injection. Taken together, these findings clearly indicate that isl1 is required early in the CPC population to guarantee normal heart development. A potential later role of isl1 as part of the SHF lineage in Xenopus has not been investigated so far and would require the design of hormone-inducible isl1 constructs which, in contrast to MO-mediated knock-down experiments, allow for a temporal controlled interference.
Danio rerio
The zebrafish heart has two chambers, a ventricle and an atrium. The earliest progenitors of the zebrafish heart are detected bilaterally of the body axis at the onset of gastrulation (5 h post-fertilization (hpf); Fig. 3c) (Lee et al. 1994; Scott and Yelon 2010). These cardiac progenitor cell populations will fuse to form the cardiac cone around 21 somites stage (21ss). This cone undergoes a process called cardiac jogging which finally results in a bent, two-chambered primary heart tube at 48 hpf. Cardiac differentiation is already initiated at segmentation stages when the early cardiac progenitors are still located bilaterally to the midline of the embryo (Schoenebeck and Yelon 2007). The first sign of differentiation is the onset of cardiac myosin light chain 2 (cmlc2) expression at 14ss. Myocardial cell differentiation commences in cells that will later form the ventricle. Differentiation continues in cells of the atrium and subsequently extends to cells of the venous pole. Cells at the arterial pole will differentiate at a different time point (de Pater et al. 2009).
In zebrafish, expression of Isl1 can be detected by immunohistochemistry from early somite stages on, however, the predominant expression domains are neural structures (Korzh et al. 1993). The neural expression was confirmed by using a transgenic zebrafish line expressing GFP under the control of the isl1 promoter (Higashijima et al. 2000). The analysis of isl1 expression in cardiac lineages, however, is difficult in this transgenic reporter line. Thus, it has been assumed for a long time that isl1 is not expressed in cardiac precursor cells in zebrafish.
Two recent studies also employed immunohistochemistry techniques and do provide evidence for Isl1 expression in the heart of zebrafish (de Pater et al. 2009; Hami et al. 2011). From 14ss on, low expression of Isl1 is detected in an area ventral and lateral to the trigeminal placode, similar to the expression domain of GFP in cmlc2::GFP transgenic fish in which GFP is expressed under the control of the cardiac myosin light chain 2 promoter (Hami et al. 2011). Analyses of isl1 expression on sections show that isl1 is also present in the splanchnic mesoderm adjacent to the cardiac mesoderm and in a few cardiomyocyte progenitors located in a region which gives rise to the arterial pole at later stages. At 23ss, Isl1 is localized in vascular endothelium, in the endoderm dorsal to the heart cone and in a subset of cardiomyocytes (de Pater et al. 2009). This demonstrates that Isl1 is expressed in cardiac cells, when the heart cone is forming. Additionally, Isl1 is detected in the mesoderm, where yet undifferentiated heart cells are residing. Starting around 24 hpf (approximately 30ss), Isl1-positive cells are present in the mesenchyme of the branchial arches next to the future arterial pole. This expression persists during heart tube extension and looping and ceases soon thereafter (de Pater et al. 2009). In summary, Isl1 is expressed in a population of cells that can be considered to be equivalent to the SHF in other species. This is also confirmed by lineage labeling experiments showing a contribution of Isl1-positive cells to the arterial pole of the heart (Hami et al. 2011). It remains unclear, however, whether Isl1 is also expressed earlier in a common CPC population in zebrafish.
Given the new insight that Isl1 is also expressed in cardiac progenitor cells, the potential function of Isl1 in zebrafish cardiogenesis was recently studied. Using ENU mutagenesis, an isl1 mutant fish line was generated (islK88X) (de Pater et al. 2009). In this mutant, a single base mutation results in a premature stop codon, which generates a truncated Isl1 protein that lacks the entire homeodomain and part of the LIM domains. Regarding the overall appearance, isl1 mutant fish are initially indistinguishable from wild-type siblings, except for being immotile, most probably due to a dysfunction of motor neurons. Isl1 mutant fish die between 5 and 7 days post-fertilization. Closer analysis of the heart revealed that loss of Isl1 function results in arrhythmia as well as bradycardia. However, in contrast to the findings in Xenopus, neither a knock-down of Isl1 in zebrafish using morpholino oligonucleotides nor the isl1 mutant line exhibited a downregulation of early cardiac markers, e.g., nkx2.5 (de Pater et al. 2009). Although isl1 mutant embryos developed substantially fewer cardiomyocytes at the venous pole, no obvious phenotype was observed at the arterial pole as could have been expected from the Isl1 expression. Most importantly, the authors already point out that this finding may indicate an evolutionary difference between teleosts and amniotes with respect to recruiting cells to the venous and the arterial pole, respectively. Unlike in the mouse where Isl1 is required for the recruitment of cells to both poles of the forming heart (Cai et al. 2003), the function of zebrafish Isl1 appears to be more restricted.
This may be explained by the fact that cardiomyocyte differentiation at the arterial pole does not require Isl1 (de Pater et al. 2009) which leaves Isl1 with the role of a molecular marker for the SHF in fish rather than an essential functional component. This notion is substantiated by a recent publication ascribing a functional role for latent TGF-β binding protein 3 (ltbp3) in zebrafish SHF development (Zhou et al. 2011). The authors describe similarities between the ltbp3 morphant phenotype and the SHF phenotypes in isl1 mutant mouse embryos. This important piece of data indicates that the crucial role of mammalian isl1 in the SHF lineage is taken on by ltbp3 in zebrafish. Nevertheless, the phenotype of isl1 mutant fish is interesting as parts of the heart are affected for which no isl1 expression has been demonstrated so far.
Ciona intestinalis
Tunicates are chordates and represent a group of sessile marine invertebrates. Based on extensive molecular phylogenetic studies, they are considered to represent the sister group of vertebrates in the phylum of chordates (Delsuc et al. 2006; Bourlat et al. 2006; Blair et al. 2005). Ciona larvae develop characteristic features of chordates including a notochord, a nerve chord, and a tail. During metamorphosis, the tail degenerates and the adult organs are formed from the remaining larval structures of the head. This also includes the heart, which consists of a simple V-shaped myocardial tube devoid of an endocardial layer, located in a pericardial sac. At either end, the myocardial tube connects to two vessels located on the dorsal and the ventral body side, respectively, feeding in an open circulatory system. The myocardial tube and the vessels are connected by a group of undifferentiated, proliferating, non-contracting cells, termed growth zone. The myocardial tube is lined with a sheet of undifferentiated cells, which form the so-called undifferentiated line. The Ciona heart shows the exciting property of life-long regeneration with new cardiomyocytes forming from the cells of the growth zone and possibly also from the undifferentiated line. For a more detailed description of Ciona heart development, we refer the reader to recent reviews (Davidson 2007; Levine 2010).
Cell fate-mapping in Ciona demonstrated that the cardiac lineage derives from the bilaterally located B7.5 cells (Fig. 4), which express Ci-Mesp, the Ciona Mesp1 ortholog (Satou et al. 2004). The more anteriorly located progeny of the B7.5 cells represent the trunk ventral cells (TVCs) that give rise to the heart, whereas the more posteriorly located progeny of the B7.5 cells will form the anterior tail muscles (Fig. 4). Consistent with their cardiac fate, the trunk ventral cells express the transcription factors NK4 (the Tinman/Nkx2.5 ortholog), FoxF, GATAa, and Hand-like. Descendants of B7.5 cells not only generate the heart as indicated by a MesP > H2B::CFP transgene but also give rise to the atrial siphon muscle (ASM) after metamorphosis (Stolfi et al. 2010). The two muscle types can be discriminated by the expression of the myosin heavy chain isoforms MHC2 and MHC3 (Ogasawara et al. 2002). A reporter construct driven by a 3.2-kb regulatory region of the single Ciona isl1 gene is expressed in the ASM in juveniles. Precursor cells of the heart, however, are located more ventrally and medially to the progenitors of the ASM. Interestingly, this arrangement resembles the relative position of cells of the FHF and SHF lineage in vertebrates, but a contribution of Isl1-positive cells to the tunicate heart could not be demonstrated. This finding suggests that both, vertebrates and tunicates, posses a population of cardiopharyngeal mesodermal cells that arises from Mesp1-positive progenitor cells which give rise to jaw and siphon muscles.
Fig. 4.
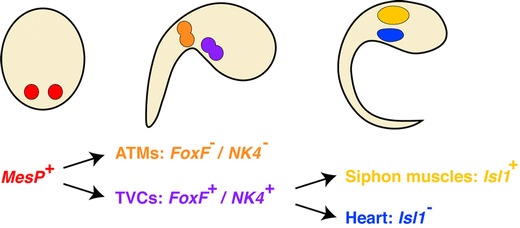
Early heart development in C. intestinalis. The heart derives from the bilateral B7.5 blastomeres indicated in red. TVC: trunk ventral cells, ATM: anterior tail muscles. Adopted from Stolfi et al. (2010)
D. melanogaster
From an evolutionary perspective, it is very interesting that the Drosophila ortholog of isl1, tailup (tup), was demonstrated to hold a crucial role in fly heart development. The fly heart, also called the dorsal vessel, is a simple tube that consists in principle of two main cardiac cell types. The contractile myocardial cells form the lumen of the dorsal vessel and are surrounded by a number of pericardial cells. The heart tube is subdivided into the posterior heart proper and the anterior aorta with a valve separating these two “chambers.” Localized bilaterally at the anterior end of the aorta are cell clusters of the forming lymph glands, which are the hematopoietic organ in Drosophila larvae. During embryonic heart development, Tup expression is detected in all myocardial cells of the heart tube, in a subset of pericardial cells, as well as in a subset of lymph gland cells. Tup expression is also detected early in the cardiac mesoderm where it co-localizes with some Tinman (Tin)-expressing and Even-skipped (Eve)-positive pericardial cells (Mann et al. 2009). The Tup/Isl1 expression pattern supports a role for this factor in the development of different cardiac lineages. Analyses of tup mutant embryos revealed defects for all examined molecular markers for the different heart and lymph gland cell types (Mann et al. 2009; Tao et al. 2007). It could be demonstrated that the early expression of the crucial cardiac factors Tinman (Tin), Pannier (Pnr), and Dorsocross (Doc) was severely affected in tup mutants and vice versa; Tup expression was strongly downregulated in embryos mutant either for tin, pnr, or Doc (Mann et al. 2009). These findings were the first genetic evidence for the interdependency of essential cardiac factors including tup. Moreover, additional experiments set up to demonstrate genetic interactions showed indeed that tup and tin, as well as tup and pnr cooperate during cardiogenesis (Mann et al. 2009).
An additional interesting piece of data came from the analysis of the Drosophila hand enhancer by Tao et al. (2007). The authors identified and confirmed Tup binding sites in the hand enhancer and demonstrated the importance of the presence of these sites for hand expression in vivo. This finding is interesting with respect to mouse heart development where the hand2 gene functions in SHF progenitors and is required for proper cardiogenesis (Tsuchihashi et al. 2011). However, a regulation of hand2 through Isl1 has not yet been investigated. Taken together, the described data are clear evidence for the requirement of tup/isl1 in early steps of Drosophila cardiogenesis and add tup/isl1 to the core set of ancestral cardiac transcription factors.
Evidences for an Isl1-positive common cardiac progenitor
Isl1 was initially hypothesized to be a specific marker for the SHF and to solely function in the development of SHF lineages. However, accumulating data strongly indicate that Isl1 is already expressed in the common cardiac progenitor cell population and has an important function in heart development. In chicken and Xenopus, it was clearly demonstrated that isl1 transcripts are present in this progenitor pool which splits into FHF and SHF lineages during development (Yuan and Schoenwolf 2000; Brade et al. 2007; Abu-Issa and Kirby 2008). Also, as demonstrated by immunohistochemistry, Isl1 expression in mouse embryos commences before E7.5 (Prall et al. 2007) in regions that give rise to both FHF and SHF lineages, providing more evidence for the existence of an initially single heart field. Moreover, by applying a similar Isl1–Cre lineage analysis as was performed by Cai et al. (2003) but using a different floxed locus (Gata flap), it was shown that the overall contribution of isl1-positive cells to different heart compartments is much broader (Ma et al. 2008). This finding reinforces the above listed observations that isl1 marks a much earlier, common cardiovascular progenitor cell as opposed to solely being a marker of the second heart field. Also, results obtained from murine and human embryonic stem (ES) cell cultures indicate that Isl1 is one of the first cardiac transcription factors that marks a common progenitor, which gives rise not only to the FHF and SHF lineages but also to cells of the vascular system. In particular, Isl1-positive progenitors can also develop into endothelial and smooth muscle cells (Moretti et al. 2006, 2010; Bu et al. 2009). Consistently, a knock-down of Isl1 in Xenopus not only results in cardiac but also in endothelial marker gene expression defects (Brade et al. 2007). A recent study suggested that an early function of Isl1 is required for endothelial differentiation in ES cells, whereas slightly later Isl1 is required for cardiomyocyte development (Bondue et al. 2011). Importantly, even in Drosophila, the isl1 ortholog tup is expressed early in the cardiac mesoderm and loss of function results in heart defects encompassing the different heart cell types as well as lymph gland cells (Mann et al. 2009; Tao et al. 2007). With respect to these comparable findings across species, it is interesting that neither in zebrafish nor in Amphioxus or lamprey has Isl1 expression been unambiguously shown to be present in the CPC pool of cells (Jackman et al. 2000; de Pater et al. 2009; Hami et al. 2011; Kokubo et al. 2010). This could be due to technical limitations to detect isl1 transcripts or Isl1 protein in the cardiac mesoderm of these organisms.
Since, in Drosophila and Xenopus, Isl1 is expressed in the cardiac mesoderm and was shown to be important for cardiogenesis, several questions arise. Has Isl1 been integrated independently into the cardiac gene regulatory networks of Drosophila (or protostomes in general) and of vertebrates? Here, it is of particular interest that the Drosophila dorsal vessel could be considered as a specialized blood vessel with the ability to contract (as described above) and which is equally related to vertebrate blood vessels (most probably to veins) and vertebrate hearts. Moreover, since, in vertebrates, Isl1 is not only found in the common cardiac progenitor but also in endothelial and smooth muscle precursor cells, it is tempting to speculate that Isl1 might label a subpopulation of mesothelial cells. Also of interest is the question whether the function of Isl1 during early cardiogenesis became dispensable in tunicates and fishes (and possibly other species). Maybe the role of Isl1 in early cardiac (and vascular) development in tunicates and fishes has not been fully understood so far. Further analyses of these issues will help to clarify these questions.
Isl1 within a cardiac gene regulatory network
Hardly anything is known about the regulation of isl1 expression in the common cardiovascular progenitor cell population in vertebrates. Contradicting data are available with respect to a potential regulation of isl1 through Mesp1. A recent gain and loss of function study suggested that Isl1 is not regulated by Mesp1, blatantly contradicting previous results showing an upregulation of Isl1 expression upon Mesp1 overexpression (Lindsley et al. 2008; Bondue et al. 2011; 2008). This issue therefore is not yet solved. A dual reporter approach described an enhancer, 3–6-kb downstream of the mouse isl1 locus, that faithfully reproduces isl1 expression in the heart of E7.5–E10.5 transgenic mouse embryos and contains conserved potential binding sites for Foxo/Foxd, Tcf1, and GATA transcription factors (Kappen and Salbaum 2009). However, investigation of the interactions of isl1 with the other cardiac core transcription factors in the CPC requires further detailed investigations. In Drosophila, however, it has already been shown, as discussed earlier in this review, that tup/isl1 expression depends on tin (nkx2.5 ortholog), Doc (Tbx factor ortholog), and pnr (GATA factor ortholog) and vice versa. Loss-of-function studies in Xenopus also suggest an involvement of Isl1 in the early cardiac regulatory network (Brade et al. 2007). Thus, it is tempting to speculate that these four classes of transcription factors are wired to form a stable gene regulatory network within the common cardiac progenitor cell population.
More is known about the regulation of isl1 expression in the second heart field in the mouse. The Isl1-positive cardiac progenitors in the pharyngeal mesoderm are subjected to regulations from several signaling pathways, including the Wnt, BMP, and FGF signaling pathways. There is a general consensus that Wnt signaling functions at various stages of heart development to promote proliferation of cardiac progenitors and blocking their differentiation (reviewed in Gessert and Kühl 2010). In this context, it is interesting to note that Isl1 and FGF10 (both endogenous markers for the second heart field) are direct targets of Wnt/β-catenin signaling (Cohen et al. 2007; Lin et al. 2007; Qyang et al. 2007), implicating Wnt and FGF signaling in the proliferation of Isl1-positive progenitors. Furthermore, FGF8 (an additional endogenous marker for the second heart field) has been shown to be required for the elongation of the heart and development of the right ventricle and the OFT. FGF8 also controls Isl1 expression in the pharyngeal mesoderm, thus connecting Isl1 expression not only to FGF signaling but also to retinoic acid and Tbx1 signaling (Sirbu et al. 2008; Ryckebusch et al. 2008; Ilagan et al. 2006; Park et al. 2006; Xu et al. 2004). Notch signaling inhibits Wnt/β-catenin signaling in the cardiac precursors, while the Notch target gene Hes1 is expressed in a pharyngeal domain overlapping with Isl1 expression and is required for proliferation in the second heart field and outflow tract extension, thus integrating Notch signaling in a cardiac differentiation-promoting position within the cardiac transcription network (Kwon et al. 2009; Rochais et al. 2009). Interestingly, Isl1 was found to directly regulate the expression of Myocd, a transcriptional cofactor of SRF known to promote the development of ventricular cardiomyocytes to functions as a molecular switch between vascular smooth muscle and skeletal muscle differentiation (Hoofnagle et al. 2011; Wang et al. 2001; Li et al. 2003; Long et al. 2007).
More data on Isl1 regulation have come from the evaluation of mutant mouse embryos phenocopying the Isl1 -/- heart phenotype. Mef2c -/- and Foxh1 -/- embryos both lack the outflow tract and the right ventricle, with Mef2c being a direct target of both Isl1 and Foxh1 (while Isl1 transcription does not depend on Foxh1) (Dodou et al. 2004; von Both et al. 2004). BMP4 signaling has been shown to be required for OFT development (Liu et al. 2004) while a conditional deletion of the BMP receptor BMPR1a using Isl1–Cre affected both the outflow tract and the right ventricle (Yang et al. 2006). Interestingly, Isl1 expression was found to be upregulated in Isl1-Cre/BMPR1a embryos, a negative effect that was subsequently shown to be mediated by microRNAs from the BMP4-regulated miR-17-92 s cluster (Wang et al. 2010). Last but not least, shh mutant embryos are also characterized by defective OFT and SHF development as Shh signaling from the pharyngeal endoderm is required for correct Isl1 expression and for correct development of SHF derivatives (Goddeeris et al. 2007; Washington Smoak et al. 2005; Lin et al. 2006). This network appears to be conserved across species as in zebrafish; Isl1 expression in the second heart field also depends on Shh function and Tbx1 expression (Hami et al. 2011). In summary, these data describe the image of a complex second heart field regulatory network with Isl1 in its center.
Towards an evolutionary analysis of cardiac gene regulatory networks
Taken together, the data from different organisms suggest two distinct functions of Isl1 during cardiogenesis. Early during heart development, Isl1 is part of the gene regulatory network required for the establishment of a common cardiac progenitor cell population. Data from Drosophila and Xenopus suggest a conserved network of regulatory interactions involving Isl1 and members of the GATA, Nkx, and Tbx families of transcription factors (Fig. 5a). Likely, a similar network also exists in higher vertebrates, e.g., the mouse. Further studies are required to elucidate these interactions.
Fig. 5.
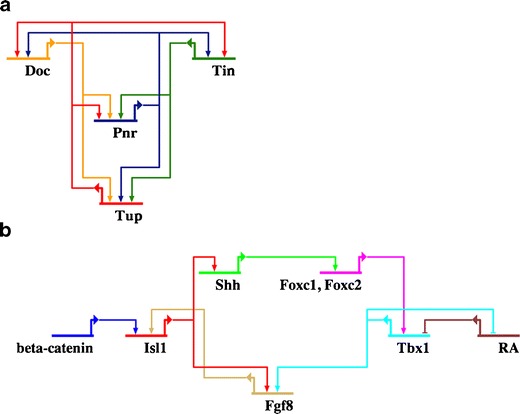
Gene regulatory networks in cardiac cells including Isl1. a Interaction of key cardiac transcription factors in the early cardiac mesoderm in Drosophila. doc: dorsocross, tup: tailup, pnr: pannier, tin: Tinman. b Interaction of signaling molecules and transcription factors including Isl1 in the pharyngeal mesoderm/SHF in the mouse. Shh: sonic hedgehog, RA: retinoic acid, Fgf8: fibroblast growth factor 8
In addition, Isl1 plays an important second role in the cardiac development of deuterostomes (Fig. 1). During embryonic development of chordates, Isl1 labels a population of pharyngeal mesodermal cells contributing to the heart as well as to the musculature of the head. In chordates such as Ciona, the corresponding cell population only gives rise to the atrial siphon muscle but not to the heart. In craniates such as the zebrafish and Xenopus, these cells only contribute to the arterial pole of the heart. In contrast, they also contribute to the venous pole in amniotes. Data from the mouse and the zebrafish, again, argue for a conserved regulatory network in this population of cells involving factors such as Isl1, Tbx1, and Shh (Fig. 5b).
So far, the available data provide significant insight into the gene regulatory networks that govern cardiogenesis. However, more research is needed to obtain a comprehensive understanding of the molecular networks present in the common CPC population and in the pharyngeal mesoderm across species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 46 kb)
Acknowledgment
Work in our lab focuses on heart development in different species including mouse (IOS), Xenopus (MK, SK, PP), zebrafish (MP), and Drosophila (PP) is funded by the DFG (IOS, MK, PP), the European Union (MP), and Stiftung für Herzforschung (MP). We thank Franziska Herrmann for providing the cartoons shown in Fig. 5. We apologize to all authors whose work we could not cite due to space limitation.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol. 2007;23:45–68. doi: 10.1146/annurev.cellbio.23.090506.123331. [DOI] [PubMed] [Google Scholar]
- Abu-Issa R, Kirby ML. Patterning of the heart field in the chick. Dev Biol. 2008;319(2):223–233. doi: 10.1016/j.ydbio.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Shah P, Hedges SB. Evolutionary sequence analysis of complete eukaryote genomes. BMC Bioinformatics. 2005;6:53. doi: 10.1186/1471-2105-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118(3):719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3(1):69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Bondue A, Tannler S, Chiapparo G, Chabab S, Ramialison M, Paulissen C, Beck B, Harvey R, Blanpain C. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J Cell Biol. 2011;192(5):751–765. doi: 10.1083/jcb.201007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, Lander ES, Thorndyke M, Nakano H, Kohn AB, Heyland A, Moroz LL, Copley RR, Telford MJ. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444(7115):85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- Brade T, Gessert S, Kuhl M, Pandur P. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev Biol. 2007;311(2):297–310. doi: 10.1016/j.ydbio.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460(7251):113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- Budelmann BU, Schipp R, von Boletzky S (1997) Cephalopoda, In: Microscopic anatomy of invertebrates. Editors: Harrison FW, Kohn AJ, Volume 6a, Molluscs. Wiley-Liss, New York, 119–414
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–889. doi: 10.1016/S1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest. 2007;117(7):1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B. Ciona intestinalis as a model for cardiac development. Semin Cell Dev Biol. 2007;18(1):16–26. doi: 10.1016/j.semcdb.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pater E, Clijsters L, Marques SR, Lin YF, Garavito-Aguilar ZV, Yelon D, Bakkers J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136(10):1633–1641. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439(7079):965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131(16):3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Gajewski K, Zhang Q, Choi CY, Fossett N, Dang A, Kim YH, Kim Y, Schulz RA. Pannier is a transcriptional target and partner of Tinman during Drosophila cardiogenesis. Dev Biol. 2001;233(2):425–436. doi: 10.1006/dbio.2001.0220. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez V, Schoenwolf GC. Primitive-streak origin of the cardiovascular system in avian embryos. Dev Biol. 1993;159(2):706–719. doi: 10.1006/dbio.1993.1276. [DOI] [PubMed] [Google Scholar]
- Genead R, Danielsson C, Andersson AB, Corbascio M, Franco-Cereceda A, Sylven C, Grinnemo KH. Islet-1 cells are cardiac progenitors present during the entire lifespan: from the embryonic stage to adulthood. Stem Cells Dev. 2010;19(10):1601–1615. doi: 10.1089/scd.2009.0483. [DOI] [PubMed] [Google Scholar]
- Gessert S, Kühl M. Comparative gene expression analysis and fate mapping studies suggest an early segregation of cardiogenic lineages in Xenopus laevis. Dev Biol. 2009;334(2):395–408. doi: 10.1016/j.ydbio.2009.07.037. [DOI] [PubMed] [Google Scholar]
- Gessert S, Kühl M. The multiple phases and faces of Wnt signaling during cardiac differentiation and development. Circ Res. 2010;107:186–199. doi: 10.1161/CIRCRESAHA.110.221531. [DOI] [PubMed] [Google Scholar]
- Goddeeris MM, Schwartz R, Klingensmith J, Meyers EN. Independent requirements for Hedgehog signaling by both the anterior heart field and neural crest cells for outflow tract development. Development. 2007;134(8):1593–1604. doi: 10.1242/dev.02824. [DOI] [PubMed] [Google Scholar]
- Hami D, Grimes AC, Tsai HJ, Kirby ML. Zebrafish cardiac development requires a conserved secondary heart field. Development. 2011;138(11):2389–2398. doi: 10.1242/dev.061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Mandal L (2006) The blood/vascular system in a phylogenetic perspective. BioEssays 28(12):1203–1210 [DOI] [PubMed]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20(1):206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoofnagle MH, Neppl RL, Berzin EL, Teg Pipes GC, Olson EN, Wamhoff BW, Somlyo AV, Owens GK. Myocardin is differentially required for the development of smooth muscle cells and cardiomyocytes. Am J Physiol Heart Circ Physiol. 2011;300(5):H1707–H1721. doi: 10.1152/ajpheart.01192.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan R, Abu-Issa R, Brown D, Yang YP, Jiao K, Schwartz RJ, Klingensmith J, Meyers EN. Fgf8 is required for anterior heart field development. Development. 2006;133(12):2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- Jackman WR, Langeland JA, Kimmel CB. islet reveals segmentation in the Amphioxus hindbrain homolog. Dev Biol. 2000;220(1):16–26. doi: 10.1006/dbio.2000.9630. [DOI] [PubMed] [Google Scholar]
- Jamieson BGM (1992) Oligochaeta. In: Microscopic anatomy of invertebrates. Editors: Harrison FW, Gardiner SL, Volume 7, Annelida. Wiley-Liss, New York, 217–322
- Kappen C, Salbaum JM. Identification of regulatory elements in the Isl1 gene locus. Int J Dev Biol. 2009;53(7):935–946. doi: 10.1387/ijdb.082819ck. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattar P, Friedrich FW, Bonne G, Carrier L, Eschenhagen T, Evans SM, Schwartz K, Fiszman MY, Vilquin JT. Distinction between two populations of islet-1-positive cells in hearts of different murine strains. Stem Cells Dev. 2011;20(6):1043–1052. doi: 10.1089/scd.2010.0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo N, Matsuura M, Onimaru K, Tiecke E, Kuraku S, Kuratani S, Tanaka M. Mechanisms of heart development in the Japanese lamprey, Lethenteron japonicum. Evol Dev. 2010;12(1):34–44. doi: 10.1111/j.1525-142X.2009.00389.x. [DOI] [PubMed] [Google Scholar]
- Korzh V, Edlund T, Thor S. Zebrafish primary neurons initiate expression of the LIM homeodomain protein Isl-1 at the end of gastrulation. Development. 1993;118(2):417–425. doi: 10.1242/dev.118.2.417. [DOI] [PubMed] [Google Scholar]
- Koshiba-Takeuchi K, Mori AD, Kaynak BL, Cebra-Thomas J, Sukonnik T, Georges RO, Latham S, Beck L, Henkelman RM, Black BL, Olson EN, Wade J, Takeuchi JK, Nemer M, Gilbert SF, Bruneau BG. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature. 2009;461(7260):95–98. doi: 10.1038/nature08324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl SJ, Kühl M. Improving cardiac regeneration after injury: are we a step closer? Bioessays. 2011;33(9):669–673. doi: 10.1002/bies.201100046. [DOI] [PubMed] [Google Scholar]
- Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol. 2009;11(8):951–957. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, Platoshyn O, Yuan JX, Evans S, Chien KR. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433(7026):647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR. Islet1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135(2):193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- Lee YH, Saint-Jeannet JP. Cardiac neural crest is dispensable for outflow tract septation in Xenopus. Development. 2011;138(10):2025–2034. doi: 10.1242/dev.061614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RK, Stainier DY, Weinstein BM, Fishman MC. Cardiovascular development in the zebrafish. II. Endocardial progenitors are sequestered within the heart field. Development. 1994;120(12):3361–3366. doi: 10.1242/dev.120.12.3361. [DOI] [PubMed] [Google Scholar]
- Lescroart F, Kelly RG, Le Garrec JF, Nicolas JF, Meilhac SM, Buckingham M. Clonal analysis reveals common lineage relationships between head muscles and second heart field derivatives in the mouse embryo. Development. 2010;137(19):3269–3279. doi: 10.1242/dev.050674. [DOI] [PubMed] [Google Scholar]
- Levine M. The specification of myocardial progenitor cells in the Ciona tadpole. Heart development and regeneration. Amsterdam: Academic Press, Elsevier; 2010. [Google Scholar]
- Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2003;100(16):9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Bu L, Cai CL, Zhang X, Evans S. Isl1 is upstream of sonic Hedgehog in a pathway required for cardiac morphogenesis. Dev Biol. 2006;295(2):756–763. doi: 10.1016/j.ydbio.2006.03.053. [DOI] [PubMed] [Google Scholar]
- Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai CL, Bu L, Yang L, Martin J, Kemler R, Rosenfeld MG, Chen J, Evans SM. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci U S A. 2007;104(22):9313–9318. doi: 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley RC, Gill JG, Murphy TL, Langer EM, Cai M, Mashayekhi M, Wang W, Niwa N, Nerbonne JM, Kyba M, Murphy KM. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial–mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3(1):55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Selever J, Wang D, Lu MF, Moses KA, Schwartz RJ, Martin JF. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proc Natl Acad Sci U S A. 2004;101(13):4489–4494. doi: 10.1073/pnas.0308466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Creemers EE, Wang DZ, Olson EN, Miano JM. Myocardin is a bifunctional switch for smooth versus skeletal muscle differentiation. Proc Natl Acad Sci U S A. 2007;104(42):16570–16575. doi: 10.1073/pnas.0708253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Zhou B, Pu WT. Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity. Dev Biol. 2008;323(1):98–104. doi: 10.1016/j.ydbio.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann T, Bodmer R, Pandur P. The Drosophila homolog of vertebrate Islet1 is a key component in early cardiogenesis. Development. 2009;136(2):317–326. doi: 10.1242/dev.022533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238(1):97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- Moore AW, Barbel S, Jan LY, Jan YN. A genomewide survey of basic helix-loop-helix factors in Drosophila. Proc Natl Acad Sci U S A. 2000;97(19):10436–10441. doi: 10.1073/pnas.170301897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127(6):1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Moretti A, Lam J, Evans SM, Laugwitz KL. Biology of Isl1+ cardiac progenitor cells in development and disease. Cell Mol Life Sci. 2007;64(6):674–682. doi: 10.1007/s00018-007-6520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A, Bellin M, Jung CB, Thies TM, Takashima Y, Bernshausen A, Schiemann M, Fischer S, Moosmang S, Smith AG, Lam JT, Laugwitz KL. Mouse and human induced pluripotent stem cells as a source for multipotent Isl1+ cardiovascular progenitors. FASEB J. 2010;24(3):700–711. doi: 10.1096/fj.09-139477. [DOI] [PubMed] [Google Scholar]
- Nathan E, Monovich A, Tirosh-Finkel L, Harrelson Z, Rousso T, Rinon A, Harel I, Evans SM, Tzahor E. The contribution of Islet1-expressing splanchnic mesoderm cells to distinct branchiomeric muscles reveals significant heterogeneity in head muscle development. Development. 2008;135(4):647–657. doi: 10.1242/dev.007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop P, Faber J. External and internal stage criteria in the development of Xenopus laevis. Amsterdam: Elsevier; 1975. [Google Scholar]
- Ogasawara M, Sasaki A, Metoki H, Shin-i T, Kohara Y, Satoh N, Satou Y. Gene expression profiles in young adult Ciona intestinalis. Dev Genes Evol. 2002;212(4):173–185. doi: 10.1007/s00427-002-0230-7. [DOI] [PubMed] [Google Scholar]
- Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133(12):2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, Stennard FA, Wise N, Schaft D, Wolstein O, Furtado MB, Shiratori H, Chien KR, Hamada H, Black BL, Saga Y, Robertson EJ, Buckingham ME, Harvey RP. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128(5):947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, Caron L, Wu X, Clarke J, Taketo MM, Laugwitz KL, Moon RT, Gruber P, Evans SM, Ding S, Chien KR. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Reim I, Lee HH, Frasch M. The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development. 2003;130(14):3187–3204. doi: 10.1242/dev.00548. [DOI] [PubMed] [Google Scholar]
- Rochais F, Mesbah K, Kelly RG. Signaling pathways controlling second heart field development. Circ Res. 2009;104(8):933–942. doi: 10.1161/CIRCRESAHA.109.194464. [DOI] [PubMed] [Google Scholar]
- Ryckebusch L, Wang Z, Bertrand N, Lin SC, Chi X, Schwartz R, Zaffran S, Niederreither K. Retinoic acid deficiency alters second heart field formation. Proc Natl Acad Sci U S A. 2008;105(8):2913–2918. doi: 10.1073/pnas.0712344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126(15):3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, Satoh N. The ascidian Mesp gene specifies heart precursor cells. Development. 2004;131(11):2533–2541. doi: 10.1242/dev.01145. [DOI] [PubMed] [Google Scholar]
- Saudemont A, Dray N, Hudry B, Le Gouar M, Vervoort M, Balavoine G. Complementary striped expression patterns of NK homeobox genes during segment formation in the annelid Platynereis. Dev Biol. 2008;317(2):430–443. doi: 10.1016/j.ydbio.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, Yelon D. Illuminating cardiac development: advances in imaging add new dimensions to the utility of zebrafish genetics. Semin Cell Dev Biol. 2007;18(1):27–35. doi: 10.1016/j.semcdb.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss TM, Xydas S, Lassar AB. Induction of avian cardiac myogenesis by anterior endoderm. Development. 1995;121(12):4203–4214. doi: 10.1242/dev.121.12.4203. [DOI] [PubMed] [Google Scholar]
- Scott IC, Yelon D. Cardiac development in the zebrafish. Heart development and regeneration. Amsterdam: Academic Press, Elsevier; 2010. [Google Scholar]
- Simoes-Costa MS, Vasconcelos M, Sampaio AC, Cravo RM, Linhares VL, Hochgreb T, Yan CY, Davidson B, Xavier-Neto J. The evolutionary origin of cardiac chambers. Dev Biol. 2005;277(1):1–15. doi: 10.1016/j.ydbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Sirbu IO, Pandur P. Saving hearts through basic research. Birth Defects Res C Embryo Today. 2009;87(3):273–283. doi: 10.1002/bdrc.20159. [DOI] [PubMed] [Google Scholar]
- Sirbu IO, Zhao X, Duester G. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev Dyn. 2008;237(6):1627–1635. doi: 10.1002/dvdy.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solloway MJ, Harvey RP. Molecular pathways in myocardial development: a stem cell perspective. Cardiovasc Res. 2003;58(2):264–277. doi: 10.1016/S0008-6363(03)00286-4. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Harvey RP. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005;132(22):4897–4910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- Stolfi A, Gainous TB, Young JJ, Mori A, Levine M, Christiaen L. Early chordate origins of the vertebrate second heart field. Science. 2010;329(5991):565–568. doi: 10.1126/science.1190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J, Evans SM. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304(1):286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Wang J, Tokusumi T, Gajewski K, Schulz RA. Requirement of the LIM homeodomain transcription factor tailup for normal heart and hematopoietic organ formation in Drosophila melanogaster. Mol Cell Biol. 2007;27(11):3962–3969. doi: 10.1128/MCB.00093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh-Finkel L, Elhanany H, Rinon A, Tzahor E. Mesoderm progenitor cells of common origin contribute to the head musculature and the cardiac outflow tract. Development. 2006;133(10):1943–1953. doi: 10.1242/dev.02365. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi T, Maeda J, Shin CH, Ivey KN, Black BL, Olson EN, Yamagishi H, Srivastava D. Hand2 function in second heart field progenitors is essential for cardiogenesis. Dev Biol. 2011;351(1):62–69. doi: 10.1016/j.ydbio.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahor E, Evans SM. Pharyngeal mesoderm development during embryogenesis: implications for both heart and head myogenesis. Cardiovasc Res. 2011;91(2):196–202. doi: 10.1093/cvr/cvr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg G, Moorman AF. Concepts of cardiac development in retrospect. Pediatr Cardiol. 2009;30(5):580–587. doi: 10.1007/s00246-008-9369-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Both I, Silvestri C, Erdemir T, Lickert H, Walls JR, Henkelman RM, Rossant J, Harvey RP, Attisano L, Wrana JL. Foxh1 is essential for development of the anterior heart field. Dev Cell. 2004;7(3):331–345. doi: 10.1016/j.devcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Wada S, Tokuoka M, Shoguchi E, Kobayashi K, Di Gregorio A, Spagnuolo A, Branno M, Kohara Y, Rokhsar D, Levine M, Saiga H, Satoh N, Satou Y. A genomewide survey of developmentally relevant genes in Ciona intestinalis. II. Genes for homeobox transcription factors. Dev Genes Evol. 2003;213(5–6):222–234. doi: 10.1007/s00427-003-0321-0. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128(16):3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105(7):851–862. doi: 10.1016/S0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, Bai Y, Huang Z, Black BL, Wang F, Martin JF. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell. 2010;19(6):903–912. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington Smoak I, Byrd NA, Abu-Issa R, Goddeeris MM, Anderson R, Morris J, Yamamura K, Klingensmith J, Meyers EN. Sonic Hedgehog is required for cardiac outflow tract and neural crest cell development. Dev Biol. 2005;283(2):357–372. doi: 10.1016/j.ydbio.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Buckingham M. The formation of the embryonic mouse heart: heart fields and myocardial cell lineages. Ann N Y Acad Sci. 2010;1188:15–24. doi: 10.1111/j.1749-6632.2009.05078.x. [DOI] [PubMed] [Google Scholar]
- Witman N, Murtuza B, Davis B, Arner A, Morrison JI. Recapitulation of developmental cardiogenesis governs the morphological and functional regeneration of adult newt hearts following injury. Dev Biol. 2011;354(1):67–76. doi: 10.1016/j.ydbio.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Xavier-Neto J, Davidson B, Simoes-Costa MS, Castro RA, Castillo HA, Sampaio AC, Azambuja AP. Evolutionary origins of hearts. Heart development and regeneration. Amsterdam: Academic Press, Elsevier; 2010. [Google Scholar]
- Xu H, Morishima M, Wylie JN, Schwartz RJ, Bruneau BG, Lindsay EA, Baldini A. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131(13):3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- Yang L, Cai CL, Lin L, Qyang Y, Chung C, Monteiro RM, Mummery CL, Fishman GI, Cogen A, Evans S. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 2006;133(8):1575–1585. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Schoenwolf GC. Islet-1 marks the early heart rudiments and is asymmetrically expressed during early rotation of the foregut in the chick embryo. Anat Rec. 2000;260(2):204–207. doi: 10.1002/1097-0185(20001001)260:2<204::AID-AR90>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Cashman TJ, Nevis K, Obregon P, Carney SA, Liu Y, Gu AH, Mosimann C, Sondalle S, Peterson R, Heideman W, Burns CE, and Burns CG (2011) Latent TGF-b binding protein 3 identifies a second heart field in zebrafish. Nature. 474, 645–648 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 46 kb)


