Abstract
Oxidative modifications can have significant effects on protein structure in solution. Here, the structures and stabilities of oxidized ubiquitin ions electrosprayed from an aqueous solution (pH 2) are studied by ion mobility spectrometry-mass spectrometry (IMS-MS). IMS-MS has proven to be a valuable technique to assess gas-phase and in many cases, solution structures. Herein, in vitro oxidation is performed by Fenton chemistry with Fe(II)/hydrogen peroxide. Most molecules in solution remain unmodified whereas ~20% of the population belongs to an M+16 Da oxidized species. Ions of low charge states (+7 and +8) show substantial variance in collision cross section distributions between unmodified and oxidized species. Novel and previously reported Gaussian conformers are used to model cross section distributions for +7 and +8 oxidized ubiquitin ions, respectively, in order to correlate variances in observed gas-phase distributions to changes in populations of solution states. Based on Gaussian modeling, oxidized ions of charge state +7 have an A-state conformation which is more populated for oxidized relative to unmodified ions. Oxidized ubiquitin ions of charge state +8 have a distribution of conformers arising from native-state ubiquitin and higher intensities of A- and U-state conformers relative to unmodified ions. This work provides evidence that incorporation of a single oxygen atom to ubiquitin leads to destabilization of the native state in an acidic solution (pH ~2) and to unfolding of gas-phase compact structures.
Keywords: ubiquitin, structures, ion mobility, protein folding, oxidation, ion mobility-mass spectrometry
Introduction
In biological systems, reactive oxygen species (ROS) are introduced during cellular processes and from exogenous factors such as ultraviolet radiation, environmental toxins, and chemotherapeutics.1,2 Elevated levels of oxidative stress-an overproduction of ROS and/or a deficiency of antioxidants-can result in irreversible modifications to proteins, lipids, carbohydrates, and DNA.2, 3 Oxidative stress has been heavily implicated in aging and neurodegenerative disorders such as Alzheimers and Parkinson’s Diseases, especially at the protein level.4 – 8 Several studies have shown that protein oxidation can affect structure, stability, and function. 9 – 14 Better understanding of the effects that oxidation has on protein tertiary structure can provide insight to processes that lead to enzymatic inactivation for proteins implicated in disease pathogenesis. In this work, we explore the effect of oxidation on the structure of ubiquitin using ion mobility spectrometry coupled with mass spectrometry (IMS-MS) with the aim of correlating the observed gas-phase cross section distributions to conformers that exist in solution.
Since the advent of “soft” ionization methods such as electrospray ionization (ESI),15 a range of MS-based techniques have been applied to elucidate biomolecular structures in the absence of solvent.16–22 Gas-phase conformations can provide information complementary to solution protein structures and dynamics that are measured by traditional techniques such as circular dichroism (CD) and nuclear magnetic resonance (NMR) spectroscopies. Gas-phase studies may contribute to fundamental understanding of the roles of intramolecular, solvent-molecule, and solvent-solvent interactions in establishing structures. IMS-MS has proven to be a powerful technique to assess the conformations of proteins.23,24 IMS determines the overall shape of molecules by measuring the time ions require to travel through a drift tube which is filled with an inert buffer gas under the influence of a uniform electric field. The ion’s drift time depends on its collision cross section (Ω) and charge.
Structural transitions of biomolecules from solution to the gas phase have been extensively investigated and several studies reveal that gas-phase ions retain, to some extent, their solution-structure elements up to milliseconds after ESI. 25 – 30 Recently, Clemmer and coworkers have proposed that different solution structures can be distinguished based on variances in the cross sections of their gas-phase conformations, even when gas-phase structures are dissimilar from their solution states due to solvent evaporation.31,32 Their work describes a method for determination of the populations of solution states for peptides (i.e., bradykinin31) and small proteins (i.e., ubiquitin32) by measuring IMS distributions generated from various solution conditions.
Ubiquitin is a widely studied protein with a 76 amino acid sequence containing a single methionine residue on the N terminus.33,34 Studies of the effects of oxidation on the conformation of ubiquitin have been reported and present conflicting results regarding whether oxidation influences the native-state conformation.9,10,35–38 It is suggested that ubiquitin is destabilized and expanded at low pH (~2.4) when methionine is oxidized to methionine sulfoxide based on analyses with gel electrophoresis and CD spectroscopy.10 In particular, two solution-phase isomers of methionine sulfoxide have lower mobilities on the gel relative to unmodified ubiquitin reflecting changes in solution conformation. Similarly, photochemically-oxidized ubiquitin results in conformational changes from the native state upon monoxidation (and additional oxidations) measured by CD.35 On the other hand, γ-induced oxidation of ubiquitin does not lead to any protein unfolding of the native state as also measured by CD.36 A recent IMS-MS investigation of radical-induced electrical oxidation of ubiquitin has measured similar collision cross sections for monooxidized and unmodified ubiquitin ions of charge states +5 and +6, which are formed upon electrospraying a 2.5–5 μM solution in a 10 mM ammonium acetate buffer.38 The observed structures for monooxidized and unmodified ions were compact in conformation for +5 ions.38 The distributions for ions of charge state +5 were identical for monooxidized and unmodified ions based on a single compact conformer, whereas ions of charge state +6 favored the more compact structure of two observed conformers for monooxidized species. Those IMS-MS experiments were performed on a travelling-wave IMS-MS instrument and reported no significant change to ubiquitin structures due to oxidation.38 The work presented here seeks to provide further insight and clarification to the effects of oxidation on ubiquitin conformers.
The native structure of ubiquitin consists of five stranded β-sheets and α and 310 helices.39 Native-state (N) ubiquitin is favored in aqueous solution across the pH range ~1.2 to 8.4 based on NMR data.40 At low pH (~2) in a 40:60 water:methanol solution, a partially folded state (A) of ubiquitin emerges and is stabilized.41,42 The N-terminal portion of the A state retains native-like secondary structural features, whereas, the C-terminal half has a more extended and helical structure.41–46 There is other evidence which supports the A state as folding intermediates of ubiquitin.45,47–49 When the methanol content increases or in the presence of denaturants, an unfolded state (U) is present.50,51 In the absence of solvent, ubiquitin ions have three types of gas-phase IMS structures: compact states (Ω <1120 Å2), partially-folded states (1120<Ω<1500 Å2), and elongated states (Ω >1500 Å2).52 The distribution of these states can be heavily influenced by the initial solution conditions prior to ESI.32,53
In the current study, we examine the effects of protein oxidation on gas-phase conformers of mobility-separated ubiquitin N, A, and U states. Fenton chemistry [i.e., Fe(II)/hydrogen peroxide (H2O2)] is used to oxidize ubiquitin in solution and generates an M+16 Da species which corresponds primarily to methionine oxidation to methionine sulfoxide. We note that other M+16 Da positional isomers, in substantially lower concentrations, have been reported under these and other oxidizing conditions.35,37,54 These studies examine the gas-phase structures of monooxidized ubiquitin produced from an acidic solution (pH 2) by probing IMS profiles for ions of charge states +7 to +13 with a traditional IMS-MS instrument that is capable of resolving powers as high as ~150 (as assessed by t/Δtfwhm). 55 It is observed that relatively low charge state (+7 and +8) ions of oxidized ubiquitin generate different drift time distributions from that of the unmodified ions. Novel and previously established Gaussian functions32 are employed to model the IMS distributions of oxidized ions for charge states +7 and +8, respectively, and to interpret their structural variances in the solution phase. The results presented herein provide valuable insight to the effects of a single oxygen addition on the solution and gas-phase structures of ubiquitin.
Experimental
In vitro oxidation of ubiquitin
Bovine ubiquitin was purchased from Sigma-Aldrich (St. Louis, MO). In vitro oxidation (10 mg·mL−1) was performed in 10 mM sodium phosphate buffer (pH=7.4) with 10 mM H2O2 and 1 mM FeCl2 at 37 °C for 2 hours. The oxidation reaction was quenched by flash freezing with liquid nitrogen and samples were stored at −80 °C for further use. Samples were desalted with an HLB cartridge (Waters, Milford, MA) according to manufacturer’s instructions.
LTQ-Orbitrap Velos MS and MS/MS
Unmodified only or oxidized ubiquitin protein was solubilized (0.25 mg·mL−1) in 49:49:2 (v:v:v) water:methanol:acetic acid. MS analysis was performed on a LTQ-Orbitrap Velos mass spectrometer (Thermo-Fisher Scientific, Waltham, MA) with direct infusion by a syringe pump. The following electrospray ionization parameters (i.e., source voltage 4.25 kV; capillary temperature 300.00 °C and flow rate 3 μL·min−1) were used. The Orbitrap settings included resolving power 100,000; m/z range 600–2000 for parent ion scans, 3 microscans, and a total number of scans for parent and fragmentation ions of 30 and 100, respectively. MS/MS settings used an isolation width of 1 m/z and normalized collision energy of 35%.
IMS-MS measurements
IMS theory and methods have been discussed in detail elsewhere23,24,55–60 and only a brief description is provided here. Ubiquitin ions were produced upon electrospraying aqueous solutions containing 0.5 mg·mL−1 in vitro oxidized ubiquitin with a TriVersa NanoMate autosampler (Advion, Ithaca, NY) unless otherwise noted. Solutions were prepared by adding formic acid to the desired pH value. Electrosprayed ions were introduced into a capillary tube and accumulated in an hourglass ion funnel.61 Packets of ions (150 μs wide) were pulsed periodically into a drift tube (~183 cm in length) which was filled with ~3.0 torr helium buffer gas (300K). A uniform electric field (~10 V·cm−1) was applied to the drift tube and ions were separated according to differences in their overall size and charge state. Upon exiting the drift tube, ions were extracted into a differential pumping region and focused into a time-of-flight (TOF) mass spectrometer for flight time detection. As described previously, drift times (tD) and flight times were collected in a single experimental sequence termed as a nested fashion.62 Flight times were converted to m/z values using a multipoint calibration.
Collision cross section calculations
Experimental drift times were converted into cross sections according to Equation 1.56
| (1) |
Here Ω is the ion cross section, ze is the ion’s charge, and kb refers to Boltzmann’s constant. Variables mI and mB are the mass of the ion and buffer gas (helium), respectively, and tD, E, and L correspond to the ion’s drift time, the applied electric field and drift length, respectively. P and T correspond to buffer gas pressure and temperature, respectively, and N is the neutral number density of the buffer gas at STP. The total drift time is calibrated by using the resident time in the first drift region that does not contain non-linear fields introduced by ion funnels.63
Data analysis
Methods for employing Gaussian functions to model collision cross section distributions for [M+8H]8+ ions generated from different water:methanol solutions have been described previously.32 Briefly, the Gaussian function is described as in Equation 2,
| (2) |
where I represents the distribution intensity at a given cross section Ω and A is the population of the represented conformer type. Ω0 and σ correspond to the center and the width of the distribution of structures within each conformation type, respectively. Similar analysis was applied to the cross section distributions of [M+7H]7+ ions. The distributions were modeled by the Peak Analyzer tool of the OriginPro 8.5.0 software (OriginLab Corporation, Northampton, MA). Peak centers and and widths were fixed (based on iterative modeling) and peak heights were varied for modeling different distributions. 32 The chosen peak widths and number of Gaussian peaks can also be correlated with distributions derived from the transport equation, which models peak widths due to normal ion-gas diffusion processes in the drift tube. The appropriateness of the Gaussian parameters is shown in Supplemental Figure 1 for [M+8H]8+ ubiquitin ions, whereby the sum of numerous transport equation-derived distributions are in alignment with our chosen Gaussian distributions.
Results and Discussion
Characterization of ubiquitin oxidation by MS and MS/MS analyses
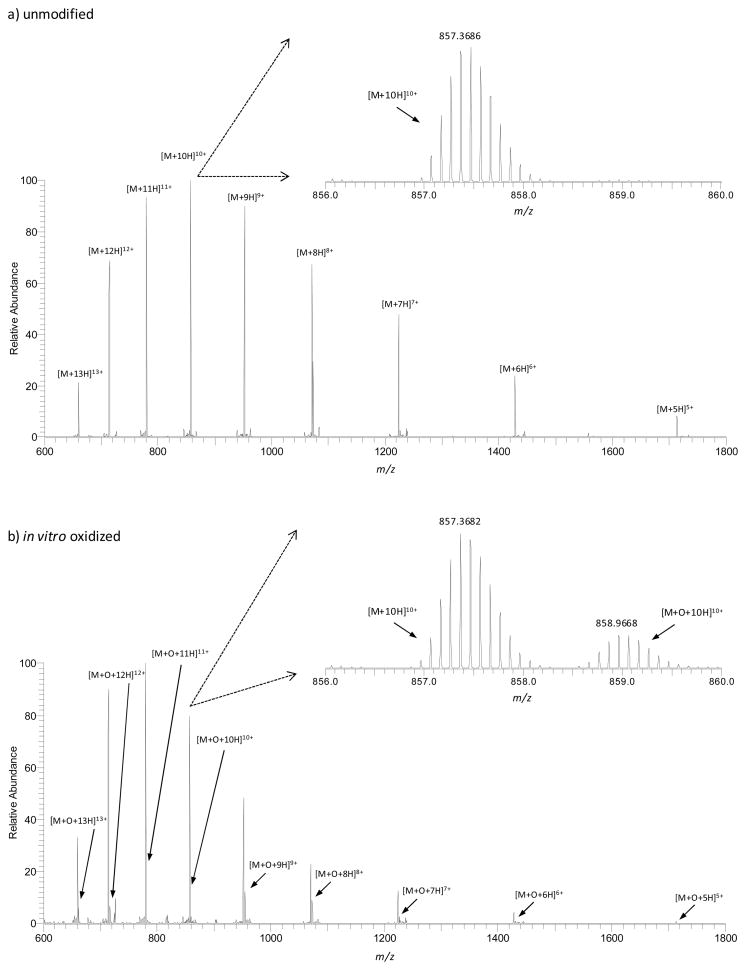
Confirmation that Fe(II)/H2O2 oxidation leads to oxidized ubiquitin species is demonstrated in Figure 1. The high resolution MS spectra for individually prepared samples of unmodified only (Figure 1a) and oxidized ubiquitin (Figure 1b) show similar charge state distributions for electrosprayed protein ions, whereby the +5 to +13 ions are observed. These charge states are typical for the denaturing solvent conditions employed.52,53 The spectrum of unmodified ubiquitin only shows a single isotopic distribution (inset of Figure 1a). The presence of oxidized peaks is shown in Figure 1b, whereby both unmodified and oxidized species occur simultaneously. The oxidized species is shifted in mass by a single oxygen atom as demonstrated for the +10 charge state ion (Figure 1b, inset). The relative intensities of these species in a deconvoluted spectrum (not shown) also shows that ~20% of ubiquitin molecules were oxidized. Higher levels of M+n16 (n>1) Da ubiquitin relative to unmodified species have been shown with other oxidizing methods.35–38
Figure 1.
High resolution mass spectra of (a) unmodified and (b) oxidized ubiquitin obtained upon ESI-LTQ-Orbitrap MS analysis. The insets show zoomed-in regions of the +10 charge state ions.
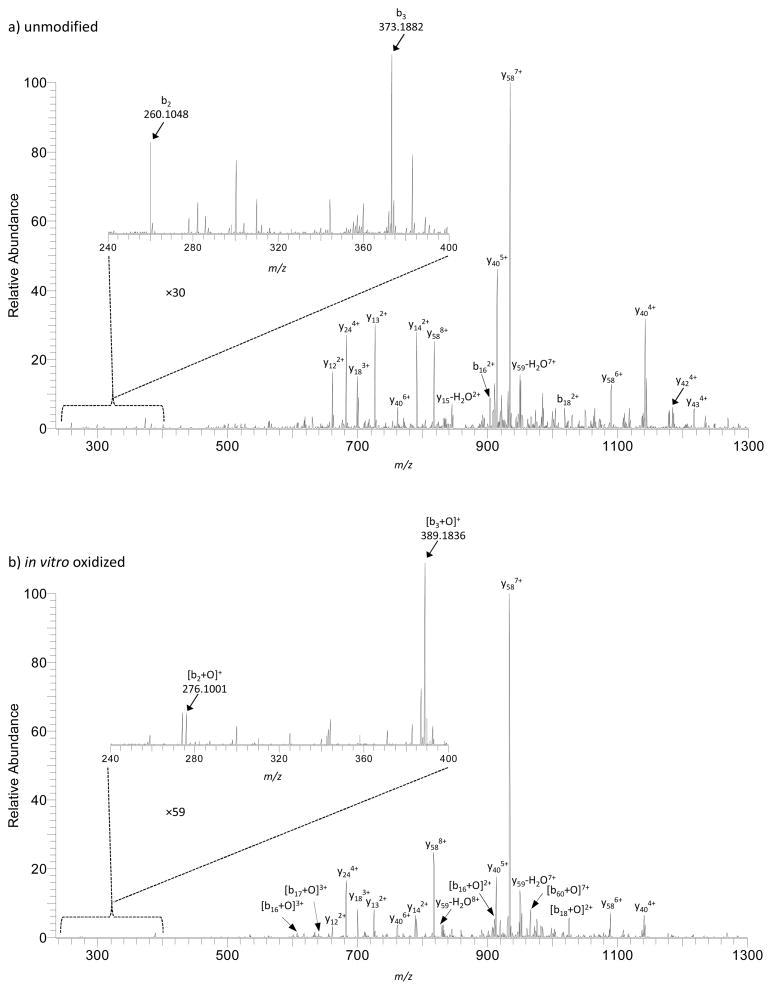
Collision induced dissociation (CID) of intact unmodified only and oxidized ubiquitin protein was performed in order to locate the modification site that incorporated the single oxygen atom. Detailed interpretation of the MS/MS spectra obtained upon isolation and CID of the +10 charge state ions locates the oxidation site to methionine. Figures 2a and 2b show many b- and y-type fragment ions that are detected for the unmodified and oxidized species, respectively. In the unmodified only sample, b-type ions at m/z 260.1048 and 373.1882 are assigned as b2 and b3 ions consistent with expected fragment ions for ubiquitin. The b2 and b3 ions in the oxidized species are shifted in mass by a single oxygen atom (i.e., m/z 276.1001 and 389.1836, respectively), which supports the predominance of an oxidized Met1. We and others report evidence for several M+16 Da positional isomers generated for oxidized ubiquitin;35,37,54 however, the most dominant isomer is the species containing methionine sulfoxide.
Figure 2.
CID MS/MS spectra obtained upon isolation (±1 m/z) of the +10 charge state ions of (a) unmodified and (b) oxidized ubiquitin species. The assigned b- and y-type fragment ions are listed in the figure. The insets show the noted magnification of the m/z range 240–400 which highlight the b2 and b3 ions.
Overview of IMS-MS distributions for unmodified and oxidized ubiquitin ions
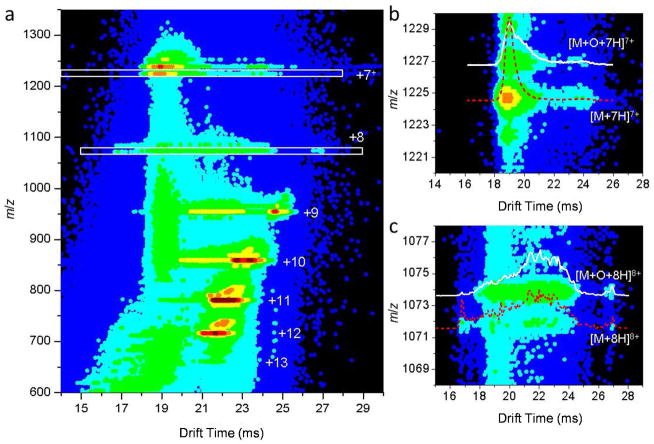
Fe(II)/H2O2 oxidized ubiquitin solution contains both unmodified and oxidized species and is used for IMS-MS investigations. The unmodified ions in this solution serve as an internal control thereby allowing any artificial effects that may occur from the sample preparation and handling process to be accounted. Figure 3a displays a nested two-dimensional (2D) IMS-MS dot plot of the observed features for the oxidized ubiquitin sample electrosprayed from a low pH (~2) solution. It is noted that higher m/z ions (M+98 Da) are also observed and correspond to ubiquitin with noncovalently bound phosphate ions.64 As observed in the figure, ions of charge state +7 are dominated by compact structures. The distribution of ions for charge state +8 is comprised of a peak of compact structures, a broad distribution of partially unfolded structures and two peaks corresponding to elongated structures. For ions of charge states +9 to +11, the distributions display both partially unfolded as well as elongated structures and charge states +12 and +13 are mainly elongated structures. The distributions reported here have some differences (i.e. shifts in positions of peak centers and abundance for different conformers) from previous reports 65 of ubiquitin electrosprayed from denaturing conditions (i.e., a 49:49:2 water:acetonitrile:acetic acid solution). Such differences may be attributable to changes in solution structures influenced by the solvent environment.
Figure 3.
(a) Two-dimensional drift time (m/z) contour plot for the Fe(II)/H2O2-induced oxidized ubiquitin electrosprayed from a solution of water and formic acid (pH 2; see Experimental). Ions of charge states from +7 to +13 are observed and each charge state has been provided as labels. To observe the features of charge states +7 and +8 more clearly, corresponding regions (marked by white boxes) have been zoomed in and displayed in (b) and (c), respectively. The unmodified ions are labeled as [M+7H]7+ (b) and [M+8H]8+ (c); the oxidized ions are labeled as [M+O+7H]7+ (b) and [M+O+8H]8+ (c). The insets in (b) and (c) show the drift time distributions for the corresponding ubiquitin species as labeled, which are normalized by the integrated peak intensity.
Comparisons of IMS profiles for oxidized and unmodified ubiquitin ions show that there is no significant variance in distributions for ions of high charge states (+9 to +13, data not shown). However, gas-phase distributions of low charge state ions (+7 and +8) of oxidized ubiquitin differ from those observed for unmodified species. Thus, we focus our discussion to distributions for charge states +7 and +8 below. The zoomed 2D IMS-MS dot plots and normalized drift time distributions for +7 and +8 unmodified and oxidized ubiquitin ions are shown in Figures 3b and 3c. It is evident that [M+O+7H]7+ ions (centered at ~18.9 ms) generate a broader distribution than that of the [M+7H]7+ ions as indicated by the presence of low mobility structures with drift times ranging from ~19.5 to 21.7 ms. The distribution of [M+8H]8+ ions shows a sharp peak corresponding to ions of high mobility at ~16.8 ms in drift time, whereas the distribution of [M+O+8H]8+ ions is much less populated in this region. At higher drift times, ~17.8 – 27.7 ms, the distributions appear relatively similar for [M+8H]8+ and [M+O+8H]8+ ions; however differences in abundances of various conformer types are apparent after Gaussian modeling (discussed below).
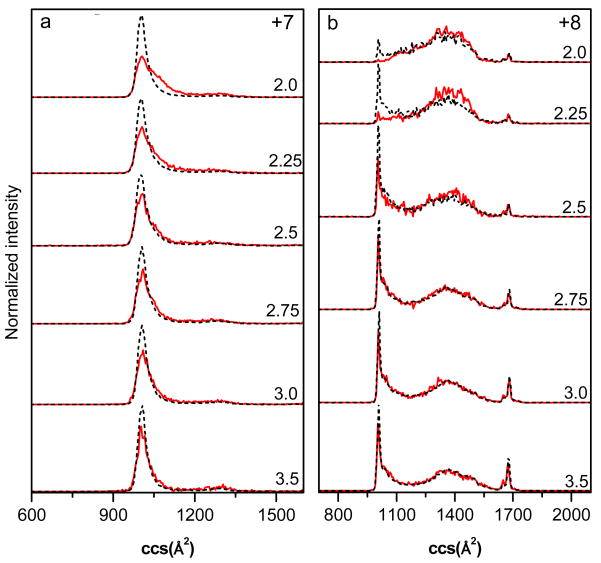
Figure 4 shows the effects of different solution pH on the collision cross sections for +7 and +8 unmodified and oxidized ions. For the +7 distribution the most intense feature is a single compact peak in which the unmodified and oxidized ions overlap at pH 3.5. As the pH of the ESI solution is lowered the [M+O+7H]7+ peak becomes broader whereas the [M+7H]7+ ions are still fairly sharp. Similarly, for the +8 distributions there are notable differences in the sharp feature observed at 1020 Å2 beginning at pH 2.25, whereby the intensity of the [M+O+8H]8+ ion is significantly lower than that of the [M+8H]8+ ion. Furthermore, it appears that there is more intensity at higher collision cross sections where the distribution is broad. In order to better understand the differences between unmodified and oxidized distributions Gaussian modeling was employed. Because the distributions have the greatest differences between oxidized and unmodified ubiquitin at solution pH 2, we have chosen this solution condition for the analysis presented below.
Figure 4.
Collision cross section (ccs) distributions for Fe(II)/H2O2-treated ubiquitin of charge states +7 (a) and +8 (b) electrosprayed from different water:formic acid solutions. The solution pH is labeled for each of the distributions. Distributions for unmodified and oxidized ubiquitin are plotted as black dashed lines and red solid lines, respectively. The distributions are normalized by the integrated peak intensity.
Modeling IMS profiles with Gaussian distributions
Gaussian functions have been used to represent gas-phase conformations across the total cross section distribution of ubiquitin [M+8H]8+ ions generated from a range (100:0 to 5:95) of water:methanol ESI solutions.32 Gaussian conformers have been assigned to specific solution states (N, A, or U) according to their population profiles as a function of solvent conditions. In order to obtain insight to population changes of solution states similar Gaussian modeling was applied to these data. Table 1 lists employed Gaussian conformers,32 whereby ten and eleven functions are used to model +7 and +8 distributions, respectively. The functions established to model +7 charge state ions are presented for the first time in these studies.
Table 1.
Gaussian function peak centers (Ω0), standard deviations (σ), and relative intensities (I) for conformers of +7 and +8 unmodified (M) and oxidized (M+O) ions.
| +7
|
+8
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ω0 (σ)a | SSb | Ic (M) | I (M+O) | Ω0 (σ) | SS | I (M) | I (M+O) | Ω0 (σ) | SS | I (M) | I (M+O) | Ω0 (σ) | SS | I (M) | I (M+O) |
|
|
|
|
|
||||||||||||
| 980 (11) | N1 | 0.000 | 0.028 | 1160 (42) | N5 | 0.028 | 0.092 | 1020 (6) | N1 | 0.033 | 0.003 | 1360 (47) | N6 | 0.246 | 0.304 |
| 1010 (17) | N2 | 0.703 | 0.326 | 1250 (36) | N6 | 0.009 | 0.031 | 1040 (25) | N2 | 0.039 | 0.000 | 1450 (49) | A1 | 0.210 | 0.253 |
| 1040 (25) | N3 | 0.179 | 0.167 | 1280 (38) | N7 | 0.000 | 0.000 | 1120 (41) | N3 | 0.091 | 0.063 | 1570 (28) | A2 | 0.012 | 0.001 |
| 1060 (32) | A | 0.017 | 0.203 | 1300 (36) | N8 | 0.026 | 0.053 | 1160 (60) | U | 0.060 | 0.079 | 1650 (11) | A3 | 0.009 | 0.010 |
| 1100 (28) | N4 | 0.030 | 0.077 | 1370 (64) | N9 | 0.008 | 0.023 | 1210 (34) | N4 | 0.099 | 0.070 | 1680 (6) | A4 | 0.014 | 0.012 |
| 1290 (42) | N5 | 0.187 | 0.206 | ||||||||||||
|
| |||||||||||||||
| SUM | N | 0.983 | 0.797 | A | 0.017 | 0.203 | SUM | N | 0.695 | 0.645 | A | 0.245 | 0.276 | ||
|
| |||||||||||||||
| U | 0.060 | 0.079 | |||||||||||||
Ω0 (σ) has unit of Å2.
Solution states (N, A, and U) assigned to the respective gas-phase conformation types.
The reported relative intensities are obtained by dividing the area of each Gaussian distribution by the total area.
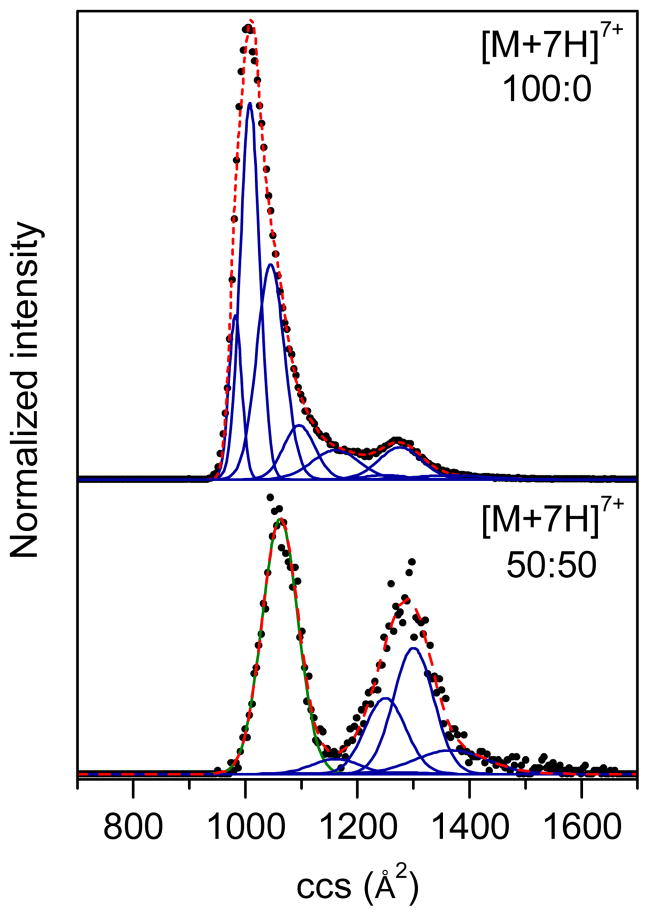
Figure 5 displays representative cross section distributions for ubiquitin [M+7H]7+ ions generated from 100:0 and 50:50 water:methanol solutions, respectively. The Gaussian functions employed are highlighted underneath the curves. It is noted that ions generated from the 100:0 water:methanol solution favor the N state and ions from the 50:50 water:methanol solution favor the A state. For [M+7H]7+ ions formed from the 100:0 water:methanol solution, the distribution is dominated by compact conformations centered at ~1010 Å2 with a small portion of more elongated structures having cross sections around 1280 Å2. When the solution is changed to 50:50 water:methanol, the 1010 Å2 peak shifts to 1060 Å2 and the 1280 Å2 peak moves to 1300 Å2. Additional studies that examine Gaussian conformers across several water:methanol solution conditions (data not shown) support the notion that the observed peaks, with the exception of the Ω = 1060±32 Å2 peak, originate from the N state. This assessment is based on the peak populations decreasing with higher amounts of methanol in the ESI solvent. The Ω= 1060±32 Å2 peak originates from the A state based on the following observations: 1) it does not exist in aqueous solution, 2) it begins to appear in an 80:20 water:methanol solution, and 3) it maintains a constant relative intensity as the percentage of methanol is ≥30%. Gaussian distributions of [M+8H]8+ ions have been discussed previously.32 Overall, the application of Gaussian modeling to the unmodified +7 and +8 ions, allows us to assign N, A, and U states to IMS distributions for comparison to oxidized ions.
Figure 5.
Gaussian models of collision cross section (ccs) distributions for unprocessed ubiquitin of charge state +7 electrosprayed from two solution conditions (100:0 and 50:50 water:methanol, pH ~2 adjusted by formic acid). The solution conditions have been labeled. The experimental data (normalized) are drawn as black circles, the Gaussian distributions employed in the modeling are depicted as blue, green and pink solid lines, representing the N, A, and U state, respectively, and the sums of the Gaussian functions are shown as red dashed lines.
Gas-phase conformers and solution states of unmodified and oxidized ubiquitin
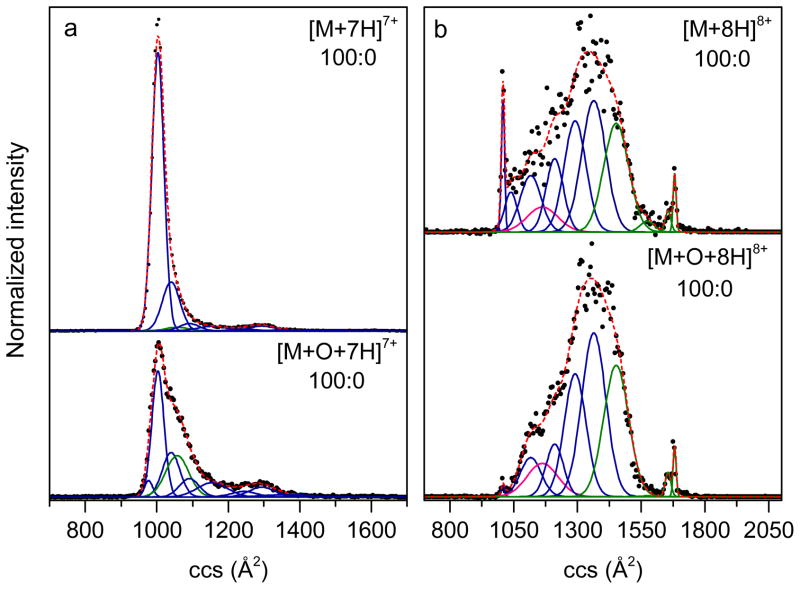
Figure 6 compares collision cross section distributions for unmodified and oxidized ubiquitin ions of charge states +7 and +8. The employed Gaussian distributions are also displayed in Figure 6. The experimental distributions are well represented by the Gaussian models with R2 of 0.994, 0.997, 0.972, and 0.988 for [M+7H]7+, [M+O+7H]7+, [M+8H]8+, and [M+O+8H]8+ ions, respectively. As displayed in Figure 6a, both unmodified and oxidized species of charge state +7 generate a compact conformer peak with the cross section centered at 1010 Å2. However, in addition to a narrow range of structures around 1010 Å2, the oxidized ions also have conformations that are more extended with cross sections ranging from 1040 Å2 to 1170 Å2. Based on modeling, the relative intensity of the A-state Gaussian conformer (Ω = 1060±32 Å2) of [M+O+7H]7+ is 20% of the total ion population (Table 1). This value is significantly higher than that for [M+7H]7+ which is 1.7%. It is possible that this A-state distribution is generated from other partially folded or extended structures which have similar cross sections as the A-state gas-phase conformer. On the other hand, this Gaussian conformer may originate from solution folding intermediates (e.g., unfolded to native) of oxidized ubiquitin, which are captured in the gas phase. For [M+O+7H]7+ ions, higher intensities for N-state conformers are observed as peaks with cross sections: Ω = 980±11 Å2, Ω = 1100±28 Å2, Ω = 1160±42 Å2, Ω = 1250±36 Å2, Ω = 1300±36 Å2, and Ω = 1370±64 Å2. The N-state peaks at Ω = 1010±17 Å2 and Ω = 1040±25 Å2 have lower intensities for [M+O+7H]7+ ions relative to [M+7H]7+. The abundance information of specific conformer peaks can give insight to protein dynamics in solution and in the gas phase. Overall, the N-state structures are less populated for [M+O+7H]7+ ions (80%) relative to [M+7H]7+ ions (98%).
Figure 6.
Gaussian models of collision cross section (ccs) distributions for Fe(II)/H2O2-treated ubiquitin of charge states +7 (a) and +8 (b) electrosprayed from a solution of water and formic acid (pH 2; see Experimental). The ccs distributions of unmodified ubiquitin are labeled as [M+7H]7+ and [M+8H]8+ for charge states of +7 and +8, respectively. The ccs distributions of oxidized ubiquitin are labeled as [M+O+7H]7+ and [M+O+8H]8+ for charge states of +7 and +8, respectively. The solution conditions (water:methanol 100:0, pH ~2 adjusted by formic acid) have been labeled. The experimental data (normalized) are shown as black circles, the Gaussian distributions employed in the modeling are drawn as blue, green and pink solid lines, representing the N, A, and U state, respectively, and the sums of the Gaussian functions are shown as red dashed lines.
The cross section distributions of ubiquitin [M+8H]8+ and [M+O+8H]8+ ions are very similar to that of previously reported ubiquitin measurements, although [M+8H]8+ ions have higher abundances of U- and A-state conformers in comparison to previously published results.32 We speculate that residual oxidizing reagents (i.e., Fe(II)/H2O2) from the sample preparation process could promote the unfolding of ubiquitin in the solution or during ESI thus leading to differences in the abundances of populations in this work to previous studies32. Specifically, there is a broad distribution ranging in cross sections from ~1040 Å2 to 1620 Å2 and three sharp features with cross sections of 1020 Å2, 1650 Å2 and 1680 Å2. The most apparent feature that is different between the distributions of [M+8H]8+ and [M+O+8H]8+ ions is the Ω = 1020±6 Å2 peak, which corresponds to a gas-phase conformer produced from the N state. The relative abundance of the Ω = 1020±6 Å2 peak for oxidized ubiquitin ions is substantially lower (0.3%) compared to that for the unmodified species (3.3%). Table 1 lists six gas-phase conformer types for ubiquitin [M+8H]8+ ions that are related with the solution N state: a compact peak (Ω = 1020±6 Å2) and five other peaks corresponding to partially unfolded conformations (Ω = 1040±25 Å2, Ω = 1120±41 Å2, Ω = 1210±34 Å2, Ω = 1290±42 Å2, and Ω = 1360±47 Å2).32 Among the six conformers, peaks with cross sections values Ω = 1020±6 Å2, Ω = 1040±25 Å2, Ω = 1120±41 Å2, and Ω = 1210±34 Å2 have lower relative intensities for [M+O+8H]8+ ions compared to that of [M+8H]8+ ions, whereas the Ω = 1290±42 Å2 and Ω = 1360±47 Å2 peaks show higher relative intensities for [M+O+8H]8+ ions. It is worthwhile to consider the origination of multiple N-state conformers in the gas phase. One interpretation is that various native forms of ubiquitin presented in solution as observed by NMR66 produce multiple gas-phase conformers as slight differences in the solution structures might be amplified upon desolvation.32 Another interpretation is that the ESI process perturbs the solution states and leads to a wide range of partially unfolded structures.32 With the second interpretation, because the largest cross section N-state conformers of the total N-state population (see Table 1) are higher in abundance for oxidized +7 and +8 ions, it appears that an oxygen atom promotes gas-phase unfolding of native ubiquitin. N-state Gaussian conformers for [M+O+8H]8+ ions are overall less populated (65%) than that of [M+8H]8+ ions (70%), suggesting that methionine oxidation also leads to destabilization of solution native-state ubiquitin. This is also supported by the detection of more abundant A- and U-state conformers for oxidized ubiquitin ions. Furthermore, Aye et al. conclude that monooxidized ubiquitin species result in a distribution of partially folded and unfolded conformers that expose buried residues increasing susceptibility to further M+n16 Da oxidations.35 Results from their solution-phase studies are consistent with our gas-phase observations of solution conformers.
The acidity of the ESI solution, as well as organic content (i.e., methanol, acetonitrile) can greatly influence the observed solution10 and gas-phase conformers. Solution studies show that the oxidation of methionine to methionine sulfoxide leads to destabilization and expansion of the structure at low pH ~2.4.10 Our results in the gas phase are consistent with those studies. Whereas solution studies observe two sulfoxide isomers that are mobility-separated on an electrophoretic gel,10 our studies provide evidence for many conformers of the methionine sulfoxide species that are mobility-separated in a drift tube. We note that although methionine is the predominant monooxidized species, other lower concentration M+16 Da isomers exist54 and may also contribute to the broad range of A-state structures observed. The acid present in low pH conditions of the ESI solvent may influence protein structure by binding to the protein in a noncovalent fashion10 or through affecting the stability of hydrogen bonding interactions. The crystal structure of ubiquitin describes the sulfur atom in methionine as participating in hydrogen bonding with the proton in the NH group of Lys63.39 We hypothesize that this hydrogen bond contributes to the stabilization of the interactions between ubiquitin β-strands 1 and 5. When methionine is oxidized to methionine sulfoxide, the weakening of this hydrogen bond leads to destabilization of the interactions between β-strand 1 and 5 and contributes to unfolding of protein tertiary structure. The presence of multiple partially-folded gas-phase intermediates of the +8 charge state ions implies that a range of structures may be captured in solution or present after the ESI process. Additionally, the broad distribution of +8 A and U states implies significant unfolding of oxidized ubiquitin.
Conclusions
IMS-MS analysis has been applied to the structural characterization of Fe(II)/H2O2 oxidized ubiquitin in the gas phase. These oxidative conditions lead to a low abundant (~20%) M+16 Da species, consisting of methionine sulfoxide at the N-terminal single methionine residue. Collision cross section distributions of oxidized ubiquitin ions electrosprayed from an aqueous solution of pH from 2.0 to 3.5 have been reported. Compared to unmodified ubiquitin ions from the solution of pH 2.0, oxidized ions of charge states +7 and +8 generate distributions of relatively lower intensity for conformations that arise from the solution N state. Based on these IMS-MS analyses, methionine oxidation reduces the stability of native-state ubiquitin in acidic solution conditions. Moreover, N-state conformers of larger cross sections are more populated for oxidized ions relative to unmodified ions which implies that the activation energy required to produce more extended gas-phase states from the native structure is decreased for oxidized ubiquitin. These studies demonstrate that under acidic solution conditions (pH ~2) the incorporation of a single oxygen atom destabilizes the native structure and facilitates ubiquitin unfolding in the gas phase. Observation of these effects was possible due to the high-resolution obtainable in the mobility separation using a traditional IMS drift tube instrument and the ability to detect low abundance oxidatively-modified ions. IMS-MS is a useful technique for gaining insight to the effects of oxidation on protein gas-phase structure and subsequently solution states.
Supplementary Material
Acknowledgments
The authors acknowledge funding of this work provided by the Society of Analytical Chemists of Pittsburgh Starter Grant Award and University of Pittsburgh Start-Up Funds (R.A.S.R), grants from the NIH (1RC1GM090797-02, D.E.C.) and funds from the Indiana University METACyt initiative (D.E.C.) that is funded by a grant from the Lilly Endowment.
Footnotes
Supporting Information Available
Supplementary Figure S1. Transport-derived distributions of ubiquitin [M+8H]8+ ions.
This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Finkel T, Holbrook NJ. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 2.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Davies MJ. Biochim Biophys Acta. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 4.von Zglinicki T. Trends Biochem Sci. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 5.Halliwell B. J Neurochem. 2006;97:1634–1658. doi: 10.1111/j.1471-4159.2006.03907.x. [DOI] [PubMed] [Google Scholar]
- 6.Butterfield DA, Galvan V, Lange MB, Tang H, Sowell RA, Spilman P, Fombonne J, Gorostiza O, Zhang J, Sultana R, et al. Free Radic Biol Med. 2010;48:136–144. doi: 10.1016/j.freeradbiomed.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aluise CD, Robinson RAS, Beckett TL, Murphy MP, Cai J, Pierce WM, Markesbery WR, Butterfield DA. Neurobiol Dis. 2010;39:221–228. doi: 10.1016/j.nbd.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson RAS, Lange MB, Sultana R, Galvan V, Fombonne J, Gorostiza O, Zhang J, Warrier G, Cai J, Pierce WM, et al. Neuroscience. 2011;177:207–222. doi: 10.1016/j.neuroscience.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breslow E, Chauhan Y, Daniel R, Tate S. Biochem Biophys Res Commun. 1986;138:437–444. doi: 10.1016/0006-291x(86)90300-1. [DOI] [PubMed] [Google Scholar]
- 10.Bamezai S, Banez MAT, Breslow E. Biochemistry. 1990;29:5389–5396. doi: 10.1021/bi00474a026. [DOI] [PubMed] [Google Scholar]
- 11.Gao J, Yin DH, Yao Y, Sun H, Qin Z, Schöneich C, Williams TD, Squier TC. Biophys J. 1998;74:1115–1134. doi: 10.1016/S0006-3495(98)77830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DalleDonne I, Milzani A, Colombo R. Biochemistry. 1999;38:12471–12480. doi: 10.1021/bi990367k. [DOI] [PubMed] [Google Scholar]
- 13.Liu D, Ren D, Huang H, Dankberg J, Rosenfeld R, Cocco MJ, Li L, Brems DN, Remmele RL., Jr Biochemistry. 2008;47:5088–5100. doi: 10.1021/bi702238b. [DOI] [PubMed] [Google Scholar]
- 14.Burkitt W, Domann P, O’Connor G. Protein Sci. 2010;19:826–835. doi: 10.1002/pro.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 16.Chowdhury SK, Chait BT. Biochem Biophys Res Commun. 1990;173(3):927–931. doi: 10.1016/s0006-291x(05)80874-5. [DOI] [PubMed] [Google Scholar]
- 17.Loo JA, Edmonds CG, Smith RD. Science. 1990;248:201–204. doi: 10.1126/science.2326633. [DOI] [PubMed] [Google Scholar]
- 18.Winger BE, Light-Wahl KJ, Rockwood AL, Smith RD. J Am Chem Soc. 1992;114:5897–5898. [Google Scholar]
- 19.Clemmer DE, Hudgins RR, Jarrold MF. J Am Chem Soc. 1995;117:10141–10142. [Google Scholar]
- 20.Vachet RW, Asam MR, Glish GL. J Am Chem Soc. 1996;118:6252–6256. [Google Scholar]
- 21.Zubarev RA, Kelleher NL, McLafferty FW. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 22.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clemmer DE, Jarrold MF. J Mass Spectrom. 1997;32:577–592. [Google Scholar]
- 24.Bohrer BC, Merenbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. Annu Rev Anal Chem. 2008;1:293–327. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loo JA. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 26.Loo JA, He JX, Cody WL. J Am Chem Soc. 1998;120:4542–4543. [Google Scholar]
- 27.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 28.Ruotolo BT, Robinson CV. Curr Opin Chem Biol. 2006;10:402–408. doi: 10.1016/j.cbpa.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 29.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea J-E, Ruotolo BT, Robinson CV, et al. Nat Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyttenbach T, Bowers MT. J Phys Chem B. 2011;115:12266–12275. doi: 10.1021/jp206867a. [DOI] [PubMed] [Google Scholar]
- 31.Pierson NA, Chen L, Valentine SJ, Russell DH, Clemmer DE. J Am Chem Soc. 2011;133:13810–13813. doi: 10.1021/ja203895j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H, Pierson NA, Valentine SJ, Clemmer DE. J Phys Chem B. 2012;116:3344–3352. doi: 10.1021/jp210797x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldstein G, Scheid M, Hammerling U, Boyse EA, Schlesinger DH, Niall HD. Proc Natl Acad Sci USA. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rechsteiner M. Ubiquitin. Plenum Press; New York: 1988. [Google Scholar]
- 35.Aye TT, Low TY, Sze SK. Anal Chem. 2005;77:5814–5822. doi: 10.1021/ac050353m. [DOI] [PubMed] [Google Scholar]
- 36.Venkatesh S, Tomer KB, Sharp JS. Rapid Commun Mass Spectrom. 2007;21:3927–3936. doi: 10.1002/rcm.3291. [DOI] [PubMed] [Google Scholar]
- 37.McClintock C, Kertesz V, Hettich RL. Anal Chem. 2008;80:3304–3317. doi: 10.1021/ac702493a. [DOI] [PubMed] [Google Scholar]
- 38.Downard KM, Maleknia SD, Akashi S. Rapid Commun Mass Spectrom. 2012;26:226–230. doi: 10.1002/rcm.5320. [DOI] [PubMed] [Google Scholar]
- 39.Vijay-Kumar S, Bugg CE, Cook WJ. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 40.Lenkinski RE, Chen DM, Glickson JD, Goldstein G. Biochim Biophys Acta. 1977;494:126–130. doi: 10.1016/0005-2795(77)90140-4. [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson KD, Mayer AN. Arch Biochem Biophys. 1986;250:390–399. doi: 10.1016/0003-9861(86)90741-1. [DOI] [PubMed] [Google Scholar]
- 42.Harding MM, Williams DH, Woolfson DN. Biochemistry. 1991;30:3120–3128. doi: 10.1021/bi00226a020. [DOI] [PubMed] [Google Scholar]
- 43.Brutscher B, Brüschweiler R, Ernst RR. Biochemistry. 1997;36:13043–13053. doi: 10.1021/bi971538t. [DOI] [PubMed] [Google Scholar]
- 44.Stockman BJ, Euvrard A, Scahill TA. J Biomol NMR. 1993;3:285–296. doi: 10.1007/BF00212515. [DOI] [PubMed] [Google Scholar]
- 45.Cordier F, Grzesiek S. Biochemistry. 2004;43:11295–11301. doi: 10.1021/bi049314f. [DOI] [PubMed] [Google Scholar]
- 46.Kony DB, Hünenberger PH, van Gunsteren WF. Protein Sci. 2007;16:1101–1118. doi: 10.1110/ps.062323407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sorenson JM, Head-Gordon T. Proteins Struct Funct Genet. 2002;46:368–379. [PubMed] [Google Scholar]
- 48.Zhang J, Qin M, Wang W. Proteins Struct Funct Bioinf. 2005;59:565–579. doi: 10.1002/prot.20430. [DOI] [PubMed] [Google Scholar]
- 49.Jackson SE. Org Biomol Chem. 2006;4:1845–1853. doi: 10.1039/b600829c. [DOI] [PubMed] [Google Scholar]
- 50.Jourdan M, Searle MS. Biochemistry. 2001;40:10317–10325. doi: 10.1021/bi010767j. [DOI] [PubMed] [Google Scholar]
- 51.Mohimen A, Dobo A, Hoerner JK, Kaltashov IA. Anal Chem. 2003;75:4139–4147. doi: 10.1021/ac034095+. [DOI] [PubMed] [Google Scholar]
- 52.Valentine SJ, Counterman AE, Clemmer DE. J Am Soc Mass Spectrom. 1997;8:954–961. doi: 10.1016/S1044-0305(99)00079-3. [DOI] [PubMed] [Google Scholar]
- 53.Li J, Taraszka JA, Counterman AE, Clemmer DE. Int J Mass Spectrom. 1999;185/186/187:37–47. [Google Scholar]
- 54.Gu L, Robinson RAS. Positional Isomers of Fe(II)/H2O2 Oxidized Ubiquitin Characterized by Top-Down and Bottom-Up Mass Spectrometry . submitted. [Google Scholar]
- 55.Koeniger SL, Merenbloom SI, Valentine SJ, Jarrold MF, Udseth HR, Smith RD, Clemmer DE. Anal Chem. 2006;78:4161–4174. doi: 10.1021/ac051060w. [DOI] [PubMed] [Google Scholar]
- 56.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. Wiley; New York: 1988. [Google Scholar]
- 57.StLouis RH, Hill HH, Eiceman GA. Crit Rev Anal Chem. 1990;21:321–355. [Google Scholar]
- 58.Hoaglund-Hyzer CS, Counterman AE, Clemmer DE. Chem Rev. 1999;99:3037–3079. doi: 10.1021/cr980139g. [DOI] [PubMed] [Google Scholar]
- 59.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. J Phys Chem. 1996;100:16082–16086. [Google Scholar]
- 60.Wyttenbach T, von Helden G, Batka JJ, Carlat D, Bowers MT. J Am Soc Mass Spectrom. 1997;8:275–282. [Google Scholar]
- 61.Tang K, Shvartsburg AA, Lee H-N, Prior DC, Buschbach MA, Li F, Tolmachev AV, Anderson GA, Smith RD. Anal Chem. 2005;77:3330–3339. doi: 10.1021/ac048315a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Anal Chem. 1998;70:2236–2242. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 63.Koeniger SL, Merenbloom SI, Sevugarajan S, Clemmer DE. J Am Chem Soc. 2006;128:11713–11719. doi: 10.1021/ja062137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chowdhury SK, Katta V, Beavis RC, Chait BT. J Am Soc Mass Spectrom. 1990;1:382–388. doi: 10.1016/1044-0305(90)85018-H. [DOI] [PubMed] [Google Scholar]
- 65.Koeniger SL, Clemmer DE. J Am Soc Mass Spectrom. 2007;18:322–331. doi: 10.1016/j.jasms.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 66.Lange OF, Lakomek N–A, Farès C, Schröder GF, Walter KFA, Becker S, Meiler J, Grubmüller H, Griesinger C, de Groot BL. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.