Abstract
Metazoan replication-dependent histone mRNAs have a conserved stem-loop (SL) at their 3′-end. The stem–loop binding protein (SLBP) specifically recognizes the SL to regulate histone mRNA metabolism, and the 3′-5′ exonuclease 3′hExo trims its 3′-end after processing. We report the crystal structure of a ternary complex of human SLBP RNA binding domain, human 3′hExo, and a 26-nucleotide SL RNA. Only one base of the SL is recognized specifically by SLBP, and the two proteins primarily recognize the shape of the RNA. SLBP and 3′hExo have no direct contact with each other, and induced structural changes in the loop of the SL mediate their cooperative binding. The 3′ flanking sequence is positioned in the 3′hExo active site, but the ternary complex limits the extent of trimming.
Metazoan replication-dependent histone mRNAs have a conserved stem-loop (SL) structure at their 3′-end. (1, 2), distinct from the polyadenylate tail found on all other known eukaryotic mRNAs (3, 4). SLBP (5), also known as hairpin binding protein (6), is a central regulator of histone mRNA metabolism. SLBP and the U7 snRNP (7) are required for the 3′-end processing of histone pre-mRNAs (Fig. S1). SLBP is also required for the export, stability and translation of mature mRNAs. The 3′-5′ exonuclease 3′hExo (also known as Eri-1) forms a tight ternary complex with SL and SLBP. 3′hExo can trim three nucleotides in vitro from the processed histone mRNA 3′-end, and SLBP protects against further trimming (8-11). 3′hExo is also involved in microRNA homeostasis (12) and 5.8S rRNA 3′-end maturation (13, 14). The stem-loop RNA consists of a six base-pair stem and a four base loop, as well as flanking sequences at both ends (Fig. S1). SLBP (31 kD) has high affinity for the SL (Kd 1–10 nM) (15-19). It contains a ~70 residue RNA binding domain (RBD, Figs. 1A, S2). 3′hExo (40 kD) consists of an N-terminal SAP domain (~60 residues) followed by a nuclease domain (~220 residues) that belongs to the DEDDh superfamily (Figs. 1A, S3) (9-11, 20).
Figure 1.
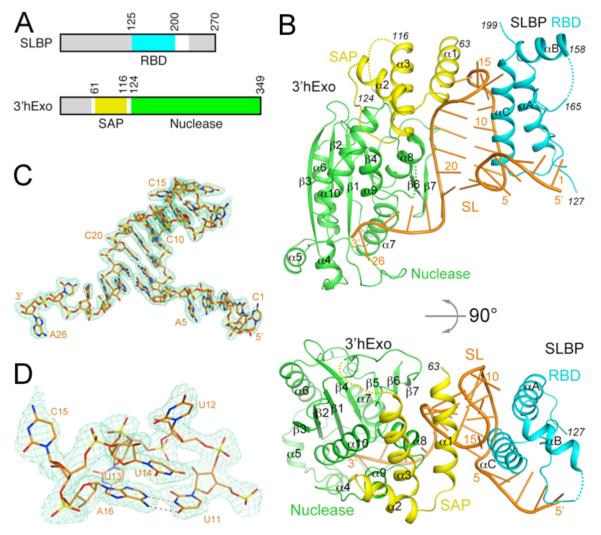
Structure of human SLBP RBD, human 3′hExo, and SL RNA ternary complex. (A). Domain organizations of human SLBP and human 3′hExo. Residues not included in the expression constructs are shown in gray. (B). Schematic drawings of the structure of the ternary complex of human SLBP RBD (cyan), human 3′hExo (SAP domain in yellow and nuclease domain in green), and 26-nucleotide SL RNA (orange). (C). Simulated annealing omit Fo–Fc electron density (light green) for the SL RNA at 2.6 Å resolution, contoured at 3σ. (D). Closeup of the loop region of the SL RNA. All the structure figures were produced with PyMOL (www.pymol.org).
We report here the crystal structure at 2.6 Å resolution of the ternary complex of human SLBP RBD, human 3′hExo (SAP and nuclease domains), and a 26-nucleotide SL with consensus sequence (Figs. 1A, 1B, Table S1) (21). Clear electron density was observed for all 26 nucleotides of the SL (Fig. 1C). The stem (nucleotides 6-11 and 16-21) has a slightly flattened classical A-form structure (Fig. S4, Table S2). Of the four nucleotides in the loop, the first (U12), second (U13) and fourth (C15) bases are flipped out (Fig. 1D). In the 3′ flanking sequence, nucleotides 22-25 continue the helical structure of the stem, but the base of the last nucleotide (A26) is flipped by ~180° relative to C25 (Fig. 1C). The riboses of all four nucleotides in the loop (12-15) and C25 are in the 2′ endo configuration, and the RNA backbone adopts sharp turns at these nucleotides.
The structure of SLBP RBD contains three helices (αA-αC). Helices αA and αC interact with the 5′ flanking sequence, 5′ arm of the stem and the loop of the RNA (Fig. 1B), consistent with earlier data (9, 15, 17). In particular, helix αC is positioned closest to the SL and may function as a ruler that can measure the length of the stem, with residues near its N-terminus (conserved 177-KYSRR-181 motif, Fig. S2) contacting the 5′-end of the stem and the 5′ flanking sequence and its C-terminal region contacting the loop.
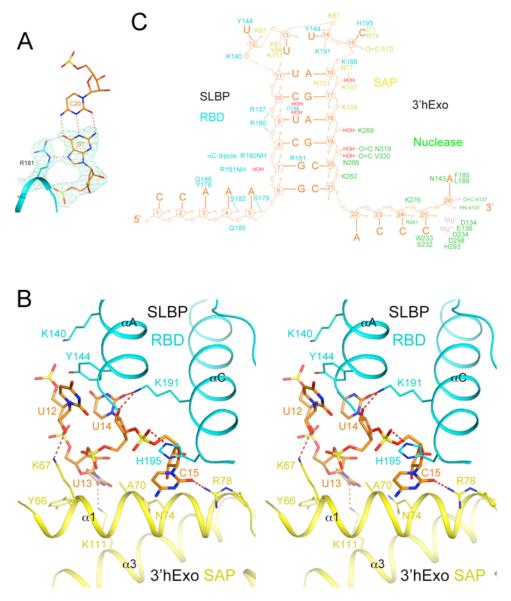
The only direct recognition between SL and SLBP is through the guanine base of the second nucleotide of the stem (G7), via two hydrogen bonds with the side-chain guanidinium group of Arg181 (Fig. 2A). The side chain of Tyr144 (αA) is π-stacked with the first and the third base, and the side chain of His195 (αC) with the fourth base of the loop (Fig. 2B). Other interactions are primarily between the RNA backbone and the SLBP RBD (Figs. 2C, S5).
Figure 2.
Interactions between the SL RNA and SLBP RBD and 3′hExo. (A). Specific recognition of G7 in the second base pair of the stem (orange) by hydrogen bonding (dashed lines in red) with the side chain of Arg181 (cyan) of SLBP. Simulated annealing omit Fo–Fc electron density for G7 and Arg181 is also shown, contoured at 5σ. (B). Interactions between the loop of the SL RNA (orange) with the SLBP RBD (cyan) and the 3′hExo SAP domain (yellow). (C). Schematic drawing summarizing the interactions between SL and SLBP RBD (cyan) and 3′hExo.
Nucleotides 3-5 in the 5′ flanking sequence, also implicated in binding to SLBP (9, 15, 17), have interactions with the RBD (Fig. S6). Besides residues Tyr178 and Ser179, the connection between αA and αC is not in direct contact with the RNA (Fig. 1B). This segment contains the conserved 171-TPNK-175 sequence, and Thr171 phosphorylation produces a 7-fold enhancement in the affinity for SL (18). This residue is located near the side chains of Lys146 (αA), Tyr151 (αA) and Trp190 (αC), and its phosphorylation may affect the positioning of the αA and αC helices (Fig. S6). Tyr151 is part of the conserved YDRY motif (Fig. S2), and the Y151F mutant has ~10-fold lower affinity for SL (22).
3′hExo contacts the loop, the 3′ arm of the stem, and the 3′ flanking sequence of the SL (Fig. 1B), as suggested by earlier studies (9-11). The SAP domain contains three helices (α1-α3) and interacts primarily with the loop of the SL through α1 (Figs. 1B, 2B). The U13 base interacts with the side chain of Tyr66 (α1) and Lys111 (α3), and the C15 base has a hydrogen bond to the side chain of Arg78 (α1). Additional interactions are with the backbone of the RNA (Figs. 2C, S7).
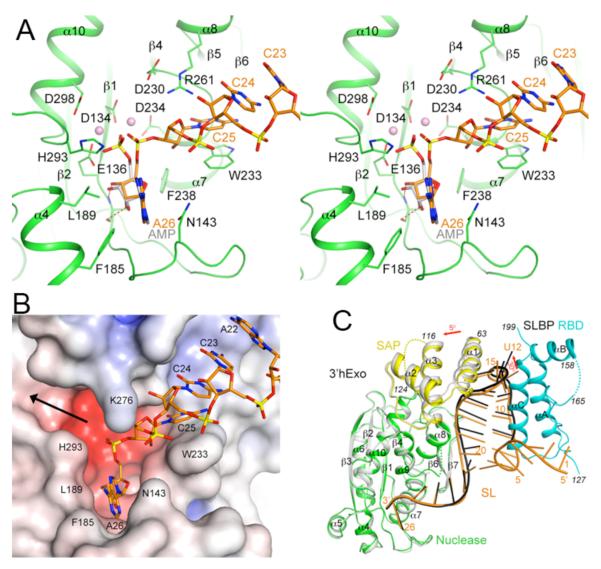
Nucleotides 24-26 at the 3′-end of the SL are located in the active site of the nuclease domain of 3′hExo (Fig. 1B). The C25 base is π-stacked with that of C24 on one face and the side chain of Trp233 on the other (Fig. 3A), thereby breaking the helical pattern of the RNA. The side chain of Arg261 is located close to the base and ribose of both C24 and C25 (Fig. 3A). In comparison, the first two nucleotides of the 3′ flanking sequence (A22-C23) do not make direct contacts with 3′hExo (Fig. 3B). The binding mode of the last nucleotide (A26) is similar to that of AMP in the nuclease domain reported earlier (Fig. 3A) (20). The phosphate group of A26 is located near the cluster of acidic side chains that coordinate two metal ions for catalysis.
Figure 3.
The 3′ flanking sequence of the SL RNA is located in the 3′hExo active site. (A). Interactions between the 3′ flanking sequence of the SL RNA (orange) with the active site of 3′hExo nuclease domain (green). The bound positions of AMP (gray) and two metal ions (pink spheres) to the nuclease domain of 3′hExo as observed earlier are also shown (20). (B). Molecular surface of the active site region of 3′hExo colored based on electrostatic potential. The SL RNA is shown as a stick model (orange). The black arrow indicates another opening from the active site, through which 3′hExo may accommodate longer RNA molecules. (C). Overlay of the structures of the ternary (SL-SLBP-3′hExo, in color) and binary (gray for 3′hExo and black for SL) complexes. The superposition is based on the nuclease domain of 3′hExo.
The crystal also contained a 3′hExo-SL binary complex (Figs. 3C, S8). SLBP RBD has low solubility and some of it precipitated during the preparation of the complex. The nuclease domains of 3′hExo in the two complexes have essentially the same conformation (rms distance 0.4 Å). The SAP domain shows a small movement (~5° rotation), together with a movement of the RNA (Fig. 3C). However, the first eight nucleotides of the SL, including three at the base of the stem, are disordered in this binary complex (Fig. 3C). The structure of this binary complex is similar to that of 3′hExo in complex with a stem-loop RNA without any flanking sequences reported earlier (PDB entry 1ZBH), although the SAP domain in that crystal comes from another 3′hExo molecule of a domain-swapped dimer (Fig. S9).
Transversion of the second base pair of the stem leads to >200-fold reduction in affinity for SLBP, while transversion of the first, third, fourth or fifth base pair had <5-fold effect (17), consistent with the structural observations (Figs. 2A, S5). Mutation of Arg181 in SLBP also inhibited SL binding in yeast three-hybrid assays (23, 24). In comparison, transversion of the second base pair had little effect on 3′hExo binding (9), also consistent with the structure (Table S3). To further validate the structural observations, we introduced mutations in the SL-SLBP RBD and SL-3′hExo interfaces and determined their effects on the formation of the binary and ternary complexes. Overall, the mutagenesis results are in good agreement with the structure (Fig. S10, Table S4).
The modes of SL recognition by the RBD of SLBP and the SAP domain of 3′hExo appear to be distinct from other RNA binding proteins. The SLBP RBD does not have a close structural homolog in the PDB. While the SAP domain has structural similarity to a domain in the recombination endonuclease VII (25), it does not share a common mode of nucleic acid interaction with that domain.
Our studies suggest that SLBP RBD and 3′hExo recognize the overall shape of the SL (especially its loop) rather than the sequence, which is also supported by observations from the single transversion studies (9, 17). At the same time, the sequence of the SL plays a role in determining its shape. The first (U12) and third (U14) nucleotides of the loop are highly conserved (Fig. S11) and contribute to the specificity of recognition (9, 17). C. elegans SLBP is more selective for a C at the first position of the loop, while human SLBP binds RNAs with C or U at the first position with comparable affinity (16). Tyr144 of human SLBP is replaced by an Arg residue in C. elegans SLBP, and this may result in a distinct mechanism of recognizing the C in the first nucleotide of the loop (Fig. 2B).
There are no direct contacts between SLBP RBD and 3′hExo in the ternary complex (Figs. 1B, S12). The RBD and SAP domain are arranged on opposite sides of the loop, and they approach each other most closely there. Cooperative binding between the two proteins (9-11) is likely due to induced structural changes in the loop, such that binding of one protein induces a conformation of the loop that promotes the binding of the other protein. In structures of the SL alone in solution (26, 27), the conformation of the loop region is different from that in the complex observed here (Fig. S13).
3′hExo has primarily bipartite interactions with the SL. The SAP domain recognizes the loop while the nuclease domain binds the 3′ flanking sequence (Fig. 1B). Disruption of interactions at either of these two sites leads to reduced binding (9, 10). Nucleotide A26 would be the leaving group for the 3′-5′ exonuclease activity (Fig. 3A), which does not show sequence preference (9) as neither C25 nor A26 is recognized specifically. While 3′hExo can remove the last 3 nucleotides of the SL (9), further degradation is not possible as the 3′-end of the shortened SL can no longer reach the active site of 3′hExo in the ternary complex (Fig. 3B), thereby explaining how SLBP protects histone mRNAs from excessive trimming by 3′hExo.
Besides recognizing the SL RNA, another function of SLBP is the recruitment of U7 snRNP and stabilization of its interaction with the histone pre-mRNA for 3′-end processing (Fig. S1) (22, 28). The 20 residues immediately C-terminal to the RBD of SLBP are required for this processing (28). These residues are present in the recombinant SLBP used in the current structural studies, but they are disordered. A second region required for processing is located in helix αB of the RBD, especially the YDRY motif (Figs. 1B, S6) where mutation of the DR residues to QC did not affect binding but abolished processing (22). Our structure shows that these two regions are likely located close to each other (Fig. S6), and therefore also identifies a surface feature of SLBP that is involved in histone pre-mRNA 3′-end processing (Fig. S14).
Supplementary Material
Acknowledgments
We thank Neil Whalen, Stuart Myers, Rick Jackimowicz and Howard Robinson for access to the X29A beamline at the NSLS. This research was supported in part by grants from the NIH to LT (GM077175) and to WFM and ZD (GM029832). The structure has been deposited at the PDB (accession code 4HXH).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References
- 1.Dominski Z, Marzluff WF. Gene. 2007;396:373. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marzluff WF, Wagner EJ, Duronio RJ. Nat. Rev. Genet. 2008;9:843. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao J, Hyman L, Moore CL. Microbiol. Mol. Biol. Rev. 1999;63:405. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandel CR, Bai Y, Tong L. Cell. Mol. Life Sci. 2008;65:1099. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang ZF, Whitfield ML, Ingledue TC, III, Dominski Z, Marzluff WF. Genes Develop. 1996;10:3028. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 6.Martin F, Schaller A, Eglite S, Schumperli D, Muller B. EMBO J. 1997;16:769. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mowry KL, Steitz JA. Science. 1987;238:1682. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- 8.Mullen TE, Marzluff WF. Genes Develop. 2008;22:50. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominski Z, Yang X-C, Kaygun H, Dadlez M, Marzluff WF. Mol. Cell. 2003;12:295. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- 10.Yang X-C, Purdy M, Marzluff WF, Dominski Z. J. Biol. Chem. 2006;281:30447. doi: 10.1074/jbc.M602947200. [DOI] [PubMed] [Google Scholar]
- 11.Yang X-C, Torres MP, Marzluff WF, Dominski Z. Mol. Cell. Biol. 2009;29:4045. doi: 10.1128/MCB.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas MF, et al. Blood. 2012;120:130. doi: 10.1182/blood-2011-11-394072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ansel KM, et al. Nature Struct. Mol. Biol. 2008;15:523. doi: 10.1038/nsmb.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabel HW, Ruvkun G. Nature Struct. Mol. Biol. 2008;15:531. doi: 10.1038/nsmb.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams AS, Marzluff WF. Nucl. Acid Res. 1995;23:654. doi: 10.1093/nar/23.4.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel F, Schumperli D, Muller B. RNA. 2000;6:1539. doi: 10.1017/s135583820000056x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battle DJ, Doudna JA. RNA. 2001;7:123. doi: 10.1017/s1355838201001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borchers CH, et al. Proc. Natl. Acad. Sci. USA. 2006;103:3094. doi: 10.1073/pnas.0511289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang M, Lam TT, Tonelli M, Marzluff WF, Thapar R. Biochem. 2012;51:3215. doi: 10.1021/bi2018255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Y, Patel DJ. J. Mol. Biol. 2004;343:305. doi: 10.1016/j.jmb.2004.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Detailed materials and methods can be found in the supplementary information.
- 22.Dominski Z, Erkmann JA, Greenland JA, Marzluff WF. Mol. Cell. Biol. 2001;21:2008. doi: 10.1128/MCB.21.6.2008-2017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin F, Michel F, Zenklusen D, Muller B, Schumperli D. Nucl. Acid Res. 2000;28:1594. doi: 10.1093/nar/28.7.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaeger S, Eriani G, Martin F. FEBS Lett. 2004;556:265. doi: 10.1016/s0014-5793(03)01433-9. [DOI] [PubMed] [Google Scholar]
- 25.Biertumpfel C, Yang W, Suck D. Nature. 2007;449:616. doi: 10.1038/nature06152. [DOI] [PubMed] [Google Scholar]
- 26.DeJong ES, Marzluff WF, Nikonowicz EP. RNA. 2002;8:83. doi: 10.1017/s1355838202013869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanier K, et al. RNA. 2002;8:29. doi: 10.1017/s1355838202014061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominski Z, Zheng LX, Sanchez R, Marzluff WF. Mol. Cell. Biol. 1999;19:3561. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.