Abstract
Recent years have witnessed a sea change in our understanding of transcription regulation: whereas traditional models focused solely on the events that brought RNA polymerase II (Pol II) to a gene promoter to initiate RNA synthesis, emerging evidence points to the pausing of Pol II during early elongation as a widespread regulatory mechanism in higher eukaryotes. Current data indicate that pausing is particularly enriched at genes in signal-responsive pathways. Here the evidence for pausing of Pol II from recent high-throughput studies will be discussed, as well as the potential interconnected functions of promoter-proximally paused Pol II.
Introduction
Higher organisms have evolved sophisticated mechanisms for responding in an integrated and balanced manner to a variety of developmental, environmental and nutritional cues by precisely modulating transcription output. In response to both intra- and extracellular cues, organisms must execute complex programs that require exquisite regulation of both the timing and level of gene expression. These different transcriptional regulatory programs are orchestrated by the concerted action of sequence-specific transcription factors that recruit the transcription machinery. The enzyme that transcribes messenger RNA from protein-encoding genes is Pol II. With the help of a constellation of accessory factors, Pol II executes a series of distinct steps: it binds to promoters; initiates RNA synthesis and then pauses in early transcriptional elongation. The paused Pol II remains stably associated with the nascent RNA and is fully capable of resuming elongation, however further signals are needed to elicit the transition to a productive elongation complex. Once this maturation occurs, the polymerase processively progresses through the gene, terminates and eventually re-initiates transcription. To understand how developmental and homeostatic transcriptional programs operate requires that we know the transcription factors that are involved and their targets. But just as important is an understanding of the mechanisms by which the interplay between Pol II and regulatory factors leads to highly specific, yet readily modulated transcription profiles.
Traditional models of eukaryotic gene regulation were based largely on studies in S. cerevisiae, which emphasized primarily the recruitment step in the transcription cycle and assumed that little regulation occurred after the formation of a Pre-Initiation Complex (PIC). However, recent findings in metazoan systems have revealed that much of transcription regulation occurs well after the recruitment of Pol II and the transcription machinery to a gene promoter, through controlling pausing and the efficiency of early elongation. Thus, we are in the midst of a paradigm shift in our understanding of gene regulation as it applies to higher eukaryotic systems.
Here, we focus on the promoter-proximal pausing of Pol II and its regulated escape into productive elongation1. In this Review, we use the shorthand of calling promoter-proximal pausing simply Pol II pausing, although (as discussed below) there is evidence that the polymerase can pause during productive elongation as well. We describe the basic biochemical properties of paused Pol II and recent evidence from genome-wide studies indicating that this type of regulation is widespread in metazoans2–9. We then discuss how Pol II pausing can influence chromatin structure at promoters to facilitate gene activity, and how pausing might lead to rapid or synchronous transcriptional responses when cells are exposed to an activation signal. We also highlight how regulation of early elongation can interplay with factors regulating Pol II recruitment and the RNA processing machineries to finely modulate transcription in response to distinct signals occurring during development, homeostasis, and disease.
Discovery of promoter-proximal Pol II pausing
Although pausing has only recently been recognized to be a prevalent regulatory strategy, evidence that transcription elongation could be a rate limiting step in gene expression surfaced more than thirty years ago. A number of studies in mammalian cell culture in the late 1970’s and early 80’s indicated that transcription, once initiated, did not obligatorily produce a full-length transcript10, 11. Insight into when this post-initiation block occurred came from in vivo analyses of the uninduced Drosophila melanogaster heat shock (Hsp) genes in the Lis laboratory (using UV protein-DNA crosslinking12, nuclear-run on13, permanganate footprinting14 and analysis of the short, capped RNAs15) (Box 1). These studies revealed that transcriptionally engaged polymerase accumulates just downstream of the Hsp promoters, associated with 20–60 nucleotide-long nascent RNA13, 15. The properties of these promoter-associated Pol IIs were strikingly similar to those ascribed by Roberts and colleagues to E. coli RNA polymerases that pause at the start of the lambda late gene transcription unit16. Thus, the Lis group referred to the promoter-proximal Pol II found at the Hsp genes as ‘paused’17.
Box 1. Methods used to detect paused polymerase.
Chromatin immunoprecipitation (ChIP)
This technique involves protein-DNA crosslinking coupled with immunoprecipitation. When an antibody targeting Pol II is used, ChIP can identify regions of DNA that are bound by Pol II. We note that several antibodies are available that recognize different phosphorylation states of the Pol II CTD, including the early elongation form characterized by predominant Ser7/Ser5-P and the productive elongation form that is also phosphorylated at Ser291. However, because our knowledge is incomplete concerning how the many reported modifications of the CTD affect the affinity of these antibodies for their target epitope and because we do not know the complete modification status of Pol II at every step of the transcription cycle, we caution against using phospho-CTD antibodies as the sole method for establishing the presence of a paused Pol II.
Advantages: A snapshot of Pol II distribution can be achieved through rapid cross-linking of whole cells. Analysis of individual genes is straightforward using quantitative PCR. ChIP is readily adapted for high-throughput genome-wide studies, either by hybridizing immunoprecipitated DNA to an array (ChIP-chip) or through high-throughput sequencing of Pol II-bound DNA (ChIP-seq).
Disadvantages: Low spatial resolution and sensitivity. ChIP signal and specificity is highly dependent upon the antibody used.
Permanganate footprinting
Detects locally melted regions of DNA, such as those arising from paused polymerase, by selectively modifying unpaired thymines within a stable, open transcription bubble. Modified thymines are then converted to strand breaks that are visualized by Ligation-Meditated (LM) PCR.
Advantages: Can be performed directly on whole cells or tissues. Achieves nucleotide-level resolution for mapping paused polymerase. Does not require antibodies.
Disadvantages: Low throughput, since the readout involves LM-PCR on individual genes. As a result, the application is limited to genes where good primers for primer extension and LM PCR can be designed, making permanganate footprinting challenging in mammalian systems.
Nuclear run-on assays and Global Run On (GRO)-Seq
Run-on assays detect elongation-competent RNA polymerases through their ability to incorporate a label into nascent RNA in isolated nuclei. GRO-seq is a genome-wide nuclear run-on method that enables high resolution mapping of transcriptionally-engaged Pol II. Transcriptionally engaged Pol II are allowed to elongate for ~100 nucleotides in the presence Br-UTP. The RNAs are then base hydrolyzed to ca. 100 nucleotides in length and RNAs are affinity purified using anti-BrU beads and specific linkers are added to the 5’ and 3’ ends before submitting samples to Next-Generation Sequencing. The specific 5’ primer allows the orientation of the RNAs to be determined, while three affinity purifications at various points in the sample preparation provide a very low background.
Advantages: Specifically reveals transcriptionally engaged and active polymerase, with high sensitivity and low background. Adaptable for high-throughput genome-wide applications. Can be used in various organisms.
Disadvantages. Technically challenging and requires preparation of nuclei. Resolution for mapping of paused polymerase is reduced by the necessity to allow polymerase to run-on and incorporate labelled nucleotides into RNA.
Short, capped RNA (scRNA) analysis and scRNA-seq
Direct isolation and identification of short RNA species derived from promoter-proximal Pol II. Initial use of this technique isolated RNAs produced at individual genes using complementary sequence specific probes15, 98. Extending this technique genome-wide by scRNA-seq7 involves isolation of nuclei, size selection of short (<100nt) RNA species, and enzymatic degradation of RNAs lacking the 5’-cap prior to directional linker addition and high-throughput sequencing. This strategy allows for highly sensitive detection of RNA produced by promoter-proximal Pol II, including RNA species generated by Pol II that pauses only transiently or terminates transcription prematurely.
Advantages. Sequencing of short capped RNAs pinpoints the start site of transcription and the final nucleotide added by paused polymerase at single nucleotide resolution.
Low background and high sensitivity assay well suited for high-throughput genome-wide applications. Does not require antibodies, cell treatment or labelling. Can be used in various organisms.
Disadvantages. Technically challenging and requires preparation of nuclei. Does not distinguish between RNA species that remain associated with paused Pol II and those that have been released through transcription termination.
Importantly, additional work performed in the late 1980’s and early 90’s revealed that other promoters displayed paused Pol II. In fact, a large fraction of Drosophila genes investigated in detail (6 out of 10 genes) showed characteristics of Pol II pausing17, 18. Moreover, a handful of mammalian genes, including key cell regulatory genes such as human c-myc and Fos, showed an enrichment of engaged Pol II just downstream of the transcription start site (TSS) that was effectively indistinguishable from that seen at the Drosophila Hsp genes19–21. Pol II was also found to accumulate on promoters of the HIV LTR, although this regulatory system displayed several features that distinguished it from pausing at endogenous genes. First, the nascent RNA transcribed at the HIV LTR forms a functionally important unique secondary structure4, 22 and secondly, the HIV LTR produces an abundant, 59 nucleotide-long RNA that results from premature termination of the early elongation complex22. In contrast, there is no current evidence suggesting high levels of promoter-proximal termination by Pol II at endogenous genes. Nonetheless, these studies of multiple gene systems provided evidence of regulation after recruitment of Pol II to a gene promoter, begging the question of how widespread these ‘alternate’ mechanisms of gene regulation might be.
Pol II pausing is widespread in metazoans
The findings described above, while appreciated by the field, were eclipsed by studies of transcription in the powerful yeast model system that demonstrated that recruitment of Pol II to promoters was a major mode of gene regulation and provided no compelling evidence for promoter-proximally paused Pol II23, 24. The interest in promoter-associated polymerase was recently reignited by the ability to carry out Pol II chromatin immunoprecipitation (ChIP) assays genome-wide using ChIP-chip or ChIP-seq techniques (See Box 1 on methods for detecting Pol II). These studies have provided evidence for widespread post-recruitment regulation of gene expression in metazoans.
Identifying and defining promoter-associated Pol II
The global localization of Pol II occupancy in multiple species has revealed that Pol II exhibits a variety of distributions along genes that provide insights into the mechanics of their regulation. In yeast, Pol II usually displays a relatively uniform distribution across the transcription unit25, as expected from models where, once recruited, Pol II experiences few regulatory barriers. In striking contrast, Pol II in Drosophila4, 6, 9 and mammalian cells26, 27 is frequently distributed non-uniformly on the bodies of genes. In these higher eukaryotes, a large fraction of genes display Pol II signal that is concentrated near transcription start sites (TSSs), indicating that polymerases recruited to these promoters are not released efficiently downstream into the gene. However, initial genome-wide studies employed Pol II ChIP to localize the polymerase, which in itself is not sufficient to distinguish between species that are paused during early elongation and those that are blocked at another post-recruitment step in the transcription cycle (Figure 1, Box 1: see ChIP). Therefore, elucidating the true status of poised, promoter-associated Pol II (Figure 1e) required the development and use of additional assays.
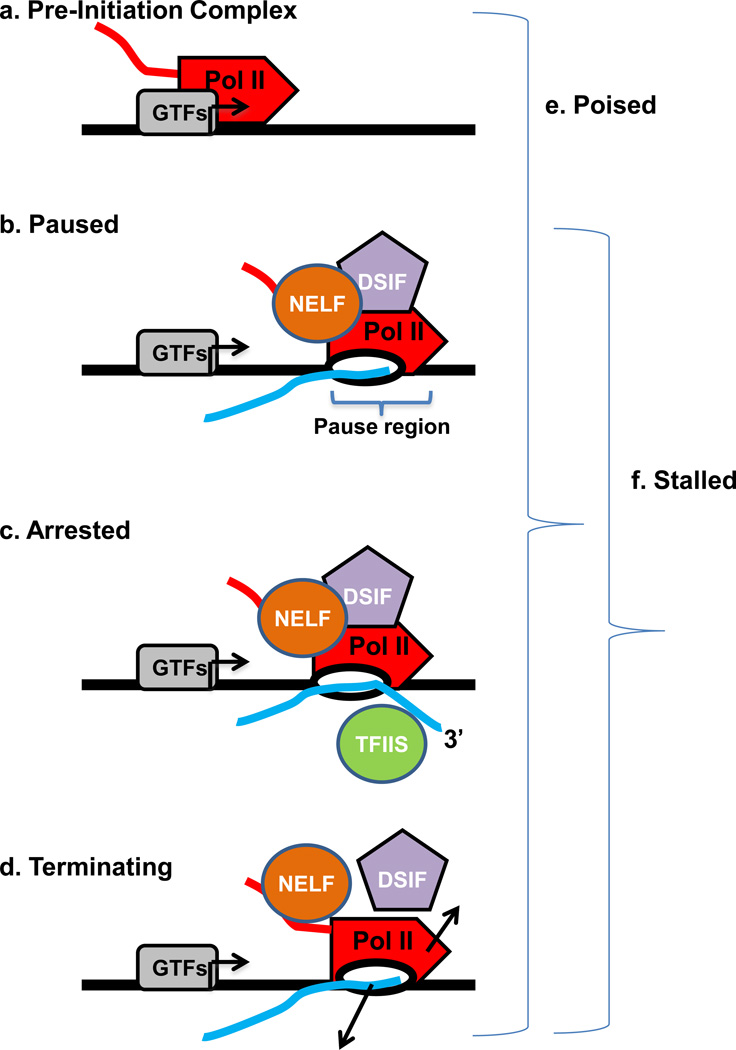
Figure 1. Defining the terms used to describe promoter-associated Pol II complexes.
- Pre-Initiation Complex: an entry form of Pol II in complex with general transcription factors, where the polymerase is bound to the promoter DNA but has not yet initiated RNA synthesis.
- Paused: an early elongation complex that has transiently halted RNA synthesis. Paused polymerase is fully competent to resume elongation, remaining stably engaged and associated with the nascent RNA. The 3’-end of the RNA may have ‘frayed’ slightly from the Pol II active site in a manner that would slow further RNA synthesis, but the RNA is properly aligned with the active site. Two protein complexes, DSIF and NELF reduce the rate of elongation and facilitate the establishment of the stably paused state.
- Arrested: a stably engaged elongation complex wherein the polymerase has backtracked along the DNA template, such that the RNA 3’-end is displaced from the active site. Re-start of an arrested complex usually requires the transcript cleavage factor TFIIS, which induces Pol II to cleave the nascent RNA at the active site, creating a new 3’-end that is properly aligned with the Pol II active site and releasing a short (2–9 nt) 3’ RNA.
- Terminating: an unstable elongation complex that is in the process of dissociating from the DNA template and releasing the nascent RNA. The released Pol II could have the potential to rapidly re-initiate transcription and “re-cycle” at the promoter.
- Poised: very generic term that simply indicates that Pol II is located near the TSS, but does not specify anything about its transcriptional status. Can include any of the above complexes (a–d).
- Stalled: term indicating that Pol II is engaged in transcription, but that makes no assumptions about its ability to resume synthesis. This term includes paused, arrested and terminating complexes (b–d, above).
Defining the status of promoter-proximal Pol II is important for understanding the regulation of polymerase release from the promoter region into productive synthesis. For example, a recruited polymerase that fails to initiate RNA synthesis and is trapped as a PIC (Figure 1a) would involve significantly different mechanisms for release than a Pol II that had synthesized a short transcript but was blocked in early elongation. Moreover, early elongation complexes that accumulate downstream of promoters can be present in several conformations that are not all competent to resume RNA synthesis. Paused Pol II can be readily induced to re-start transcription (Figure 1b), whereas arrested and terminating elongation complexes cannot (Figure 1c–d) and require either rescue or re-initiation in order to generate a productive transcript.
The first genome-wide ChIP study of Pol II distribution in human primary lung fibroblasts26 in 2005 referred to promoter-proximal accumulation of Pol II as PICs (Figure 1a), because the peak of Pol II mapped near the TSSs and because extensive studies in vitro had firmly established the concept of a PIC as an intermediate that occurs early in the transcription cycle. However, in 2007, genomic analysis of Pol II in human ES cells revealed that the Pol II accumulation was accompanied by chromatin signatures of gene activity, suggesting that these Pol II had undergone transcription initiation27. Concurrent ChIP-chip analyses in Drosophila S2 cells and early embryos also identified a widespread accumulation of promoter-associated Pol II, and followed up with permanganate footprinting (Box 1) to investigate whether or not the observed Pol II had paused during elongation through the promoter-proximal region6, 9. This permanganate footprinting demonstrated the presence of stably melted DNA located 20–60 bases downstream of the TSSs of dozens of genes4, 6, 9, which is diagnostic of a transcriptionally engaged polymerase. In addition, depletion of a negative elongation factor that induces pausing (NELF, discussed below) released many of these Pol II complexes from promoter regions, further supporting their designation as engaged, but paused species6. However, it was unclear what fraction of these promoter-proximal elongation complexes was competent to resume RNA synthesis, because permanganate footprinting cannot distinguish between paused, arrested and terminating complexes (Figure 1b–d). Thus, these Pol II species were initially referred to as ‘stalled’4, 6, 9, which is a general term that includes all of these different forms of engaged Pol II (Figure 1f).
Global nuclear run-on assays (GRO-seq, see Box 1) in human primary lung fibroblasts in 2008 confirmed that many of the promoter-associated Pol II molecules are indeed paused, by demonstrating that they are largely capable of resuming transcription in vitro following treatment with the detergent sarkosyl2. Sarkosyl is thought to remove pause-inducing factors from the elongation complex, allowing for RNA synthesis to continue. Importantly, arrested or terminating elongation complexes cannot be induced to “run-on” in this assay, such that the peak of signal observed near promoters by GRO-seq clearly represents Pol II in a paused state2, 5, 28.
Furthermore, although backtracking and arrest of early elongation complexes (Figure 1c) were found to occur commonly in vitro using metazoan transcription systems29, promoter-proximal backtracked complexes were found to be rapidly rescued from arrest in vivo. Indeed, genomic analyses of short, capped RNAs generated by paused Pol II in Drosophila demonstrated that such backtracking is efficiently followed by TFIIS-mediated cleavage of the extruded RNA7. Thus, current evidence suggests that a large fraction of promoter-associated Pol II is in a stably paused state that is competent to resume RNA synthesis. However, more detailed analyses of Pol II status, and in particular the contribution of premature transcription termination to the promoter-proximal Pol II signal, will be required to conclusively address this issue.
Patterns of paused Pol II
Many metazoan genes display significantly higher levels of Pol II on their promoters than on their gene body, but this ratio of promoter to gene body Pol II density, termed the pausing index, can vary dramatically among genes (examples shown in Figure 2). Due to this broad spectrum of pausing indices, and the inherent difficulties in applying a discrete threshold to continuous data sets, calculations of the fraction of genes that display Pol II pausing in mouse embryonic stem (ES) cells have produced estimates ranging from ~30% to ~90% 5, 8. Therefore, rather than reflecting a biological difference, the reported differences in prevalence of pausing are likely a consequence of using different methods and different statistical criteria to define Pol II pausing. Notably, when GRO-seq and a consistent data analysis method are used to measure Pol II occupancy in human primary lung fibroblasts, mouse ES cells, or Drosophila cell culture, a similar fraction of genes are found to display paused Pol II in all cases: ~30% of all genes2, 5, 28. Thus, despite the fact that the definition of what constitutes ‘pausing’ is highly variable among the different groups studying this phenomenon, the proportion of genes that exhibit Pol II pausing appears to be relatively constant across species and developmental stages reported to date.
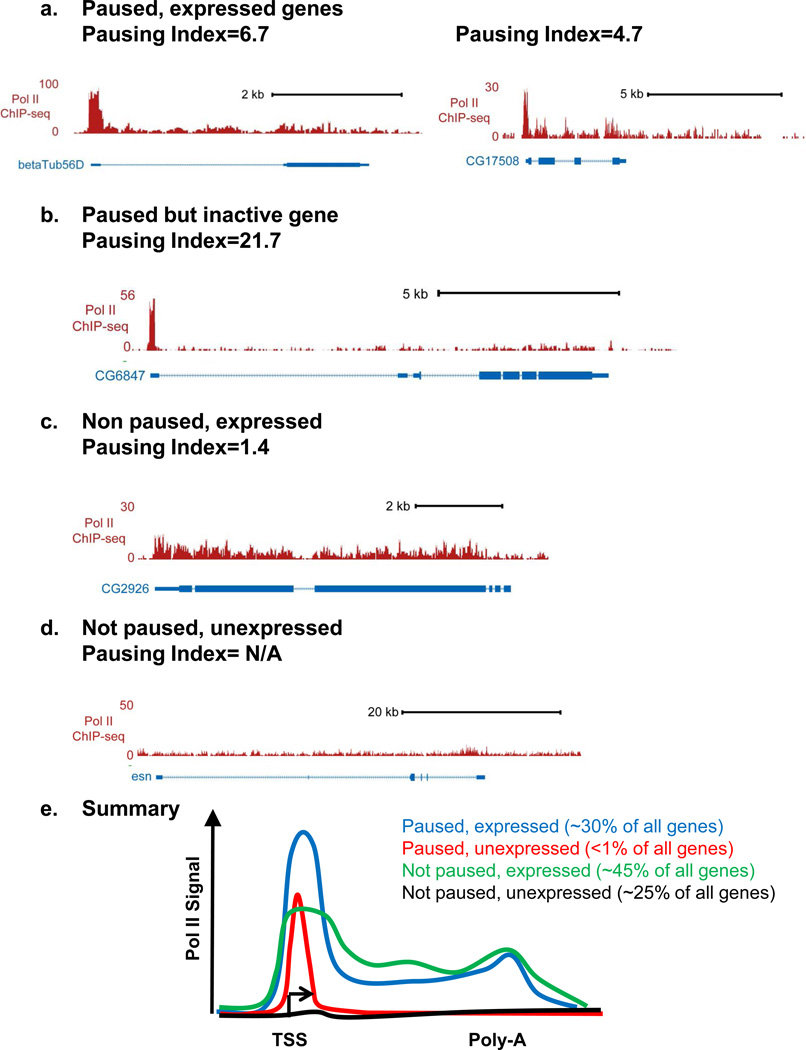
Figure 2. Patterns of Pol II distribution across gene regions.
- Two paused, but active genes with differing pausing indices
- A paused but inactive gene
- A Non-paused gene that is expressed
- A Non-paused, unexpressed gene. We note that a pausing index cannot reliably be calculated for genes that lack significant Pol II promoter signal.
- Shown are profiles of Pol II signal that exemplify the four major groups of genes. We note that the “paused, unexpressed” group is significantly under-represented in vivo, suggesting that most paused genes display some basal RNA synthesis. Approximate percentages of genes that fall into each category are given2, 5, 28.
Interestingly, in all systems evaluated thus far, genes that exhibit pausing are enriched in signal-responsive pathways including development, cell proliferation, and stress or damage responses. This enrichment is of particular interest in pluripotent cells such as ES cells, where pausing has been suggested to play a role in cell differentiation27. Underscoring that the presence and level of paused Pol II can be highly regulated, the specific genes that are paused in various cell types and under varying conditions such as cell stress or cell cycle regulation can differ dramatically5, 30
Genomic analysis of Pol II distribution also indicates that pausing occurs at genes across the spectrum of expression levels3, 7, 8. In fact, recent global analyses of Pol II distribution by GRO-seq indicate that very few paused genes are transcriptionally inactive (<1%)2, 5. This argues strongly against a common perception that Pol II pausing is predominantly a mechanism to silence gene expression31, 32. It is consistent with previous data on paused Pol II: for example, all of the traditionally defined paused genes (Drosophila Hsps, β-tubulin, mammalian c-myc, Fos) exhibit considerable basal expression, and the Drosophila heat shock genes continue to undergo pausing during activation1. Based on these data, we argue that pausing should be considered a mechanism for tuning expression from active genes and perhaps poising them for future changes in expression, rather than as a means of gene inactivation.
We note that Pol II can reduce its elongation velocity and/or pause during productive synthesis as well, although the factors involved and the mechanisms governing pausing within the gene appear to be distinct from those regulating promoter-proximal Pol II33. Slowing of productive elongation is best characterized at the 3’-end of genes, where considerable accumulation of Pol II is observed just downstream of the Poly-A site (Figure 2e)2, 34. This slowing of Pol II at the end of the transcription unit is thought to facilitate the coupling of RNA cleavage with transcription termination35. Likewise, pausing within exons has been reported36 where it is proposed to play a role in promoting splicing. Accordingly, evidence suggests that Pol II elongation rates can impact alternative splicing, with slower elongation favouring inclusion of exons with inherently weak splice sites37. As such, we now appreciate that gene expression can be regulated at virtually every step in the transcription cycle, from PIC formation through productive elongation and RNA processing.
Mechanisms of Pol II pausing and release
The establishment of paused polymerase requires both bringing Pol II to promoters, and stably retaining the early elongation complex within the pause region. These depend on the intrinsic strength of the core promoter38 and specific transcription factors that recruit chromatin remodelling proteins and the transcription machinery (Figure 3a–b; e.g. transcription factor TF1). After formation of the PIC, the promoter DNA is locally unwound, allowing the polymerase to initiate RNA synthesis and undergo promoter escape, wherein it releases many of the contacts with promoter-bound general transcription factors (GTFs)39. During this process, the GTF TFIIH phosphorylates Serine residues within the C-terminal heptapeptide repeat domain (CTD) of the Pol II largest subunit. The early elongation complex then extends the nascent RNA as it moves downstream into the gene. However, detailed analyses of early elongation have demonstrated that this process is fraught with difficulty29.
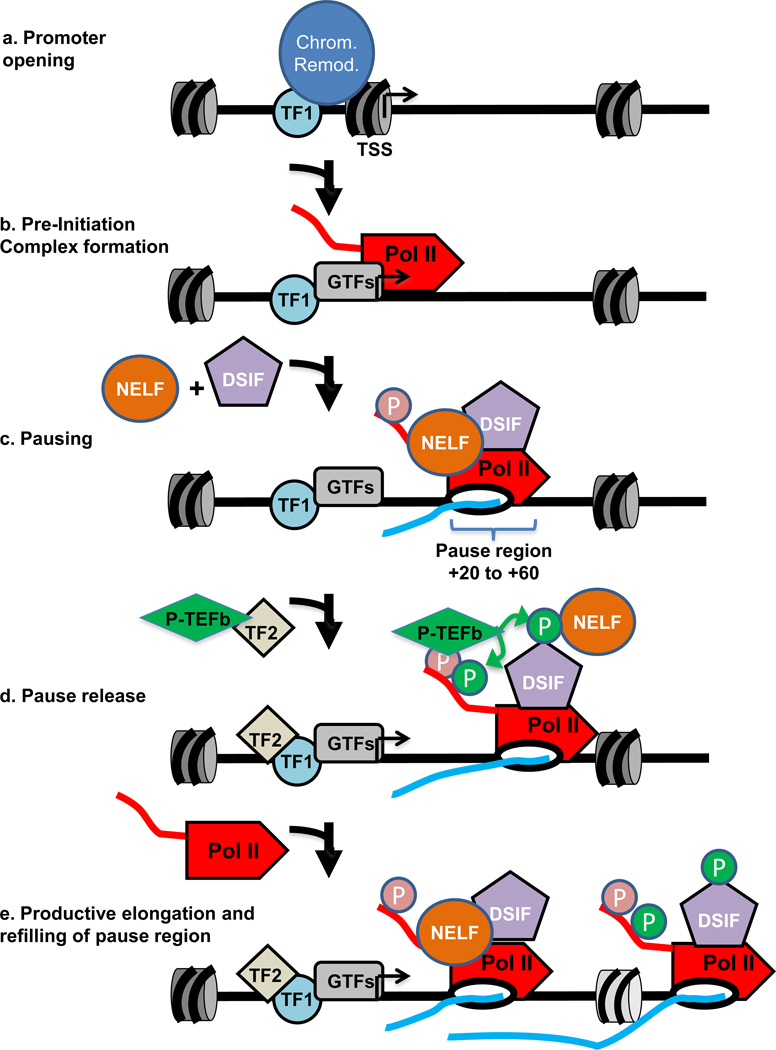
Figure 3. Establishment and release of paused Pol II.
- Promoter opening often involves the binding of a sequence specific transcription factor (shown here as TF1, light blue circle) that brings in chromatin remodelers (blue oval) to remove nucleosomes from around the TSS and render the promoter accessible for recruitment of the transcription machinery.
- Pre-Initiation Complex (PIC) formation involves the recruitment of a set of general transcription factors (GTFs, grey oval) and Pol II which is also facilitated by the binding of specific transcription factors (also shown as TF1 for simplicity). This step precedes the initiation of RNA synthesis.
- Pol II pausing occurs shortly after transcription initiation and involves the association of pausing factors DSIF and NELF. The paused Pol II is phosphorylated on its CTD (shown in pink). The region wherein pausing takes place is shown by a bracket.
- Pause release is triggered by the recruitment of the P-TEFb kinase (green diamond), either directly or indirectly by a transcription factor (shown here as TF2, tan diamond). P-TEFb kinase phosphorylates the DSIF/NELF complex to release paused Pol II and also targets the CTD (shown in green). Phosphorylation of DSIF/NELF dissociates NELF from the elongation complex and transforms DSIF into a positive elongation factor that associates with Pol II throughout the gene.
- In the presence of both TF1 and TF2, escape of the paused Pol II into productive elongation is followed rapidly by entry of another Pol II into the pause site, allowing for efficient RNA production. When the gene is activated, some nucleosome disruption is likely, as depicted by the lighter colouring of the downstream nucleosome.
Work done largely in the Handa and Price labs in the early 90’s demonstrated that Pol II elongates inefficiently through the promoter-proximal region, displaying a strong tendency to halt or terminate within the first 100 nucleotides29, 40, 41. These studies provided key mechanistic insights into Pol II pausing by revealing that the block in early transcription elongation results in part from the association of two pause-inducing factors with the early elongation complex (Figure 3c). These factors, called DRB sensitivity inducing factor (DSIF)41 and negative elongation factor (NELF)42 are together sufficient to inhibit early elongation in a purified system, indicating that they work directly on the polymerase to help establish the paused elongation complex. Consistent with the lack of evidence for Pol II pausing in S. cerevisiae, homologs of the pause-inducing NELF proteins are absent in yeast, but are conserved from Drosophila to man43.
Despite the clear importance of DSIF/NELF in establishing paused polymerase, growing evidence suggests that these factors are not alone in affecting the residence time of promoter-associated Pol II. Recent in vitro work suggests that additional factors, such as Gdown1 and the general transcription factor TFIIF may also influence the stability or lifetime of the paused polymerase, perhaps by affecting the susceptibility of the early elongation complex to premature termination44. Although it remains unclear whether termination in the promoter-proximal region occurs in vivo, Pol II ChIP-seq studies have provided evidence for premature termination within transcribed units (i.e. downstream of +500)45, suggesting that the processivity of elongating Pol II is continually subject to regulation. Thus, much yet remains to be learned about how the efficiency of early elongation is regulated at the mechanistic and biochemical level.
The maturation of paused Pol II to a productively elongating form requires the kinase activity P-TEFb40, 46, 47. P-TEFb phosphorylates the repressive DSIF/NELF complex, causing NELF to dissociate from Pol II and transforming DSIF to a state that promotes Pol II elongation (Figure 3d)33. P-TEFb also performs additional phosphorylation of Serine residues within the Pol II CTD, creating a platform for binding of RNA processing and chromatin modifying factors that facilitate productive RNA synthesis33, 48. Given its key role in pause release, there is considerable interest in understanding how P-TEFb is targeted to particular gene promoters (shown in Figure 3d as TF2). Befitting the diversity of genes that exhibit Pol II pausing, a large repertoire of factors have been reported to perform this activity, including the acetylated histone-binding protein Brd449, 50, DNA-binding transcription activators such as c-myc and NF-κB8, 51, 52 and the Med26 component of the Mediator complex53. Moreover, P-TEFb is found to be associated with a large number of other elongation factors and chromatin modifying proteins in the “Super Elongation Complex”, suggesting that these factors work together to stimulate productive elongation54–57.
Interestingly, although only a subset of genes appear to accumulate high levels of paused Pol II, most Drosophila or mammalian promoters display a detectable enrichment of polymerase near the promoter compared with the gene body3, 8. In addition, analysis of the location of factors that regulate the establishment and release of pausing suggest that transient Pol II pausing is a general feature of the transcription cycle. For example, the vast majority of active promoters are bound by the pause-inducing factors DSIF and NELF3, 8. The levels of DSIF/NELF at promoters corresponds extremely well with total promoter Pol II, suggesting that these factors associate with most early elongation complexes. Further, treatment of cells with the specific P-TEFb inhibitor, Flavopiridol, blocks the entry of most Pol II into productive synthesis in both Drosophila and mammals8, 58, indicating that polymerase release from the promoter region typically requires the activity of P-TEFb. Taken together, these data suggest that the early elongation complex comes under control of DSIF/NELF at most genes, and that the escape of Pol II into productive elongation involves release of this repressive complex by P-TEFb. Thus, we envision that the rate of P-TEFb recruitment would be critical for determining both gene expression levels and the appearance of paused Pol II. At many genes, P-TEFb recruitment to promoters may immediately follow transcription initiation, leading to rapid release of polymerase into the gene. However, at other genes, P-TEFb recruitment may be a much slower event, permitting accumulation of paused Pol II.
Functions of paused Pol II
Given the prevalence of paused Pol II at genes within critical developmentally- and environmentally-responsive pathways, identifying the functional roles of paused Pol II has become an active topic of research. We discuss models for four functions below, some of which may be interconnected (Figure 4).
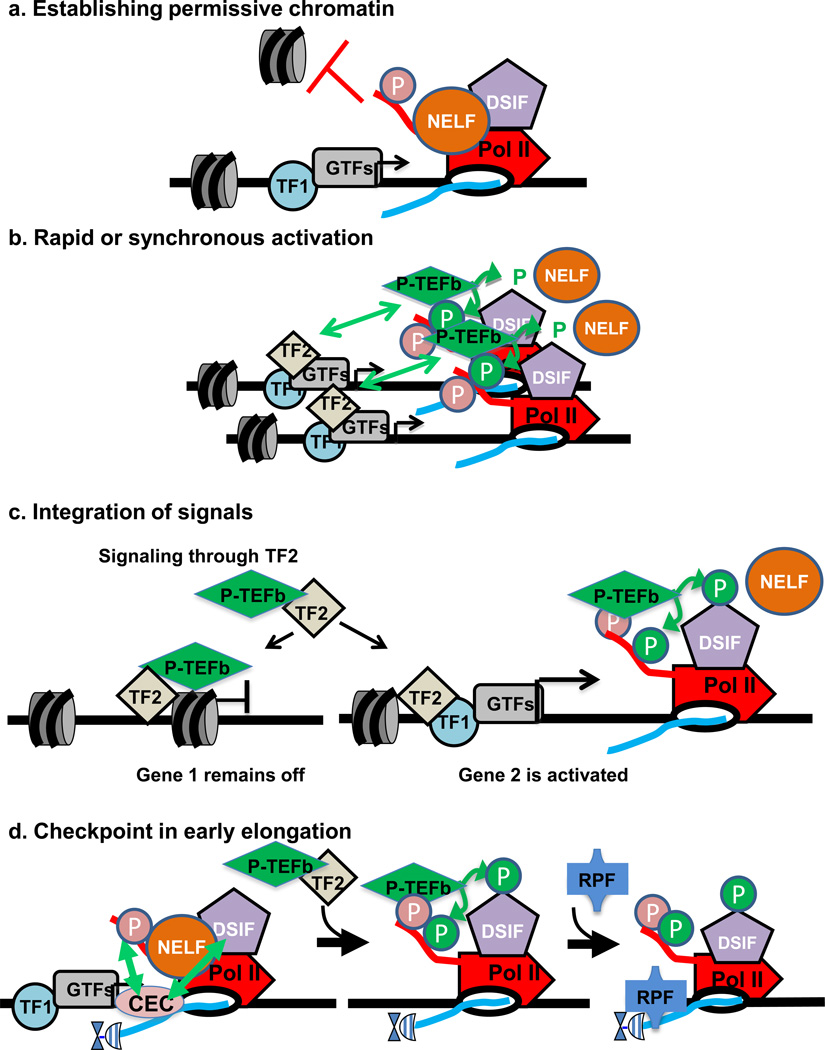
Figure 4. Illustrations of the main hypotheses for the functions of Pol II pausing.
- Establishing permissive chromatin: in the absence of paused Pol II, nucleosomes occlude the promoter region and inhibit gene expression, as shown in the left hand panel. Once, these nucleosomes are remodelled, paused Pol II helps to maintain the nucleosome-deprived structure by blocking nucleosome assembly over promoter sequences (depicted at right). Pausing thus would keep the promoter region accessible for activator and transcription factor binding.
- Rapid or synchronous gene activation: at a gene with paused Pol II, gene activation could proceed simply through recruitment of P-TEFb, thereby triggering the rapid release of paused Pol II into productive elongation. If a number of genes harbouring paused Pol II were all activated by the same signal and associated transcription factor (shown as TF2), then these genes could activated simultaneously and in a rapid fashion.
- Integrating multiple regulatory signals: Pausing represents a separate step in the transcription cycle for factors to act, and allows for combinatorial control between transcription factors that recruit the transcription machinery (TF1) and those that trigger pause release (TF2), where both would be necessary for gene activation. In this example, signalling through TF2 such that it binds DNA and recruits P-TEFb would not lead to activation of a gene that was not paused (i.e. lacks TF1), but would stimulate synthesis from Gene 2 that was loaded with paused polymerase.
- Checkpoint in early elongation: on the left, arrows depict interactions between the Capping Enzyme Complex (CEC) and DSIF/NELF as well as the Ser5-P modification of the CTD of Pol II, which is thought to stimulate capping activity. The hat represents the 5’ RNA cap. Center, P-TEFb dependent phosphorylation events release paused Pol II, and create a platform for binding of RNA processing factors (RPF) on the Ser2-P CTD of Pol II, as shown at right.
Establishing permissive chromatin
The wrapping of promoter DNA around histone proteins to form nucleosomes can present a barrier to transcription by rendering critical recognition sequences inaccessible. As a result, the remodelling of promoter chromatin to remove or displace nearby nucleosomes is often required to permit recruitment of the transcription machinery and gene expression (shown in Figure 3a)59. Whereas many genes, especially those in yeast60, 61, have been shown to temporally couple this nucleosome remodelling with gene activation, genes with paused polymerase have been shown to undergo nucleosome removal to open promoters prior to and independently from gene activition62, 63. Moreover, paused genes have been shown to persist in a nucleosome-deprived, regulatory factor accessible state that is dependent on the presence of the paused Pol II (Figure 4a)3, 64–66.
A relationship between the paused polymerase and the lack of promoter nucleosomes is apparent at the Drosophila heat shock genes62, 63, whose promoter regions were shown to be nucleosome-deprived even in the un-induced state. Further studies of Hsp70 transgenes indicated that promoter-proximal mutations that affected the levels of paused Pol II also disrupted the binding of HSF to its target sites during heat shock and subsequent gene activation64, 67. Notably, this work suggested that pausing could help maintain an open and accessible promoter structure to facilitate binding by regulatory transcription factors as well as the transcription machinery.
The link between paused Pol II and maintenance of a nucleosome-deprived promoter has recently been demonstrated at a genome-wide level in Drosophila3. Genes with paused polymerase were globally shown to possess low levels of promoter nucleosome occupancy which was dependent on the presence of promoter-associated Pol II: depletion of the pause-inducing factor NELF, which considerably reduced promoter Pol II levels at highly paused genes, led to a concomitant increase in promoter nucleosome occupancy at these genes3, 65. Thus, paused promoters display a dynamic competition for promoter binding between nucleosomes and Pol II. Importantly, genes affected in this way often decreased their expression levels upon NELF-depletion and loss of paused Pol II.
Interestingly, the underlying DNA sequence appears to contribute to the requirement for promoter-proximal Pol II to prevent nucleosome assembly over many TSSs. Packaging of DNA into nucleosomes requires that the underlying sequences are somewhat flexible and amenable to making regular bends as they wrap around the histone proteins, and it has been shown that certain sequences are particularly well- or ill- suited for this purpose68, 69. Strikingly, genes with high levels of paused Pol II in Drosophila possess promoter sequences that are very nucleosome friendly and predicted to promote chromatin assembly3. Genes at which less paused Pol II is present, tend to disfavour nucleosome assembly3. Likewise, paused Pol II is enriched in mammals at CpG-island promoters2 which tend to possess open chromatin70. Although currently a subject of debate, evidence suggests that mammalian promoters with moderately high CG content intrinsically favour nucleosome formation71–73 suggesting that, like in Drosophila, the transcription machinery helps maintain accessible chromatin architecture around these promoters.
Thus, it is tempting to speculate that highly-regulated promoters, many of which exhibit paused Pol II, have evolved DNA sequences that enable a dynamic competition between paused Pol II and nucleosomes74. For example, it has been proposed that the presence of paused Pol II poises genes in ES cells for expression during development, in part by altering promoter chromatin27. As such, the loss of paused Pol II later in development could enable nucleosome occlusion and permanent gene repression. The formation of repressive chromatin may be further enhanced by the recruitment of Polycomb Repression Complexes, PRC1 and PRC2. Notably, in ES cells, bivalent genes75 that contain both PRC complexes have significantly less paused Pol II than genes that lack Polycomb5, 76. Likewise, in Drosophila, mutations in a key component of the PRC2 complex that would presumably create a more accessible chromatin structure allows an increase in Pol II recruitment and pausing on thousands of promoters in the early embryo77. Importantly, the chromatin opening function of paused Pol II would be connected to the other potential functions of pausing. For example, the presence of paused Pol II might allow genes that are transcribed at lower basal levels to be continually accessible and primed for bursts of transcription activation in response to specific cues or for generating synchronous transcriptional responses to signalling (see below).
A pausing framework for rapid and/or synchronous activation
While rapid gene activation at many genes involves mechanisms that are independent of pausing78, 79 the presence of Pol II is an appealing way to generate an accessible promoter region that can be bound quickly by activators and coactivators. Importantly, the presence of paused polymerase would allow a promoter to be readily switched from experiencing long-lived pausing to undergoing productive elongation simply through the binding of transcription activators that associate with P-TEFb (Figure 4b), bypassing a number of potentially slow or stochastic steps involved in PIC formation. The open promoter and scaffold of GTFs that remain after Pol II escape80 may ensure continuous rapid entry of a succession of Pol II complexes on the activated gene (Figure 3b). Moreover, the nucleosome-deprived status of paused promoters is likely to facilitate transcription factor binding, resulting in more efficient, reliable activation81.
In support of a role for paused Pol II in rapid activation, pausing has been observed at Drosophila genes that are rapidly induced, like the Hsps1 and a number of genes involved in early embryonic development6, 9, 82, leading to the idea that pausing facilitates synchronous changes in gene expression83. Consistent with this, many mammalian genes with paused Pol II (c-myc, fos, junB, TNF- α) have fast, transient expression kinetics20, 84, 85.
However, not all rapidly induced genes display paused Pol II prior to activation86–88, nor are the majority of paused genes highly inducible. In fact, recent work surveying the prevalence of paused genes across several signal transduction networks in Drosophila and murine ES cells revealed that pausing was more enriched at promoters encoding the constitutively expressed components of signal transduction pathways (e.g. receptors, kinases, transcription factors) than at the inducible downstream targets of these pathways89. Moreover, pausing was shown to regulate network activity largely through affecting the basal expression of signal transduction machineries89. Thus, the role of pausing within stimulus-responsive networks is not limited to poising inducible genes for activation. Instead, Pol II pausing can regulate the expression of key molecules such as transcription factors and signalling proteins, thereby tuning cellular responsiveness to external cues.
Integrating multiple regulatory signals
Pausing represents an additional regulatory step in the transcription cycle, beyond Pol II recruitment. Accordingly, this could allow activators that influence pause release to work together with factors that stimulate recruitment to exert combinatorial control of transcription levels (Figure 4c)30, 90. Indeed, most promoters contain binding sites for multiple transcription activators. Importantly, some activators specifically function to recruit general transcription factors (GTFs) or establish a paused Pol II (e.g. Transcription factors Sp190 and GAGA factor4; shown as TF1 in Figure 4c), some factors bring P-TEFb to the promoter (e.g. c-myc and HIV Tat8, 52, 90; shown as TF2), and others appear to both recruit and release paused Pol II (e.g. NF-κB and herpes virus VP-16 protein51, 90). Thus, the particular combination of transcription activators that bind near any promoter would determine the rates of Pol II recruitment and pause release, thereby defining the rate-limiting step for transcription. In this way, cellular events that altered the levels or activity of individual transcription factors could be integrated on a gene-by-gene basis, depending on the sequence context and associated factors on the promoter and enhancer regions.
Checkpoint for coupling elongation and RNA processing
Pol II coordinates the efficient processing of nascent RNA: adding a Cap to the 5’-end, coupling splicing events to transcription, and facilitating the 3’-end processing of RNAs. By coupling RNA processing to the status and activity of Pol II itself, the cell ensures that nascent RNA is properly protected from degradation and efficiently matures into a functional mRNA. Pol II is phosphorylated on its CTD at various positions providing a binding platform to recruit an entourage of protein factors that can execute both early and later events of RNA processing91. Phosphorylation of Ser5 within the CTD creates a binding platform for interaction with the 5’ capping enzyme (Figure 4d), and stimulates the activity of this enzyme92. In vivo, 5’ capping occurs as the nascent RNA is extended from 20 to 30 nucleotides in length and the bulk of RNAs associated with paused Pol II are capped7, 15. This was determined initially by detailed analysis of the Hsp genes15 and extended by recent global analyses7 in Drosophila. Interactions have also been reported between the RNA capping machinery and the pause-regulatory factor DSIF93, 94. Thus, pausing may provide both a kinetic “window of opportunity” as well as an interaction surface to facilitate addition of the 5’-methyl cap to the nascent RNA prior to the transition to productive elongation.
As mentioned above, phosphorylation of paused Pol II by P-TEFb provides a binding platform for complexes that carry out 3’-end processing95. As such, the requirement for P-TEFb activity to phosphorylate the DSIF-NELF complex and trigger pause release also may ensure that Pol II does not proceed into the gene before it is appropriately modified for binding by the RNA processing factors (Figure 4d). While rigorous tests of pausing as an obligatory checkpoint for Pol II CTD modification are lacking, the fact that the P-TEFb kinase phosphorylates both DSIF-NELF and Pol II might functionally couple pause release to this Pol II modification.
Conclusions & Perspectives
In the last few years, a new picture of transcription regulation has emerged: genome-wide data in metazoans now points to the widespread importance of Pol II pausing in transcription regulation. Indeed, escape of paused Pol II into productive elongation is regulated during environmental stress6, immunological signalling85 and development96.
Studies of pausing over the decades coupled with an explosion of interest in recent years have led to considerable understanding of the characteristics and function of paused Pol II. Nonetheless, three major categories of questions remain. The first concerns the pervasiveness and patterns of pausing in eukaryotes. Studies underway in many labs will sample a broad swath of additional cell types and organisms in addition to Drosophila, mouse and human studied thus far. These studies should identify common features of genes regulated by this mechanism, as well as revealing cell type- or condition-specific patterns of paused Pol II. Quantitative genome-wide studies should also assess if paused polymerases constitute nearly all promoter-associated Pol II or if there are promoters with significant amounts of other forms: e.g. PICs or arrested Pol II, indicative of alternative modes of regulation (Figure 1).
The second category deals with mechanistic questions designed to understand pausing in molecular terms. We know several factors41, 42 and DNA elements7, 82 that are involved in stabilizing the paused state, but the full repertoire of factors and their interactions remains to be determined. Moreover, we know very little about how these factors interact to mediate efficient pausing. How stable are paused Pol II complexes, what are the relative levels of termination and escape to productive elongation and how might this balance be controlled? It will also be important to further elucidate how P-TEFb is either directly or indirectly targeted to promoters, and how its kinase activity is regulated. Several mechanisms for P-TEFb recruitment have been documented, but surely more are to be discovered33. Future work should also elucidate exactly how the Pol II paused complex is transformed into a productively elongating machine. These events need to be examined in living cells with optical and biochemical methods that provide detailed information on the position and dynamics of paused Pol II and the accompanying protein and DNA interactions. Improvements in inhibitor discovery and in the already powerful molecular (Box 1) and microscopic technologies97 provoke optimism that these challenging mechanistic goals will be achieved.
The third category contains questions addressing the function of this regulation. We emphasize in this review varying levels of evidence for four potential roles of pausing (Figure 4). These proposed functions will be clarified by further rigorous tests that include global studies as well as targeted analysis of specific genes and phenotypic analysis following systematic disruption of pausing. It will be interesting to define whether pausing serves different roles at different functional classes of genes and how these putative roles are interconnected. For example, the transient checkpoint established by pausing could be particularly useful at highly active genes, to ensure that the nascent RNA is properly processed. On the other hand, the opening of chromatin structure by paused Pol II could both fine-tune the basal expression of signalling proteins89 and facilitate a rapid transcriptional response64. In this review, we highlight our current but incomplete understanding of Pol II pausing at promoters and its role in gene regulation. After decades of research and numerous cycles of simplifying and confounding theories and observations, we now have a framework and many of the tools needed to understand mechanistically transcription and its regulation genome-wide.
Online 'at-a-glance' summary.
Pausing is now recognized to be a pervasive feature of promoters in mammals and Drosophila and the evidence supporting this from genome-wide studies is presented
The nomenclature of different promoter-associated Pol II species is explicitly defined in an effort to provide consistency in the future literature
The known mechanistic features of Pol II pausing and its release to productive elongation are described. Most genes are associated with factors that establish and release paused Pol II and therefore appear to progress through this step, although only a subset of genes appear to be directly regulated by pausing.
Multiple lines of evidence support the idea that Pol II and nucleosomes compete for promoter binding and suggest that a critical role of paused Pol II involves maintenance of accessible promoter chromatin architecture.
While pausing has been connected to extremely rapid and synchronous activation of genes, pausing is also highly associated with constitutively expressed genes which encode signalling and transcription factors. Pausing provides a mechanism to tune these key genes to cellular and external regulatory cues.
Pausing provides a point of regulation that is distinct from Pol II recruitment and initiation, and this may facilitate the integration of multiple cellular signals. Distinct signals that act through diverse targeted transcription factors can regulate different steps in the transcription pathway and provide a highly modulated transcriptional response at individual genes.
Pol II Pausing and release occurs at a point when 5’-end RNA processing and phosphorylation of the Pol II C-terminal domain occurs. We speculate that by coupling RNA processing to the status and activity of Pol II itself, the cell ensures that nascent RNA is properly protected from degradation and efficiently matures into a functional mRNA.
Acknowledgements
We thank the members of the Lis and Adelman labs for their helpful discussions on this review. Funding for this work was provided by NIH grant GM25232 to J.T.L. and the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01 ES101987) to K.A.
Biographies
John Lis is the Barbara McClintock Professor of Molecular Biology and Genetics at Cornell University. He did his Ph.D thesis research at Brandeis University and his postdoctoral studies at Stanford University, as a Helen Hay Whitney Foundation Fellow. Dr. Lis joined the faculty at Cornell in 1978 where his laboratory has developed and used a variety of strategies to probe the regulation of gene expression and the structure of promoters and genes in living cells. The lab’s primary model system has been the highly-inducible heat shock genes of Drosophila, and more recently genome-wide assays have been used in Drosophila and mammals to assess the generality of findings.
Karen Adelman is an investigator at the National Institute of Environmental Health Sciences (NIEHS). She received her Ph.D. from the Université de Paris VI, working at the Institut Pasteur as a National Science Foundation Pre-doctoral Fellow. Dr. Adelman did her post-doctoral research at Cornell University before joining the NIEHS in 2005. Her laboratory uses genetic, genomic and biochemical techniques to study gene regulation in Drosophila and murine systems, focusing on signalling pathways that respond to environmental and developmental cues.
Footnotes
Glossary
Pre-Initiation Complex (PIC) is an entry form of Pol II in complex with general transcription factors, where the polymerase is bound to the promoter DNA but has not yet initiated RNA synthesis.
Heat shock (Hsp) genes are a set of highly conserved genes encoding molecular chaperones. These genes are rapidly induced in cell or organisms in response to a variety of cellular stresses including a several degree increase in temperature.
The Human immunodeficiency virus (HIV) promoter resides within a long terminal repeat (LTR) region. Transcription from this promoter produces both viral proteins and new RNA genomes.
CpG-islands are regions of higher than normal CpG sequence content that are on average 1000 base pairs in length. Such regions contain ~70% of all mammalian promoters, including both genes that are highly regulated and are broadly expressed.
Polycomb group proteins regulate chromatin structure to contribute to epigenetic inheritance of a repressed state. They form several complexes, broadly defined as Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2), which are thought to compact chromatin structure.
Ligation-Meditated (LM) PCR is a technique that can be used to map precisely the ends of DNA fragments from a specific region of the genome. Small DNA linkers are added to ends of DNA samples and then primer complementary to this linker is combined with a sequence specific primer to amplify the DNA of interest by PCR. The resulting DNA can then be sequenced by any of a variety of methods, or its size examined by gel electrophoresis.
The promoters of Bivalent genes exhibit histone modifications characteristic of both gene repression and activation. These genes display very low levels of Pol II occupancy and activity, and are hypothesized to be poised for activation during development.
Contributor Information
Karen Adelman, Email: adelmank@niehs.nih.gov.
John T. Lis, Email: johnlis@cornell.edu.
References
- 1.Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- 2. Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228.. Global run-on sequencing (GRO-seq) maps the position, amount, and orientation of transcriptionally engaged RNA polymerases genome-wide, and shows peaks of promoter-proximal polymerase residing on ~30% of human genes.
- 3. Gilchrist DA, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004.. Global analyses of Pol II pausing and nucleosome occupancy reveal that Pol II and nucleosomes compete for promoter occupancy to co-ordinately regulate gene expression.
- 4. Lee C, et al. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07.. Comprehensive analysis of promoter-associated Pol II in Drosophila using ChIP-chip and permanganate demonstrates that NELF-mediated pausing of Pol II is common in Drosophila.
- 5.Min IM, et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21.. Genome-wide Pol II ChIP-chip assays, coupled with permanganate footprinting and genetic manipulation of NELF indicated that Pol II pausing is widespread in Drosophila.
- 7.Nechaev S, et al. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rahl PB, et al. c-Myc Regulates Transcriptional Pause Release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030.. Investigation of transcription factor c-myc reveals its important role in releasing paused Pol II through recruitment of P-TEFb.
- 9. Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26.. Genome-wide Pol II ChIP-chip assays indicated that Pol II pausing is widespread during early embryonic development in Drosophila.
- 10.Fraser NW, Sehgal PB, Darnell JE. DRB-induced premature termination of late adenovirus transcription. Nature. 1978;272:590–593. doi: 10.1038/272590a0. [DOI] [PubMed] [Google Scholar]
- 11.Gariglio P, Bellard M, Chambon P. Clustering of RNA polymerase B molecules in the 5' moiety of the adult beta-globin gene of hen erythrocytes. Nucleic Acids Res. 1981;9:2589–2598. doi: 10.1093/nar/9.11.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2.. Nuclear run-on assays demonstrate that the Pol II complex associated with Hsp70 promoter is transcriptionally engaged but unable to penetrate further into the gene without heat shock induction.
- 14.Giardina C, Perez-Riba M, Lis JT. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992;6:2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci U S A. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grayhack EJ, Yang XJ, Lau LF, Roberts JW. Phage lambda gene Q antiterminator recognizes RNA polymerase near the promoter and accelerates it through a pause site. Cell. 1985;42:259–269. doi: 10.1016/s0092-8674(85)80121-5. [DOI] [PubMed] [Google Scholar]
- 17.Rougvie AE, Lis JT. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990;10:6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Law A, Hirayoshi K, O'Brien T, Lis JT. Direct cloning of DNA that interacts in vivo with a specific protein: application to RNA polymerase II and sites of pausing in Drosophila. Nucleic Acids Res. 1998;26:919–924. doi: 10.1093/nar/26.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strobl LJ, Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. Embo J. 1992;11:3307–3314. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- 21.Plet A, Eick D, Blanchard JM. Elongation and premature termination of transcripts initiated from c-fos and c-myc promoters show dissimilar patterns. Oncogene. 1995;10:319–328. [PubMed] [Google Scholar]
- 22.Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 23.Stargell LA, Struhl K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 24.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 25.Steinmetz EJ, et al. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larschan E, et al. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kephart DD, Marshall NF, Price DH. Stability of Drosophila RNA polymerase II elongation complexes in vitro. Mol Cell Biol. 1992;12:2067–2077. doi: 10.1128/mcb.12.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nechaev S, Adelman K. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle. 2008;7:1539–1544. doi: 10.4161/cc.7.11.6006. [DOI] [PubMed] [Google Scholar]
- 31.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 34.Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 37.de la Mata M, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wada T, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamaguchi Y, et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8.. Biochemical assays reveal the presence and identity of the NELF complex and elucidate its role in inhibiting early transcription elongation.
- 43.Narita T, et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng B, et al. Functional Association of Gdown1 with RNA Polymerase II Poised on Human Genes. Mol Cell. 2012;45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brannan K, et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335.. Pioneering paper describing the purification and characterization of the kinase P-TEFb.
- 47.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Czudnochowski N, Bosken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun. 2012;3:842. doi: 10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- 49.Jang M, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 50.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 51.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 52.Eberhardy S, Farnham P. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi H, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mueller D, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin C, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sobhian B, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He N, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ni Z, et al. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol. 2008;28:1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 60.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 61.Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 62.Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 63.Costlow N, Lis JT. High-resolution mapping of DNase I-hypersensitive sites of Drosophila heat shock genes in Drosophila melanogaster and Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1853–1863. doi: 10.1128/mcb.4.9.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shopland LS, Hirayoshi K, Fernandes M, Lis JT. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- 65.Gilchrist DA, et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leibovitch BA, et al. GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol Cell Biol. 2002;22:6148–6157. doi: 10.1128/MCB.22.17.6148-6157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee H, Kraus KW, Wolfner MF, Lis JT. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- 68.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. Embo J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 71.Tillo D, et al. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 2010;5:e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valouev A, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilchrist DA, Adelman K. Coupling polymerase pausing and chromatin landscapes for precise regulation of transcription. Biochim Biophys Acta. 2012;1819:700–706. doi: 10.1016/j.bbagrm.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 76.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chopra VS, et al. The polycomb group mutant esc leads to augmented levels of paused Pol II in the Drosophila embryo. Mol Cell. 2011;42:837–844. doi: 10.1016/j.molcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weake V, Workman J. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 79.Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 80.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 81.Bai L, Charvin G, Siggia ED, Cross FR. Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev Cell. 2000;18:544–555. doi: 10.1016/j.devcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci U S A. 2008;105:7762–7767. doi: 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aida M, et al. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adelman K, et al. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci U S A. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kininis M, et al. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol. 2007;27:5090–5104. doi: 10.1128/MCB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin C, et al. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gilchrist DA, et al. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev. 2012;26:933–944. doi: 10.1101/gad.187781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blau J, et al. Three functional classes of transcriptional activation domain. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 2012;28:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 92.Ghosh A, Shuman S, Lima CD. Structural insights to how mammalian capping enzyme reads the CTD code. Mol Cell. 2011;43:299–310. doi: 10.1016/j.molcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mandal SS, et al. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci U S A. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 95.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Levine M. Paused RNA Polymerase II as a Developmental Checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gould TJ, Verkhusha VV, Hess ST. Imaging biological structures with fluorescence photoactivation localization microscopy. Nat Protoc. 2009;4:291–308. doi: 10.1038/nprot.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rasmussen EB, Lis JT. Short transcripts of the ternary complex provide insight into RNA polymerase II elongational pausing. J Mol Biol. 1995;252:522–535. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]