Abstract
Cancer cells are characterized in general by a decrease of mitochondrial respiration and oxidative phosphorylation, together with a strong enhancement of glycolysis, the so-called Warburg effect. The decrease of mitochondrial activity in cancer cells may have multiple reasons, related either to the input of reducing equivalents to the electron transfer chain or to direct alterations of the mitochondrial respiratory complexes. In some cases, the depression of respiratory activity is clearly the consequence of disruptive mitochondrial DNA (mtDNA) mutations and leads as a consequence to enhanced generation of reactive oxygen species (ROS). By acting both as mutagens and cellular mitogens, ROS may contribute directly to cancer progression. On the basis of our experimental evidence, we suggest a deep implication of the supercomplex organization of the respiratory chain as a missing link between oxidative stress, energy failure, and tumorigenesis. We speculate that under conditions of oxidative stress, a dissociation of mitochondrial supercomplexes occurs, with destabilization of complex I and secondary enhanced generation of ROS, thus leading to a vicious circle amplifying mitochondrial dysfunction. An excellent model to dissect the role of pathogenic, disassembling mtDNA mutations in tumor progression and their contribution to the metabolic reprogramming of cancer cells (glycolysis vs. respiration) is provided by an often underdiagnosed subset of tumors, namely, the oncocytomas, characterized by disruptive mutations of mtDNA, especially of complex I subunits. Such mutations almost completely abolish complex I activity, which slows down the Krebs cycle, favoring a high ratio of α-ketoglutarate/succinate and consequent destabilization of hypoxia inducible factor 1α (HIF1α). On the other hand, if complex I is partially defective, the levels of NAD+ may be sufficient to implement the Krebs cycle with higher levels of intermediates that stabilize HIF1α, thus favoring tumor malignancy. The threshold model we propose, based on the population-like dynamics of mitochondrial genetics (heteroplasmy vs. homoplasmy), implies that below threshold complex I is present and functioning correctly, thus favoring tumor growth, whereas above threshold, when complex I is not assembled, tumor growth is arrested. We have therefore termed “oncojanus” the mtDNA genes whose disruptive mutations have such a double-edged effect.
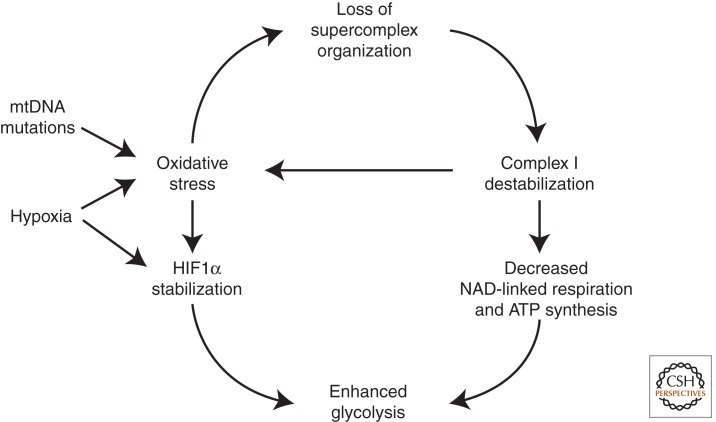
During oncogenesis, mitochondrial mutations and exogenous factors (e.g., hypoxia) induce oxidative stress. This may lead to the disorganization of mitochondrial supercomplexes, amplifying mitochondrial dysfunction.
Cancer cells are characterized by uncontrolled proliferation due to gain-of-function of oncogenes and loss of function of tumor-suppressor genes. Among the hallmarks of the cancer phenotype, there is a metabolic reprogramming (DeBerardinis et al. 2008). Cancer cells are characterized in general by a decrease of mitochondrial respiration and oxidative phosphorylation, together with a strong enhancement of glycolysis, the so-called Warburg effect, first reported by Otto Warburg (Warburg 1956). Owing to space limitations, for this particular topic we refer you to recent excellent reviews (Moreno-Sanchez et al. 2007; Mayevsky 2009; Mathupala et al. 2010; Koppenol et al. 2011). Nevertheless, it is not clear which of the two processes (i.e., decreased respiration and enhanced glycolysis) is the primary event, or even if they are concomitant consequences of a common causal event.
Although the decrease of mitochondrial oxidative phosphorylation is considered by many investigators as a universal feature of neoplastic cells, there are numerous publications in which cancer cells have been reported to show normal or even high respiratory activity (Zu and Guppy 2004; Weinberg and Chandel 2009). This apparent paradox can be explained by the hypothesis advanced by Smolkova et al (2010) that cancer progression is characterized by waves of gene expression, where initial waves suppressing mitochondrial function and stimulating glycolysis are followed by a resumption of mitochondrial oxidative phosphorylation when glutamine becomes the leading substrate for energy production and the major carbon source for anabolic processes. Each malignant tumor is characterized by a distinct metabolic phenotype reflecting the history of the different waves during the sequence of carcinogenic mutations.
The enhanced aerobic glycolysis characterizing cancer cells (and, in general, highly proliferating cells) at least in some periods of their progression toward malignancy may be vital to provide, besides energy, also the starting materials for active biosynthetic processes (Cuezva et al. 2009). Indeed, it was recognized early that incomplete activity of the Krebs cycle allows the use of citrate as a starting intermediate for the synthesis of fatty acids and cholesterol (Parlo and Coleman 1986; Kuhajda et al. 1994), whereas glycolysis may provide the majority of energy required for the anabolic processes. The supply of citrate for lipid biosynthetic processes may be provided by reductive carboxylation of α-ketoglutarate by reversal of the isocitrate dehydrogenase reaction when the major carbon source is glutamine (Holleran et al. 1995; Yoo et al. 2008) and pyruvate dehydrogenase activity is depressed by the activation of pyruvate dehydrogenase kinase (Kim et al. 2006).
During the cell cycle, the genes for glycolytic enzymes are overexpressed at the time of active DNA synthesis (Klevecz et al. 2004). Likewise, several oncogenes (such as c-myc, HIF1α, Akt) or tumor suppressors (p53) are involved in the transactivation of glycolytic enzymes (Cuezva et al. 2009). The activation of these genes may involve mitochondrial dysregulation, for example, through unbalance of Krebs cycle intermediates (see below).
DECREASED MITOCHONDRIAL RESPIRATION IN CANCER CELLS
The decrease of mitochondrial activity in cancer cells may have multiple reasons:
-
1.
Early in the 1920s, it was observed that enhanced glycolysis, as in cancer cells, inhibits respiration (the so-called Crabtree effect); it was suggested that respiratory inhibition is caused by competition of glycolysis and oxidative phosphorylation for Pi and ADP (Ibsen 1961).
-
2.
Decrease of oxidative phosphorylation (OXPHOS) may also derive from hypoxia because O2 may fall below the Km for cytochrome oxidase (Brahimi-Horn and Pouyssegur 2007) and thus induce the Pasteur effect (stimulation of phosphofructokinase by AMP when mitochondrial ATP synthesis is deficient).
-
3.
On the other hand, decreased OXPHOS becomes operative during hypoxia because of the stabilization of hypoxia inducible factor 1α (HIF1α) (Semenza 2003). This factor at normal oxygen tension undergoes hydroxylation of two key proline residues by HIF-prolyl hydroxylases (PHDs), which allow the recognition of the factor by pVHL, the product of the von Hippel–Lindau gene, that addresses the protein to ubiquitin-mediated degradation. The factor becomes stabilized during hypoxia by inactivation of PHD and binds to hypoxia-responsive elements in the DNA, stimulating a large array of genes (Semenza 2007; Kaluz et al. 2008). Besides glycolytic enzymes, HIF1α enhances the expression of pyruvate dehydrogenase kinase (PDK), thus inhibiting pyruvate dehydrogenase (PDH) complex and decreasing the input of reducing equivalents to the respiratory chain (Kim et al. 2006). The decreased input of carbon into the Krebs cycle must necessarily be limited, otherwise the lack of citrate would impair the synthesis of lipids and cholesterol that are required for the formation of new cellular membranes during tumor growth. This is the reason that glutamine becomes a preferred substrate for replenishing the Krebs cycle for both energetic and biosynthetic purposes.
It is of interest that HIF1α is also stabilized by several nonhypoxic stimuli, for example, by succinate (that inhibits prolyl hydroxylase) (King et al. 2006), but also by pyruvate and oxaloacetate (Lu et al. 2005; McFate et al. 2008) and, interestingly, by reactive oxygen species (ROS) (Patten et al. 2010), although the activating effect of ROS on HIF is still a controversial issue (Bell et al. 2008). ROS could act directly but also by stimulating PDK, thus leading to accumulation of pyruvate (Sun et al. 2009; Patten et al. 2010). Thus, derangement of the Krebs cycle may lead to changes in steady-state concentrations of important metabolites that are able to control the activity of HIF1α.
-
4.
Direct alterations of respiratory activity in cancer cells have not been frequently investigated. In general, a depression of respiratory activity is observed in cancer cells (Putignani et al. 2008; Park et al. 2009; Jahnke et al. 2010; Ma et al. 2010). In the case of Ras-transformed fibroblasts, we have observed a direct alteration of the respiratory chain, in particular, a 50% decrease of complex I content and activity, and consequently of NAD-linked respiration and ATP synthesis (Baracca et al. 2010). This seems to be due in part to decreased expression as seen by transcriptome analysis. However, we do not know yet if there are disruptive mitochondrial DNA (mtDNA) mutations as seen in oncocytic tumors (see below). The effect of the adenylate cyclase activator forskolin in reversing the changes accompanying transformation suggests that the mitochondrial changes may be caused by impairment of cAMP-dependent pathways (L Alberghina, pers. comm.).
-
5.
In some cases, the depression of respiratory activity is clearly the consequence of disruptive mtDNA mutations (see below).
Tumorigenic mtDNA mutations affect the respiratory chain complexes and enhance the production of ROS (Brandon et al. 2006); however, the pathological relevance of mtDNA mutations in cancer cells is controversial (Frezza and Gottlieb 2009). Nonetheless, a clear-cut correlation between occurrence of pathogenic mtDNA mutations and mitochondrial energetic impairment is a well-demonstrated feature of oncocytic tumors (Bonora et al. 2006a; Porcelli et al. 2010), which is dealt with in detail below.
Accumulation of NADH due to either hypoxia or alterations of the electron transfer chain inhibits isocitrate dehydrogenase and the Krebs cycle when implemented by acetyl CoA may help the exit of citrate to support lipid synthesis (fatty acids and cholesterol). On the other hand, the entry of reducing equivalents via glutamine may not be inhibited, thus sustaining a truncated Krebs cycle via glutamate, α-ketoglutarate, and the intermediates of the Krebs cycle, thus reaching malate that is converted to pyruvate by the malic enzyme after release in the cytosol. Pyruvate would contribute to stabilization of HIF1α and may also be converted to lactate by lactate dehydrogenase. The extent of this pathway of glutamine oxidation is determined by the residual activity of the respiratory chain, because both glutamate dehydrogenase and α-ketoglutarate dehydrogenase require oxidized NAD+, and both these enzymes together with succinate dehydrogenase require an intact respiratory chain for their turnover.
THE ROLE OF ROS
An overwhelming body of evidence accumulated in the last decades has shown that mitochondria have a central role in the etiology and pathogenesis of most major chronic diseases and in aging (Wallace 2005; Lenaz et al. 2006; Lenaz and Genova 2010). The involvement of mitochondria in disease, which has generated the term “mitochondrial medicine” (DiMauro et al. 2006), has been largely ascribed to their central role in production of ROS and to the damaging effect of the latter on these organelles. In particular, mtDNA mutations would induce alterations of the encoded polypeptides in the respiratory complexes, with consequent decrease of electron transfer activity, leading to further production of ROS, and thus establishing a vicious circle of oxidative stress and energetic decline (Ozawa 1997). It must be underlined that mtDNA mutations leading to increased ROS production are those allowing at least a partial assembly of the respiratory complexes. On the contrary, for instance, oncocytic tumors display homoplasmic disassembling complex I mutations, which may not be ROS-generating mutations, if one considers that the main ROS production site might be lacking as a whole (Koopman et al. 2007; Moran et al. 2010; Porcelli et al. 2010; Tuppen et al. 2010). Moreover, the mutant load ought to be considered when analyzing functional effects of mtDNA mutations. The threshold model we previously proposed implies that, below threshold, complex I is present and functioning correctly, in contrast to the case of a missense mutation leading to the coexistence of nonmutant along with mutant ROS producing complex I (Park et al. 2009).
Decreased mitochondrial activity is considered to be tumorigenic, mainly because of the enhanced ROS production. H2O2 exported to the nucleus enhances the transcription of selected genes that favor tumor progression (Wallace 2005). ROS act both as mutagens and cellular mitogens (Klaunig and Kamendulis 2004). It has been suggested that oncogenesis and neurodegeneration may share common pathogenic features as differential responses to different insults during different phases of the cell cycle, leading either to cell death or to cell proliferation (Morris et al. 2010).
Any lowering of the electron transfer chain activity, either caused by mtDNA mutations and hence structural lesions of the respiratory complexes or depending directly or indirectly on hypoxia, would enhance generation of ROS; it is known that ROS generation is enhanced in State 4 (controlled state in absence of ADP), when complex I is inhibited, and paradoxically under hypoxic conditions.
ROS may contribute to further mitochondrial dysfunction by several mechanisms: (1) direct alterations of respiratory complexes, for example, complex I, particularly on FeS clusters; (2) peroxidation of mitochondrial phospholipids, in particular, cardiolipin (Paradies et al. 2010), that are required for proper assembly and activity of mitochondrial complexes (I, III, IV); (3) (further) mtDNA mutations affecting respiratory complexes; and (4) stabilization of HIF1α (Patten et al. 2010).
In this scenario, it is easy to foresee a deep implication of supercomplex organization as the missing link between oxidative stress and energy failure (Lenaz and Genova 2007). It is tempting to speculate that under conditions of oxidative stress a dissociation of complex I–III aggregates occurs, with loss of facilitated electron channeling and resumption of a less efficient random diffusional behavior with electron transfer depending on the collisional encounters of the free ubiquinone molecules with the partner complexes.
As predicted by Lenaz and Genova (2007), dissociation of supercomplexes might have further deleterious consequences, such as disassembly of complex I and III subunits and loss of electron transfer and/or proton translocation; the consequent alteration of electron transfer may elicit further induction of ROS generation. Following this line of thought, the different susceptibility of different types of cells and tissues to ROS damage may be a consequence of the extent and tightness of supercomplex organization in their respiratory chains, which depend on phospholipid content and composition of their mitochondrial membranes. These changes may have deep metabolic consequences, as depicted in the scheme in Figure 1. An initial enhanced ROS generation due to different possible reasons and originating in different districts of the cell besides mitochondria (Lenaz and Strocchi 2009) would induce supercomplex disorganization eventually leading to possible decrease of complex I assembly; both the lack of efficient electron channeling and the loss of complex I would decrease NAD-linked respiration and ATP synthesis. Here we briefly summarize the experimental evidence pertaining to this hypothesis.
Figure 1.
The possible effects of loss of supercomplex organization on mitochondrial function during oncogenesis. Membrane phospholipid peroxidation and consequent loss of supercomplex organization may occur because of oxidative stress induced by genetic changes (i.e., mitochondrial DNA mutations) or by exogenous factors (e.g., hypoxia); the ensuing destabilization of complex I results in OXPHOS deficiency and further oxidative stress. As a consequence of these changes, cells are forced to rely on glycolysis for energy production. See text for further details.
Peroxidized Phospholipids Prevent Supercomplex Formation
Two roles of phospholipid have been distinguished: (1) a dispersive solubilization effect that can be duplicated by appropriate detergents, and (2) a catalytic effect that can be specifically fulfilled only by cardiolipin (Vik and Capaldi 1977; Fry and Green 1980, 1981; Robinson et al. 1980; Lee 2004). The phospholipids in closest vicinity to the protein surface, as well as those in the free bilayer, are actually highly mobile and free to exchange, but cardiolipin was indicated as tightly bound, being more likely buried within the protein complexes (Kang et al. 1979; Sedlak and Robinson 1999; Lange et al. 2001). Recent results seem also to indicate that cardiolipin stabilizes respiratory chain supercomplexes as well as the individual complexes. The availability of a cardiolipin-lacking yeast mutant provided the opportunity to show that mitochondrial membranes still contained the III2–IV2 supercomplex, but that it was significantly less stable than supercomplexes in the parental strain (Zhang et al. 2002; Pfeiffer et al. 2003).
Mutations of tafazzin, an acyl transferase involved in the synthesis of mature tetralinoleyl cardiolipin (Neuwald 1997), result in Barth syndrome, a cardioskeletal myopathy with neutropenia, characterized by respiratory chain dysfunction. The cardiolipin defect in Barth syndrome results in destabilization of the supercomplexes by weakening the interactions between respiratory complexes (McKenzie et al. 2006).
It is well documented that exposure of mitochondria to ROS can affect the respiratory activity via oxidative damage of cardiolipin, which is required for the optimal functioning of the enzyme complexes (Paradies et al. 2000, 2002, 2010; Petrosillo et al. 2003, 2009). We have shown that the maintenance of a I–III supercomplex after reconstitution into phospholipid vesicles is abolished if lipid peroxidation is induced before reconstitution (Bianchi et al. 2003; Genova et al. 2008).
Evidently, the distortion of the lipid bilayer induced by peroxidation and the alteration of the tightly bound phospholipids determine dissociation of the supercomplex originally present in the lipid-poor preparation.
Supercomplex Disassembly Abolishes CoQ Channeling and Resumes Less Efficient Pool Behavior
Studies of respiration in pathological conditions (Rosca et al. 2008; van Raam et al. 2008) showed that electron transfer in absence of supercomplex organization is lost even if activity of the individual complexes is normal. In reconstitution studies of complexes I and III in phospholipid vesicles (Genova et al. 2008), electron transfer between complex I and complex III (NADH cytochrome c reductase) shows rates higher than those predicted by random behavior in proteoliposomes enriched in complexes I and III at a protein-to-phospholipid ratio of 1:1 (when kinetic testing according to flux control analysis indicates the presence of a supercomplex I–III). However, at a ratio of 1:30 (when the supercomplex is dissociated), the rate of NADH cytochrome c reductase is exactly that predicted by the pool equation on the basis of the individual activities of complex I and complex III. This is a demonstration that supercomplex formation, indeed, enhances the rate of electron transfer above that occurring via a ubiquinone pool in the membrane (Bianchi et al. 2003; Genova et al. 2008), and loss of supercomplex organization resumes less efficient pool activity of the quinone.
Loss of Supercomplexes Prevents Stability and Assembly of Individual Complexes
Analysis of the state of supercomplexes in patients with an isolated deficiency of single complexes (Schagger et al. 2004) and in cultured cell models harboring cytochrome b mutations (Acin-Perez et al. 2004; Blakely et al. 2005; D’Aurelio et al. 2006; Diaz et al. 2006; McKenzie et al. 2006) provided evidence that the formation of respirasomes is essential for the assembly/stability of complex I. Genetic alterations leading to a loss of complex III prevented respirasome formation and led to secondary loss of complex I, and therefore primary complex III assembly deficiencies presented as complex III/I defects. Conversely, complex III stability was not influenced by the absence of complex I.
From these findings, supercomplex assembly emerged as a necessary step for respiration, its defect setting the threshold for respiratory impairment in mtDNA mutant cells.
Disassembly of Supercomplex Organization Enhances Generation of ROS
Indirect circumstantial evidence suggests that supercomplex assembly may limit the extent of superoxide generation by the respiratory chain (Lenaz and Genova 2010). A direct study on a mitochondrial fraction containing complexes I and III and reconstituted with different amounts of phospholipids shows that at protein:phospholipids 1:30, when the supercomplex is not formed, the production of superoxide is 20-fold higher than at a 1:1 ratio, when most units are assembled in a supercomplex (ML Genova, G Barbero, E Maranzana, et al., unpubl.).
Although the molecular structure of the individual complexes does not allow us to envision a close apposition of the matrix arm of complex I, where FMN is localized, with either complex III or IV, the actual shape of the I1III2IV1 supercomplexes from bovine heart (Schafer et al. 2007) suggests a slightly different conformation of complex I in the supercomplexes with a smaller angle of the matrix arm with the membrane arm showing a higher bending toward the membrane (and presumably complex III), in line with the notion that complex I may undergo important conformational changes (Radermacher et al. 2006).
Do Cancer Cells Lose Supercomplex Association?
The validation of the above working hypothesis requires direct studies on cancer tissues and neoplastic cells. A general decrease of respiratory chain activity is a common feature of several cancers. Nevertheless, very few studies have addressed the respiratory chain supramolecular organization.
Baracca et al. (2010) reported that down-regulation of the respiratory chain in Ras-transformed fibroblasts is operated through strong decrease of complex I activity and content, probably because of lack of correct assembly of the subunits as a consequence of altered supercomplex organization, as shown by loss of the highest-molecular-weight I1III2IV1-2 supercomplex. Significantly, ROS generation was strongly enhanced in the Ras cells. In the XTC-UC1 oncocytic cell line (see below), we have also found (Genova et al. 2008) by blue-native electrophoresis of mitochondrial proteins, a complete absence of high-molecular-weight aggregates containing either complex I or complex IV, that are instead present in a control cell line from a nononcocytic thyroid tumor. If the absence of supercomplexes comprising complex I is a consequence of its disassembly due to lack of ND1 (see below), the absence of aggregated complex IV is in line with the working hypothesis.
The loss of supercomplex assembly seems to accompany a decrease/loss of complex I activity; if this decrease is found to be common to most tumors, the consequences would depend on the extent of such a decrease. Disruptive mtDNA mutations in the ND subunits of complex I (as in oncocytomas; see below) almost completely abolish complex I activity, which presumably inactivates the Krebs cycle, that requires NAD+, favoring a high-ratio α-ketoglutarate/succinate and consequent destabilization of HIF1α. On the other hand, if complex I is partially defective (as in RAS-mutated fibroblasts), the levels of NAD+ may be sufficient to implement the Krebs cycle with higher levels of intermediates that stabilize HIF1α, thus favoring tumor malignancy.
mtDNA MUTATIONS IN CANCER: THE PECULIAR CASE OF ONCOCYTIC TUMORS
An excellent model to dissect out the role of pathogenic, disassembling mtDNA mutations in tumor progression and their contribution to the metabolic reprogramming of cancer cells (glycolysis vs. respiration) is provided by an often underdiagnosed subset of tumors, namely, the oncocytomas. These neoplasms contain a high percentage or they are completely made of cells characterized by an aberrant mitochondrial hyperplasia. They originate from epithelial tissues and occur mainly in endocrine and exocrine organs, although the oncocytic phenotype has been described in many anatomical districts (for review, see Gasparre et al. 2011a). The majority of oncocytic tumors are considered as benign, displaying low invasiveness and showing a usually favorable prognosis. In the thyroid, nonetheless, the best indicator for prognosis remains the degree of differentiation of neoplastic cells, which defines adenoma or carcinoma, regardless of the occurrence of the oncocytic phenotype.
The most striking feature of oncocytic tumors is appreciated through histochemical and ultrastructural analyses, which highlight cells packed with enlarged globular or ovate mitochondria with a stack of lamelliform, tubular, or flat cristae and occupying up to 60% of the cytoplasm (Tallini 1998; Ambu et al. 2000; Riva and Tandler 2000). Such heterogeneity in mitochondrial morphology has long been an indicator of a functional along with a structural alteration of the organelles in oncocytic cells.
The cause of the mitochondrial hyperplasia in oncocytic tumors has been suspected to consist in an aberrant mitochondrial biogenesis triggered by a mitochondrial dysfunction, which may attempt to compensate for an impaired energy production. Evidence that a compensatory mechanism is responsible for oncocytic phenotype occurrence has accumulated in recent years. It is now clear that the mitochondrial respiration deficiency originally hypothesized (Muller-Hocker et al. 1998a) is, indeed, an event occurring in oncocytic tumors and mainly concerns respiratory complex I (Zielke et al. 1998; Simonnet et al. 2003). NADH-dehydrogenase activity is, in fact, reduced concomitantly with an increase in other respiratory complex abundance and, overall, in mitochondrial mass as shown by measurements of citrate synthase activity and of mtDNA copy number (Zielke et al. 1998; Simonnet et al. 2003; Gasparre et al. 2009; Porcelli et al. 2010).
In vitro studies with transmitochondrial cell hybrids (cybrids) have shown that the defective mitochondrial respiration of thyroid oncocytic cells is transferred to normally respiring osteosarcoma cells when mitochondria, not nuclei, are imported (Bonora et al. 2006a). From this observation, as well as from studies in which the mitochondrial genome was screened for oncocytic cancer-specific genetic lesions such as small and large deletions, depletion, or point mutations (Tallini et al. 1994; Maximo et al. 1998, 2002; Maximo and Sobrinho-Simoes 2000; Costa-Guda et al. 2007; Gasparre et al. 2007, 2008, 2009; Mayr et al. 2008; Zimmermann et al. 2009; Porcelli et al. 2010; Bartoletti-Stella et al. 2011), it is now generally accepted that the genetic hallmark of the oncocytic phenotype is the occurrence of disruptive mtDNA mutations, particularly in complex I subunits. This holds true for virtually all types of oncocytic tumors analyzed to date (Gasparre et al. 2007, 2008, 2009; Porcelli et al. 2010; Bartoletti-Stella et al. 2011), and, inversely, association between the occurrence of disruptive mutations and that of oncocytic foci has been shown, for instance, in endometrial carcinoma (Guerra et al. 2011). Within the plethora of disruptive mutations, both frameshift and pathogenic missense changes are included, which ultimately lead to complex I impairment in terms of function and/or assembly. Hence, the direct consequence of these mutations impinges on respiration, whose impairment may subsequently trigger a retrograde signaling from the organelles to the nucleus, leading in a vicious circle to mitochondrial hyperplasia. An analysis of the entire mitochondrial genomes sequenced in oncocytic tumor samples has allowed identification of MTND1 as a mutational hotspot (Gasparre et al. 2007). Less importantly than MTND1, at least two of the remaining six mitochondria-coded complex I genes display a high rate of pathogenic mutations accumulation, namely, MTND4 and MTND5, which are also among the longest mitochondrial genes and richest in homopolymeric stretches, where many frameshift mutations occur. Complex IV (cytochrome c oxidase; COX) mitochondrial subunits appear to be the least involved in contributing to the mitochondrial damage leading to oncocytic transformation, although some cases have been reported of oncocytomas harboring mutations in COX genes (Maximo et al. 2002; Costa-Guda et al. 2007) or being COX-negative upon immunohistochemical analysis (Porcelli et al. 2010). These findings suggest that complex I disruption may affect, in certain cases, the organization of supercomplexes.
Although for most types of oncocytic tumors such as kidney and pituitary oncocytoma (Gasparre et al. 2008; Porcelli et al. 2010), up to 70%–100% of cases have been shown to harbor pathogenic mtDNA mutations, in heterogeneous neoplasms such as those of the thyroid, a lower percentage display the genetic features described. Although studies on microdissected tumors are currently lacking, several oncocytic cancers do not necessarily harbor mtDNA mutations, which suggests that nuclear genetic lesions may create a phenocopy. It is plausible to think that mutations in nuclear-encoded respiratory complexes subunits (38 belonging to NADH-dehydrogenase), or in chaperones, may well trigger the same bioenergetic damage and, consequently, induce the same phenotype as mtDNA mutations. Nuclear GRIM-19 (alias NDUFA13) has been one of the candidate genes, in which somatic mutations in sporadic forms of thyroid oncocytic cancer were reported. However, no GRIM-19 mutations were found to segregate in familial thyroid oncocytoma (TCO) (Maximo et al. 2005). The mitochondrial protein import system subunit TIMM44 has been reported to harbor functional mutations in familial TCO (Bonora et al. 2006b). However, preliminary investigation of about 40 oncocytic thyroid tumors has not shown a high incidence of potentially pathogenic variants in nuclear-encoded complex I subunits (E Bonora, G Gasparre, G Romeo et al., unpubl.). Further investigation of possible nuclear-coded complex I mutations in microdissected homogeneous oncocytic tissues are still warranted before the involvement of nuclear subunits can be ruled out.
ENERGETIC COMPETENCE IN ONCOCYTIC TUMORS
Mitochondrial Hyperplasia and the Compensatory Effect Hypothesis
Retrograde signaling in oncocytic tumors from the mitochondrion to the nucleus has been hypothesized to affect several pathways (Butow and Avadhani 2004). A metabolic stress in tumor cells may induce a nuclear response leading to the activation of mitochondrial biogenesis pathways to restore a defective respiration.
Increased mitochondrial biogenesis has been evaluated in oncocytic tumors as well as in the only existing model of oncocytic cancer, the XTC.UC1 cell line. In both models, a defective mitochondrial ATP synthesis and an overexpression of uncoupling proteins and of the biogenesis regulator PGC-1-related coactivator (PRC) have been reported (Savagner et al. 2001a,b, 2003; Higgins et al. 2003; Baris et al. 2004; Finley et al. 2004; Jacques et al. 2005). From these data, it was initially concluded that the defective ATP synthesis may explain the mitochondrial hyperplasia and hence justify the compensatory effect, caused by an oxidative phosphorylation coupling defect of unknown cause (Savagner et al. 2001b). Moreover, the up-regulation of mitochondrial proteins has been described both in thyroid and kidney oncocytomas (Higgins et al. 2003; Baris et al. 2004; Finley et al. 2004). In kidney oncocytomas, the increase in mitochondrial proteins and in the activity of respiratory complexes II, III, IV, and V as well as of citrate synthase seems to be a common feature, in correlation with their benign behavior (Simonnet et al. 2003). In apparent contrast, in oncocytes of both normal and hyperfunctional parathyroids, a deficiency of cytochrome c oxidase has been observed (Muller-Hocker 1992; Muller-Hocker et al. 1998b). In oncocytic cells of the same tissues, a higher mtDNA content was reported without difference in expression of mitochondrial transcription factor TFAM or mtDNA γ polymerase (POLG), suggesting a compensatory effect due to a complex IV deficiency (Muller-Hocker et al. 1998a). Although the compensatory effect has been described as a predominant feature in oncocytic tumors, the trigger for such a mechanism has not been identified. Godinot’s group reported that the presence of complex I deficiency might increase mitochondrial biogenesis to compensate a respiratory dysfunction (Simonnet et al. 2003), and our group described a mitochondrial energetic impairment in the XTC.UC1 cell line originally characterized by Zielke et al. (1998). In this cell line we reported a decrease in both complex I and III activity and a strong reduction of ATP synthesis, driven by complex I substrates, and mitochondrial membrane potential (Bonora et al. 2006a; Porcelli et al. 2009). We also showed that the energetic dysfunction can be transferred along with the mtDNA in a transmitochondrial cybrid model and hence concluded that the complex I defect must be due to an mtDNA mutation. These functional results were followed by an increased level of most respiratory chain complexes subunits, whereas complex I NDUFA9 and ND6 were strongly reduced. Moreover, we observed that complex I mitochondria-coded ND1 subunit was absent (Bonora et al. 2006a).
The feature of a mitochondrial energetic impairment and decrease in complex I subunits was also reported in a rare case of nasopharynx oncocytoma (Gasparre et al. 2009) and in a peculiar case of Warthin tumor (Porcelli et al. 2010). Altogether these data point to a dysfunction of at least respiratory complex I as the main trigger for oncocytic transformation with a compensatory increase in other mitochondrial proteins leading to the high mitochondrial proliferation in oncocytic cells.
Structural and Functional Consequences of mtDNA Mutations
The common occurrence of mtDNA mutations in complex I genes should result in incomplete or partial assembly and/or function of this enzyme depending on the mtDNA mutation type. Our group showed that the presence of homoplasmic truncating ND mutations mainly cause a loss of complex I subunits in oncocytic neoplasia (Gasparre et al. 2009). Moreover, we took advantage of XTC.UC1 cells bearing the m.3571insC ND1 mutation to clarify the impact of the homoplasmic truncating mtDNA mutations on the respiratory complex content/assembly and energetic function. In these cells, the ND1 subunit was ablated, and the levels of other complex I subunits (i.e., NDUFA9 and ND6) were significantly reduced (Bonora et al. 2006a). Similar results were extended to a survey of a large panel of pituitary, head-and-neck, and thyroid oncocytic tumors for the presence of homoplasmic mtDNA truncating mutations (Porcelli et al. 2010).
The presence of an incomplete or partially assembled complex I in oncocytic tumors raises the question of whether this may contribute to ROS generation. Lack of data on the role of ROS in oncocytic tumors is mainly because ROS measurements in tumor biopsies are not feasible, and cell models for in vitro studies are very scarce. In XTC.UC1 cells, different degrees of heteroplasmy of the 3571insC ND1 mutation did not influence ROS amounts, probably because of a differential expression of ROS detoxifying enzymes (i.e., manganese superoxide dismutase and catalase) in the presence/absence of complex (Porcelli et al. 2010). Kofler’s group speculated that a lack of complex I in oncocytic thyroid may increase ROS production inhibiting pro-apoptotic pathways (Zimmermann et al. 2009). Further studies are warranted to understand whether ROS may influence the proliferative potential and the accumulation of mutations in oncocytic tumors, where most of the mutations reported are indeed homoplasmic and complex I appears to be disassembled.
Because elevated ROS have been proposed to induce apoptosis, additional studies are required to determine the role of apoptosis in regulating the survival and proliferation of oncocytic cells. It has been reported that mtDNA mutations impairing mitochondrial energetic function may protect cells from apoptosis, providing a growth advantage (Shidara et al. 2005; Park et al. 2009), but whether mtDNA mutations prevent apoptosis in oncocytic tumors is still poorly defined.
Alteration of the oxidative metabolism resulting from mtDNA mutations may trigger the Warburg effect permitting a shift toward glycolysis, hence favoring tumor progression (Chen et al. 2007; Smolkova et al. 2010). Several observations have linked respiratory complex dysfunctions to alteration of upstream mitochondrial metabolism, that is, Krebs cycle, in cancer. Mutations in enzymes such as succinate dehydrogenase (SDH) and fumarate hydratase (FH) are actively involved in tumorigenesis through imbalance of the Krebs cycle and stabilization of HIF1α (Pollard et al. 2005; King et al. 2006). Mutations in the B, C, and D subunits of SDH have been shown to lead to the accumulation of the Krebs metabolite succinate at the expense of α-ketoglutarate. Mutations in FH lead to increase of fumarate, slowing down the Krebs cycle (Pollard et al. 2005; Pasini and Stratakis 2009). The accumulation of succinate and fumarate are thought to inhibit PHD, key activators of HIF1α degradation (Boulahbel et al. 2009).
The complex I dysfunction described in oncocytic tumors raises the question of whether a parallel may exist between these neoplasms and SDH/FH mutated cancers, and what is the role played by complex I in regulating cancer metabolism. We recently reported that in the XTC.UC1 cells bearing the homoplasmic truncating ND1 mutation, the ratio between succinate and α-ketoglutarate is opposite to that reported in SDH and FH mutated tumors, with increase of α-ketoglutarate at the expense of succinate (Porcelli et al. 2010). It is likely that the absence of a functional complex I in these cells leads to accumulation of NADH, which may, in turn, inhibit α-ketoglutarate dehydrogenase and prevent succinate production with a consequent α-ketoglutarate increase. Because α-ketoglutarate is the main reagent feeding PHD, HIF1α should undergo a chronic destabilization, even in the hypoxic tumor environment. We have, indeed, shown this to be the case in oncocytic tumors harboring homoplasmic disruptive mtDNA mutations causing disassembly of complex I (Porcelli et al. 2010).
CONCLUDING REMARKS: GENERAL IMPLICATIONS OF THE ONCOCYTIC TUMOR MODEL FOR CANCER RESEARCH
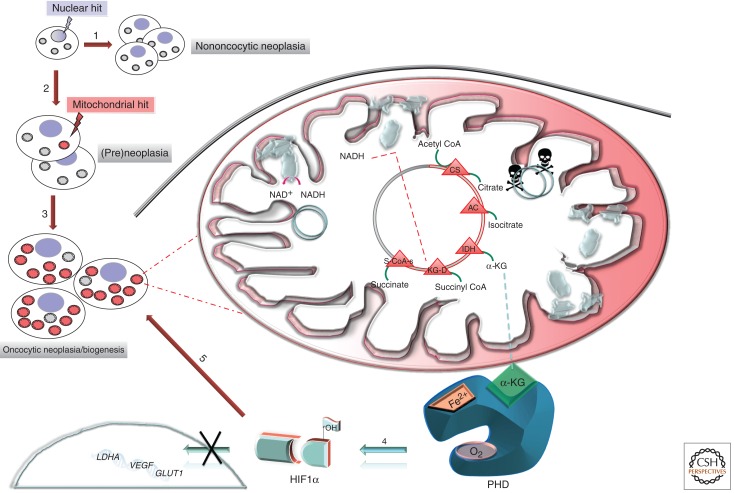
During the last few years, the oncocytic tumor model has been used to ask some basic questions regarding the functional effects of mtDNA mutations. First of all, it has been possible to establish a univocal correlation between mtDNA mutations and the corresponding biochemical phenotype thanks to the only available oncocytic cell line, the XTC.UC1, and the cybrids derived from it (Bonora et al. 2006a). Using these tools, it was realized that the mtDNA physiological polyploidy has a strong impact on the disease phenotype according to the threshold effect model. This feature of mitochondrial genetics has not always been taken into consideration when evaluating some basic questions regarding the pro-tumorigenic or metastatic potential of mtDNA mutations (Petros et al. 2005; Ishikawa et al. 2008). A further question that is closely correlated with this typical feature is the possible selective disadvantage or advantage that mtDNA mutations have in vitro as well as in vivo (Gasparre et al. 2007) as a result of the “population dynamics” that might characterize the evolution of mtDNA mutations during tumorigenesis. The possibility of a selective advantage is strongly supported by recent findings (Gasparre et al. 2009), which describe disruptive frameshift mtDNA mutations affecting the ND5 and ND1 subunits of complex I present in homoplasmy in a nasopharyngeal oncocytic tumor. In both cases, the mutations were inherited at a low degree of heteroplasmy through the germline in the patient’s family (Gasparre et al. 2009) or were germline de novo events (Pradella et al. 2011) but became homoplasmic in the patient’s oncocytic tumor. Immunohistochemistry has shown a perfect correlation between presence of a homoplasmic mutation and lack of the mitochondria-coded ND6 subunit, which suggests impaired complex I assembly. In the described nasopharynx oncocytoma, a few oncocytic areas from the same tumor, expressing ND6 and heteroplasmic for the ND5 mutation, harbored instead a de novo homoplasmic ND1 mutation. Because the shift to homoplasmy of both ND1 (somatic) and ND5 (germinal) mutations occurred exclusively in the tumor cells of the patient, we can conclude that these complex I mutations had a selective advantage that accompanied the oncocytic change. This last conclusion warrants, in turn, a revisitation of the hypotheses regarding the origin and fate of oncocytic cells. As shown in Figure 2, a general mechanism can be hypothesized in which a mitochondrial hit (either in the mtDNA or in the nuclear-coded subunits of complex I) may occur at any stage during the multistep process of tumorigenesis. This particular mitochondrial hit, which may also be inherited, will determine whether the tumor becomes oncocytic or nononcocytic. The necessity of a double hit, one mitochondrial and the main hit likely being a nuclear one conferring the transformed phenotype to a normal cell, seems to date the most plausible hypothesis to explain oncocytic transformation, as also discussed in recent reviews (Maximo et al. 2009). The implications of this model on the selective advantages or disadvantages of the oncocytic cells during tumorigenesis demand also a critical reevaluation of the functional significance of the impact of mtDNA-coded genes in cancer, in analogy to nuclear-encoded mitochondrial genes such as SDH and FH. The role of metabolic genes in tumorigenesis is still very controversial (Thompson 2009). SDH and FH are nowadays considered tumor-suppressor genes, as it has been proposed for complex I by Kofler’s group (Zimmermann et al. 2011), although exclusively on the basis, primarily, of the observation of mutations detected in oncocytic tumors. Nonetheless, in our opinion, correlation studies are not sufficient to assign complex I genes the status of tumor-suppressor genes, especially because such definition overlooks the fundamental differences between mitochondrial genetics (whose dynamics are analogous to those of population genetics) and Mendelian genetics. The typical homoplasmic complex I mutations found in oncocytic tumors and shown to generate the phenotype in vivo hamper the tumorigenic potential of cells that harbor them (Park et al. 2009; Gasparre et al. 2011b), accounting for the low-proliferative and low-aggressive behavior of most oncocytic tumors. This effect has been shown by our group to be independent from apoptosis and likely due to the lack of HIF1α stabilization described above and pictured in Figure 2. Such pseudonormoxic status may also explain why only a complete lack of complex I may determine the oncocytic phenotype. HIF1α is a pivotal intracellular oxygen sensor, whose role, when stabilized during hypoxia/anoxia, is envisioned to block mitochondrial biogenesis when O2 is limiting. The release of a HIF1α block may reinforce the induction of the compensatory mitochondrial biogenesis in complex I-lacking cells, finally leading to mitochondrial hyperplasia.
Figure 2.
The origin and fate of the oncocytic cell. Nuclear hits (light blue lightning arrow) presiding to cellular transformation contribute to the initiation stage of neoplastic or pre-neoplastic development. 1. No mtDNA mutations occur, or such mutations are detrimental to cell replication and mutated mitochondria are selected against, preventing the development of an oncocytic phenotype. 2. During cell replication a mutation (red lightning arrow) may occur in mtDNA (mitochondrial hit) either conferring a metabolic advantage by contributing to cell adaptation through the Warburg effect or remaining neutral until a mutation load threshold is reached. 3. Positive selection or random drift may set in through mitochondrial biogenesis, generating the oncocytic phenotype. The pink dotted lines lead to an enlarged dynamic view of an “oncocytic” mitochondrion schematically divided into two parts: a white area showing wild-type mtDNA molecules (gray circles) and correctly assembled complex I, and a shaded pink area with mutated mtDNA and partially or fully disassembled complex I. An mtDNA mutation disassembling complex I, such as those typical of oncocytic tumors, may not allow NADH consumption, shifting the NADH/NAD+ ratio toward NADH and leading to an increase of the α-KG/SA ratio. Such Krebs cycle alteration may foster activity of PHD with subsequent HIF1α destabilization/degradation even in low oxygen tension conditions (pseudonormoxia). 4. Inactivation of this transcription factor may functionally lead, on one hand, to the inhibition of expression of pro-tumorigenic and glycolytic genes (i.e., LDHA, GLUT1, and VEGF) and, on the other hand, to the down-regulation of glycolysis needed to compensate for the defective mitochondrial respiration (light blue arrow). This scenario does not allow the metabolic adaptation of tumor cells (Warburg effect), hampering the tumorigenic potential of cells, explaining the low-proliferative and low-aggressive features of oncocytic tumors. 5. Furthermore, HIF1α destabilization may not prevent the inhibition of mitochondrial biogenesis that should occur upon hypoxia sensing, hence feeding a vicious loop ultimately leading to the pathological mitochondrial hyperplasia of oncocytic cells (red arrow). CS, citrate synthase; AC, aconitase; IDH: isocitrate dehydrogenase; α-KG, α-ketoglutarate; KG-D, α-ketoglutarate dehydrogenase; S-CoA-s, succinyl-CoA synthase; PHD, prolyl hydroxylase; HIF1α, hypoxia-inducible factor 1α; GLUT1, glucose trasporter 1; VEGF, vascular endothelial growth factor; LDHA, lactate dehydrogenase A.
Last, we have defined the threshold mutation load needed to induce an anti-tumorigenic phenotype, which suggests that these genes are more likely to be lethality associated than canonical tumor suppressors. Mutated mtDNA genes that cause complex I disassembly may hence be anti-tumorigenic, in close dependence on their mutant load, which needs to be carefully measured (Kurelac et al. 2011). Such feature does not generally apply to nuclear genes. We have therefore proposed the denomination of “oncojanus,” reminiscent of the double-faceted Roman god Janus, to indicate the threshold-dependent double-edged properties of CI mtDNA-encoded genes (Gasparre et al. 2011b). The understanding of the different properties of mutations affecting complex I genes and the dissection of the mechanisms leading to pseudonormoxia following complex I disassembly should provide fundamental insights linking cancer metabolism to hypoxia adaptation and will likely lead to the discovery of novel approaches in cancer therapy.
ACKNOWLEDGMENTS
This work is supported by the Italian Ministry of University and Research (MIUR) grant FIRB “Futuro in Ricerca” and by Fondazione Umberto Veronesi grant “Disco trip” to G.G., by MIUR grant PRIN 2008 to A.M.P., and by the Associazione Italiana Ricerca sul Cancro (AIRC) to G.R.
Footnotes
Editors: Douglas C. Wallace and Richard J. Youle
Additional Perspectives on Mitochondria available at www.cshperspectives.org
REFERENCES
- Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C, Moraes CT, Enriquez JA 2004. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell 13: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambu R, Riva A, Lai ML, Loffredo F, Riva FT, Tandler B 2000. Scanning electron microscopy of the interior of cells in Hurthle cell tumors. Ultrastruct Pathol 24: 211–219 [DOI] [PubMed] [Google Scholar]
- Baracca A, Chiaradonna F, Sgarbi G, Solaini G, Alberghina L, Lenaz G 2010. Mitochondrial complex I decrease is responsible for bioenergetic dysfunction in K-ras transformed cells. Biochim Biophys Acta 1797: 314–323 [DOI] [PubMed] [Google Scholar]
- Baris O, Savagner F, Nasser V, Loriod B, Granjeaud S, Guyetant S, Franc B, Rodien P, Rohmer V, Bertucci F, et al. 2004. Transcriptional profiling reveals coordinated up-regulation of oxidative metabolism genes in thyroid oncocytic tumors. J Clin Endocrinol Metab 89: 994–1005 [DOI] [PubMed] [Google Scholar]
- Bartoletti-Stella A, Salfi NC, Ceccarelli C, Attimonelli M, Romeo G, Gasparre G 2011. Mitochondrial DNA mutations in oncocytic adnexal lacrimal glands of the conjunctiva. Arch Ophthalmol 129: 664–666 [DOI] [PubMed] [Google Scholar]
- Bell EL, Klimova T, Chandel NS 2008. Targeting the mitochondria for cancer therapy: Regulation of hypoxia-inducible factor by mitochondria. Antioxid Redox Signal 10: 635–640 [DOI] [PubMed] [Google Scholar]
- Bianchi C, Fato R, Genova ML, Parenti Castelli G, Lenaz G 2003. Structural and functional organization of complex I in the mitochondrial respiratory chain. Biofactors 18: 3–9 [DOI] [PubMed] [Google Scholar]
- Blakely EL, Mitchell AL, Fisher N, Meunier B, Nijtmans LG, Schaefer AM, Jackson MJ, Turnbull DM, Taylor RW 2005. A mitochondrial cytochrome b mutation causing severe respiratory chain enzyme deficiency in humans and yeast. FEBS J 272: 3583–3592 [DOI] [PubMed] [Google Scholar]
- Bonora E, Porcelli AM, Gasparre G, Biondi A, Ghelli A, Carelli V, Baracca A, Tallini G, Martinuzzi A, Lenaz G, et al. 2006a. Defective oxidative phosphorylation in thyroid oncocytic carcinoma is associated with pathogenic mitochondrial DNA mutations affecting complexes I and III. Cancer Res 66: 6087–6096 [DOI] [PubMed] [Google Scholar]
- Bonora E, Evangelisti C, Bonichon F, Tallini G, Romeo G 2006b. Novel germline variants identified in the inner mitochondrial membrane transporter TIMM44 and their role in predisposition to oncocytic thyroid carcinomas. Br J Cancer 4: 1529–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulahbel H, Duran RV, Gottlieb E 2009. Prolyl hydroxylases as regulators of cell metabolism. Biochem Soc Trans 37: 291–294 [DOI] [PubMed] [Google Scholar]
- Brahimi-Horn MC, Pouyssegur J 2007. Oxygen, a source of life and stress. FEBS Lett 581: 3582–3591 [DOI] [PubMed] [Google Scholar]
- Brandon M, Baldi P, Wallace DC 2006. Mitochondrial mutations in cancer. Oncogene 25: 4647–4662 [DOI] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG 2004. Mitochondrial signaling: The retrograde response. Mol Cell 14: 1–15 [DOI] [PubMed] [Google Scholar]
- Chen Z, Lu W, Garcia-Prieto C, Huang P 2007. The Warburg effect and its cancer therapeutic implications. J Bioenerg Biomembr 39: 267–274 [DOI] [PubMed] [Google Scholar]
- Costa-Guda J, Tokura T, Roth SI, Arnold A 2007. Mitochondrial DNA mutations in oxyphilic and chief cell parathyroid adenomas. BMC Endocr Disord 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuezva JM, Ortega AD, Willers I, Sanchez-Cenizo L, Aldea M, Sanchez-Arago M 2009. The tumor suppressor function of mitochondria: Translation into the clinics. Biochim Biophys Acta 1792: 1145–1158 [DOI] [PubMed] [Google Scholar]
- D’Aurelio M, Gajewski CD, Lenaz G, Manfredi G 2006. Respiratory chain supercomplexes set the threshold for respiration defects in human mtDNA mutant cybrids. Hum Mol Genet 15: 2157–2169 [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB 2008. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7: 11–20 [DOI] [PubMed] [Google Scholar]
- Diaz F, Fukui H, Garcia S, Moraes CT 2006. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol Cell Biol 26: 4872–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, Hirano M, Schon EA 2006. Mitochondrial medicine. Informa Healthcare, London [Google Scholar]
- Finley DJ, Zhu B, Fahey TJ 3rd 2004. Molecular analysis of Hurthle cell neoplasms by gene profiling. Surgery 136: 1160–1168 [DOI] [PubMed] [Google Scholar]
- Frezza C, Gottlieb E 2009. Mitochondria in cancer: Not just innocent bystanders. Semin Cancer Biol 19: 4–11 [DOI] [PubMed] [Google Scholar]
- Fry M, Green DE 1980. Cardiolipin requirement by cytochrome oxidase and the catalytic role of phospholipid. Biochem Biophys Res Commun 93: 1238–1246 [DOI] [PubMed] [Google Scholar]
- Fry M, Green DE 1981. Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. J Biol Chem 256: 1874–1880 [PubMed] [Google Scholar]
- Gasparre G, Porcelli AM, Bonora E, Pennisi LF, Toller M, Iommarini L, Ghelli A, Moretti M, Betts CM, Martinelli GN, et al. 2007. Disruptive mitochondrial DNA mutations in complex I subunits are markers of oncocytic phenotype in thyroid tumors. Proc Natl Acad Sci 104: 9001–9006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparre G, Hervouet E, de Laplanche E, Demont J, Pennisi LF, Colombel M, Mege-Lechevallier F, Scoazec JY, Bonora E, Smeets R, et al. 2008. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum Mol Genet 17: 986–995 [DOI] [PubMed] [Google Scholar]
- Gasparre G, Iommarini L, Porcelli AM, Lang M, Ferri GG, Kurelac I, Zuntini R, Mariani E, Pennisi LF, Pasquini E, et al. 2009. An inherited mitochondrial DNA disruptive mutation shifts to homoplasmy in oncocytic tumor cells. Hum Mutat 30: 391–396 [DOI] [PubMed] [Google Scholar]
- Gasparre G, Romeo G, Rugolo M, Porcelli AM 2011a. Learning from oncocytic tumors: Why choose inefficient mitochondria? Biochim Biophys Acta 1807: 633–642 [DOI] [PubMed] [Google Scholar]
- Gasparre G, Kurelac I, Capristo M, Iommarini L, Ghelli A, Ceccarelli C, Nicoletti G, Nanni P, De Giovanni C, Scotlandi K, et al. 2011b. A mutation threshold distinguishes the anti-tumorigenic effects of the mitochondrial gene MTND1, an oncojanus function. Cancer Res 71: 6220–6229 [DOI] [PubMed] [Google Scholar]
- Genova ML, Baracca A, Biondi A, Casalena G, Faccioli M, Falasca AI, Formiggini G, Sgarbi G, Solaini G, Lenaz G 2008. Is supercomplex organization of the respiratory chain required for optimal electron transfer activity? Biochim Biophys Acta 1777: 740–746 [DOI] [PubMed] [Google Scholar]
- Guerra F, Kurelac I, Cormio A, Zuntini R, Amato LB, Ceccarelli C, Santini D, Cormio G, Fracasso F, Selvaggi L, et al. 2011. Placing mitochondrial DNA mutations within the progression model of type I endometrial carcinoma. Hum Mol Genet 20: 2394–2405 [DOI] [PubMed] [Google Scholar]
- Higgins JP, Shinghal R, Gill H, Reese JH, Terris M, Cohen RJ, Fero M, Pollack JR, van de Rijn M, Brooks JD 2003. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol 162: 925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran AL, Briscoe DA, Fiskum G, Kelleher JK 1995. Glutamine metabolism in AS-30D hepatoma cells. Evidence for its conversion into lipids via reductive carboxylation. Mol Cell Biochem 152: 95–101 [DOI] [PubMed] [Google Scholar]
- Ibsen KH 1961. The Crabtree effect: A review. Cancer Res 21: 829–841 [PubMed] [Google Scholar]
- Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J 2008. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 320: 661–664 [DOI] [PubMed] [Google Scholar]
- Jacques C, Baris O, Prunier-Mirebeau D, Savagner F, Rodien P, Rohmer V, Franc B, Guyetant S, Malthiery Y, Reynier P 2005. Two-step differential expression analysis reveals a new set of genes involved in thyroid oncocytic tumors. J Clin Endocrinol Metab 90: 2314–2320 [DOI] [PubMed] [Google Scholar]
- Jahnke VE, Sabido O, Defour A, Castells J, Lefai E, Roussel D, Freyssenet D 2010. Evidence for mitochondrial respiratory deficiency in rat rhabdomyosarcoma cells. PLoS ONE 5: e8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaluz S, Kaluzova M, Stanbridge EJ 2008. Rational design of minimal hypoxia-inducible enhancers. Biochem Biophys Res Commun 370: 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SY, Gutowsky HS, Hsung JC, Jacobs R, King TE, Rice D, Oldfield E 1979. Nuclear magnetic resonance investigation of the cytochrome oxidase–phospholipid interaction: A new model for boundary lipid. Biochemistry 18: 3257–3267 [DOI] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV 2006. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3: 177–185 [DOI] [PubMed] [Google Scholar]
- King A, Selak MA, Gottlieb E 2006. Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene 25: 4675–4682 [DOI] [PubMed] [Google Scholar]
- Klaunig JE, Kamendulis LM 2004. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol 44: 239–267 [DOI] [PubMed] [Google Scholar]
- Klevecz RR, Bolen J, Forrest G, Murray DB 2004. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci 101: 1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman WJ, Verkaart S, Visch HJ, van Emst-de Vries S, Nijtmans LG, Smeitink JA, Willems PH 2007. Human NADH:ubiquinone oxidoreductase deficiency: Radical changes in mitochondrial morphology? Am J Physiol Cell Physiol 293: C22–C29 [DOI] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV 2011. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer 11: 325–337 [DOI] [PubMed] [Google Scholar]
- Kuhajda FP, Jenner K, Wood FD, Hennigar RA, Jacobs LB, Dick JD, Pasternack GR 1994. Fatty acid synthesis: A potential selective target for antineoplastic therapy. Proc Natl Acad Sci 91: 6379–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurelac I, Lang M, Zuntini R, Calabrese C, Simone D, Vicario S, Santamaria M, Attimonelli M, Romeo G, Gasparre G 2011. Searching for a needle in the haystack: Comparing six methods to evaluate heteroplasmy in difficult sequencecontext. Biotechnol Adv 30: 363–371 [DOI] [PubMed] [Google Scholar]
- Lange C, Nett JH, Trumpower BL, Hunte C 2001. Specific roles of protein–phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J 20: 6591–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AG 2004. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta 1666: 62–87 [DOI] [PubMed] [Google Scholar]
- Lenaz G, Genova ML 2007. Kinetics of integrated electron transfer in the mitochondrial respiratory chain: Random collisions vs. solid state electron channeling. Am J Physiol Cell Physiol 292: C1221–C1239 [DOI] [PubMed] [Google Scholar]
- Lenaz G, Genova ML 2010. Structure and organization of mitochondrial respiratory complexes: A new understanding of an old subject. Antioxid Redox Signal 12: 961–1008 [DOI] [PubMed] [Google Scholar]
- Lenaz G, Strocchi P 2009. Reactive oxygen species in the induction of toxicity. In General and applied toxicology review (ed. Ballantyne B, et al. ), Vol. 2, pp. 367–410 Wiley, Chichester, UK [Google Scholar]
- Lenaz G, Baracca A, Fato R, Genova ML, Solaini G 2006. New insights into structure and function of mitochondria and their role in aging and disease. Antioxid Redox Signal 8: 417–437 [DOI] [PubMed] [Google Scholar]
- Lu H, Dalgard CL, Mohyeldin A, McFate T, Tait AS, Verma A 2005. Reversible inactivation of HIF-1 prolyl hydroxylases allows cell metabolism to control basal HIF-1. J Biol Chem 280: 41928–41939 [DOI] [PubMed] [Google Scholar]
- Ma Y, Bai RK, Trieu R, Wong LJ 2010. Mitochondrial dysfunction in human breast cancer cells and their transmitochondrial cybrids. Biochim Biophys Acta 1797: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathupala SP, Ko YH, Pedersen PL 2010. The pivotal roles of mitochondria in cancer: Warburg and beyond and encouraging prospects for effective therapies. Biochim Biophys Acta 1797: 1225–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximo V, Sobrinho-Simoes M 2000. Mitochondrial DNA “common” deletion in Hurthle cell lesions of the thyroid. J Pathol 192: 561–562 [DOI] [PubMed] [Google Scholar]
- Maximo V, Sores P, Rocha AS, Sobrinho-Simoes M 1998. The common deletion of mitochondrial DNA is found in goiters and thyroid tumors with and without oxyphil cell change. Ultrastruct Pathol 22: 271–273 [DOI] [PubMed] [Google Scholar]
- Maximo V, Soares P, Lima J, Cameselle-Teijeiro J, Sobrinho-Simoes M 2002. Mitochondrial DNA somatic mutations (point mutations and large deletions) and mitochondrial DNA variants in human thyroid pathology: A study with emphasis on Hurthle cell tumors. Am J Pathol 160: 1857–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximo V, Botelho T, Capela J, Soares P, Lima J, Taveira A, Amaro T, Barbosa AP, Preto A, Harach HR, et al. 2005. Somatic and germline mutation in GRIM-19, a dual function gene involved in mitochondrial metabolism and cell death, is linked to mitochondrion-rich (Hurthle cell) tumours of the thyroid. Br J Cancer 23: 1892–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximo V, Lima J, Soares P, Sobrinho-Simoes M 2009. Mitochondria and cancer. Virchows Arch 454: 481–495 [DOI] [PubMed] [Google Scholar]
- Mayevsky A 2009. Mitochondrial function and energy metabolism in cancer cells: Past overview and future perspectives. Mitochondrion 9: 165–179 [DOI] [PubMed] [Google Scholar]
- Mayr JA, Meierhofer D, Zimmermann F, Feichtinger R, Kogler C, Ratschek M, Schmeller N, Sperl W, Kofler B 2008. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin Cancer Res 14: 2270–2275 [DOI] [PubMed] [Google Scholar]
- McFate T, Mohyeldin A, Lu H, Thakar J, Henriques J, Halim ND, Wu H, Schell MJ, Tsang TM, Teahan O, et al. 2008. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J Biol Chem 283: 22700–22708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie M, Lazarou M, Thorburn DR, Ryan MT 2006. Mitochondrial respiratory chain supercomplexes are destabilized in Barth syndrome patients. J Mol Biol 361: 462–469 [DOI] [PubMed] [Google Scholar]
- Moran M, Rivera H, Sanchez-Arago M, Blazquez A, Merinero B, Ugalde C, Arenas J, Cuezva JM, Martin MA 2010. Mitochondrial bioenergetics and dynamics interplay in complex I–deficient fibroblasts. Biochim Biophys Acta 1802: 443–453 [DOI] [PubMed] [Google Scholar]
- Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, Saavedra E 2007. Energy metabolism in tumor cells. FEBS J 274: 1393–1418 [DOI] [PubMed] [Google Scholar]
- Morris LG, Veeriah S, Chan TA 2010. Genetic determinants at the interface of cancer and neurodegenerative disease. Oncogene 29: 3453–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Hocker J 1992. Random cytochrome-c-oxidase deficiency of oxyphil cell nodules in the parathyroid gland. A mitochondrial cytopathy related to cell ageing? Pathol Res Pract 188: 701–706 [DOI] [PubMed] [Google Scholar]
- Muller-Hocker J, Jacob U, Seibel P 1998a. Hashimoto thyroiditis is associated with defects of cytochrome-c oxidase in oxyphil Askanazy cells and with the common deletion (4,977) of mitochondrial DNA. Ultrastruct Pathol 22: 91–100 [DOI] [PubMed] [Google Scholar]
- Muller-Hocker J, Schafer S, Copeland WC, Wiesner R, Seibel P 1998b. Immunohistochemical detection of human mtDNA polymerase γ and of human mitochondrial transcription factor A in cytochrome-c-oxidase-deficient oxyphil cells of hyperfunctional parathyroids. Virchows Arch 433: 529–536 [DOI] [PubMed] [Google Scholar]
- Neuwald AF 1997. Barth syndrome may be due to an acyltransferase deficiency. Curr Biol 7: R465–R466 [DOI] [PubMed] [Google Scholar]
- Ozawa T 1997. Genetic and functional changes in mitochondria associated with aging. Physiol Rev 77: 425–464 [DOI] [PubMed] [Google Scholar]
- Paradies G, Petrosillo G, Pistolese M, Ruggiero FM 2000. The effect of reactive oxygen species generated from the mitochondrial electron transport chain on the cytochrome c oxidase activity and on the cardiolipin content in bovine heart submitochondrial particles. FEBS Lett 466: 323–326 [DOI] [PubMed] [Google Scholar]
- Paradies G, Petrosillo G, Pistolese M, Ruggiero FM 2002. Reactive oxygen species affect mitochondrial electron transport complex I activity through oxidative cardiolipin damage. Gene 286: 135–141 [DOI] [PubMed] [Google Scholar]
- Paradies G, Petrosillo G, Paradies V, Ruggiero FM 2010. Oxidative stress, mitochondrial bioenergetics, and cardiolipin in aging. Free Radic Biol Med 48: 1286–1295 [DOI] [PubMed] [Google Scholar]
- Park JS, Sharma LK, Li H, Xiang R, Holstein D, Wu J, Lechleiter J, Naylor SL, Deng JJ, Lu J, et al. 2009. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum Mol Genet 18: 1578–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlo RA, Coleman PS 1986. Continuous pyruvate carbon flux to newly synthesized cholesterol and the suppressed evolution of pyruvate-generated CO2 in tumors: Further evidence for a persistent truncated Krebs cycle in hepatomas. Biochim Biophys Acta 886: 169–176 [DOI] [PubMed] [Google Scholar]
- Pasini B, Stratakis CA 2009. SDH mutations in tumorigenesis and inherited endocrine tumours: Lesson from the phaeochromocytoma-paraganglioma syndromes. J Intern Med 266: 19–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten DA, Lafleur VN, Robitaille GA, Chan DA, Giaccia AJ, Richard DE 2010. Hypoxia-inducible factor-1 activation in nonhypoxic conditions: The essential role of mitochondrial-derived reactive oxygen species. Mol Biol Cell 21: 3247–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, et al. 2005. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci 102: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo G, Ruggiero FM, Di Venosa N, Paradies G 2003. Decreased complex III activity in mitochondria isolated from rat heart subjected to ischemia and reperfusion: Role of reactive oxygen species and cardiolipin. FASEB J 17: 714–716 [DOI] [PubMed] [Google Scholar]
- Petrosillo G, Matera M, Moro N, Ruggiero FM, Paradies G 2009. Mitochondrial complex I dysfunction in rat heart with aging: Critical role of reactive oxygen species and cardiolipin. Free Radic Biol Med 46: 88–94 [DOI] [PubMed] [Google Scholar]
- Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H 2003. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem 278: 52873–52880 [DOI] [PubMed] [Google Scholar]
- Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, Wortham NC, Hunt T, Olpin S, Moat SJ, Hargreaves IP, et al. 2005. Accumulation of Krebs cycle intermediates and over-expression of HIF1α in tumours which result from germline FH and SDH mutations. Hum Mol Genet 14: 2231–2239 [DOI] [PubMed] [Google Scholar]
- Porcelli AM, Angelin A, Ghelli A, Mariani E, Martinuzzi A, Carelli V, Petronilli V, Bernardi P, Rugolo M 2009. Respiratory complex I dysfunction due to mitochondrial DNA mutations shifts the voltage threshold for opening of the permeability transition pore toward resting levels. J Biol Chem 284: 2045–2052 [DOI] [PubMed] [Google Scholar]
- Porcelli A, Ghelli A, Ceccarelli C, Lang M, Cenacchi G, Capristo M, Pennisi LF, Ciccarelli E, Melcarne A, Bartoletti-Stella A, et al. 2010. The genetic and metabolic signature of oncocytic transformation implicates HIF1α destabilization. Hum Mol Genet 19: 1019–1032 [DOI] [PubMed] [Google Scholar]
- Pradella LM, Zuntini R, Magini P, Ceccarelli C, Neri I, Cerasoli S, Graziano C, Gasparre G, Turchetti D 2011. Two distinct thyroid tumours in a patient with Cowden syndrome carrying both a 10q23 and a mitochondrial DNA germline deletion. J Med Genet 48: 779–782 [DOI] [PubMed] [Google Scholar]
- Putignani L, Raffa S, Pescosolido R, Aimati L, Signore F, Torrisi MR, Grammatico P 2008. Alteration of expression levels of the oxidative phosphorylation system (OXPHOS) in breast cancer cell mitochondria. Breast Cancer Res Treat 110: 439–452 [DOI] [PubMed] [Google Scholar]
- Radermacher M, Ruiz T, Clason T, Benjamin S, Brandt U, Zickermann V 2006. The three-dimensional structure of complex I from Yarrowia lipolytica: A highly dynamic enzyme. J Struct Biol 154: 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva A, Tandler B 2000. Three-dimensional structure of oncocyte mitochondria in human salivary glands: A scanning electron microscope study. Ultrastruct Pathol 24: 145–150 [DOI] [PubMed] [Google Scholar]
- Robinson NC, Strey F, Talbert L 1980. Investigation of the essential boundary layer phospholipids of cytochrome c oxidase using Triton X-100 delipidation. Biochemistry 19: 3656–3661 [DOI] [PubMed] [Google Scholar]
- Rosca MG, Vazquez EJ, Kerner J, Parland W, Chandler MP, Stanley W, Sabbah HN, Hoppel CL 2008. Cardiac mitochondria in heart failure: Decrease in respirasomes and oxidative phosphorylation. Cardiovasc Res 80: 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savagner F, Chevrollier A, Loiseau D, Morgan C, Reynier P, Clark O, Stepien G, Malthiery Y 2001a. Mitochondrial activity in XTC.UC1 cells derived from thyroid oncocytoma. Thyroid 11: 327–333 [DOI] [PubMed] [Google Scholar]
- Savagner F, Franc B, Guyetant S, Rodien P, Reynier P, Malthiery Y 2001b. Defective mitochondrial ATP synthesis in oxyphilic thyroid tumors. J Clin Endocrinol Metab 86: 4920–4925 [DOI] [PubMed] [Google Scholar]
- Savagner F, Mirebeau D, Jacques C, Guyetant S, Morgan C, Franc B, Reynier P, Malthiery Y 2003. PGC-1-related coactivator and targets are upregulated in thyroid oncocytoma. Biochem Biophys Res Commun 310: 779–784 [DOI] [PubMed] [Google Scholar]
- Schafer E, Dencher NA, Vonck J, Parcej DN 2007. Three-dimensional structure of the respiratory chain supercomplex I1III2IV1 from bovine heart mitochondria. Biochemistry 46: 12579–12585 [DOI] [PubMed] [Google Scholar]
- Schagger H, de Coo R, Bauer MF, Hofmann S, Godinot C, Brandt U 2004. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J Biol Chem 279: 36349–36353 [DOI] [PubMed] [Google Scholar]
- Sedlak E, Robinson NC 1999. Phospholipase A(2) digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry 38: 14966–14972 [DOI] [PubMed] [Google Scholar]
- Semenza GL 2003. Targeting HIF-1 for cancer therapy. Nat Rev Cancer 3: 721–732 [DOI] [PubMed] [Google Scholar]
- Semenza GL 2007. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J 405: 1–9 [DOI] [PubMed] [Google Scholar]
- Shidara Y, Yamagata K, Kanamori T, Nakano K, Kwong JQ, Manfredi G, Oda H, Ohta S 2005. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res 65: 1655–1663 [DOI] [PubMed] [Google Scholar]
- Simonnet H, Demont J, Pfeiffer K, Guenaneche L, Bouvier R, Brandt U, Schagger H, Godinot C 2003. Mitochondrial complex I is deficient in renal oncocytomas. Carcinogenesis 24: 1461–1466 [DOI] [PubMed] [Google Scholar]
- Smolkova K, Plecita-Hlavata L, Bellance N, Benard G, Rossignol R, Jezek P 2010. Waves of gene regulation suppress and then restore oxidative phosphorylation in cancer cells. Int J Biochem Cell Biol 43: 950–968 [DOI] [PubMed] [Google Scholar]
- Sun W, Zhou S, Chang SS, McFate T, Verma A, Califano JA 2009. Mitochondrial mutations contribute to HIF1α accumulation via increased reactive oxygen species and up-regulated pyruvate dehydrogenease kinase 2 in head and neck squamous cell carcinoma. Clin Cancer Res 15: 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallini G 1998. Oncocytic tumours. Virchows Arch 433: 5–12 [DOI] [PubMed] [Google Scholar]
- Tallini G, Ladanyi M, Rosai J, Jhanwar SC 1994. Analysis of nuclear and mitochondrial DNA alterations in thyroid and renal oncocytic tumors. Cytogenet Cell Genet 66: 253–259 [DOI] [PubMed] [Google Scholar]
- Thompson CB 2009. Metabolic enzymes as oncogenes or tumor suppressors. N Engl J Med 360: 813–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuppen HA, Blakely EL, Worgan L, Al-Dosary M, Saretzki G, Alston CL, Morris AA, Clarke M, Jones S, Devlin AM, et al. 2010. The p.M292T NDUFS2 mutation causes complex I–deficient Leigh syndrome in multiple families. Brain 133: 2952–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raam BJ, Sluiter W, de Wit E, Roos D, Verhoeven AJ, Kuijpers TW 2008. Mitochondrial membrane potential in human neutrophils is maintained by complex III activity in the absence of supercomplex organisation. PLoS ONE 3: e2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vik SB, Capaldi RA 1977. Lipid requirements for cytochrome c oxidase activity. Biochemistry 16: 5755–5759 [DOI] [PubMed] [Google Scholar]
- Wallace DC 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet 39: 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O 1956. On the origin of cancer cells. Science 123: 309–314 [DOI] [PubMed] [Google Scholar]
- Weinberg F, Chandel NS 2009. Mitochondrial metabolism and cancer. Ann NY Acad Sci 1177: 66–73 [DOI] [PubMed] [Google Scholar]
- Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK 2008. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem 283: 20621–20627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W 2002. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem 277: 43553–43556 [DOI] [PubMed] [Google Scholar]
- Zielke A, Tezelman S, Jossart GH, Wong M, Siperstein AE, Duh QY, Clark OH 1998. Establishment of a highly differentiated thyroid cancer cell line of Hurthle cell origin. Thyroid 8: 475–483 [DOI] [PubMed] [Google Scholar]
- Zimmermann FA, Mayr JA, Neureiter D, Feichtinger R, Alinger B, Jones ND, Eder W, Sperl W, Kofler B 2009. Lack of complex I is associated with oncocytic thyroid tumours. Br J Cancer 100: 1434–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann FA, Mayr JA, Feichtinger R, Neureiter D, Lechner R, Koegler C, Ratschek M, Rusmir H, Sargsyan K, Sperl W, et al. 2011. Respiratory chain complex I is a mitochondrial tumor suppressor of oncocytic tumors. Front Biosci 1: 315–325 [DOI] [PubMed] [Google Scholar]
- Zu XL, Guppy M 2004. Cancer metabolism: Facts, fantasy, and fiction. Biochem Biophys Res Commun 313: 459–465 [DOI] [PubMed] [Google Scholar]