Abstract
The link between perturbations in translational control and cancer etiology is becoming a primary focus in cancer research. It has now been established that genetic alterations in several components of the translational apparatus underlie spontaneous cancers as well as an entire class of inherited syndromes known as “ribosomopathies” associated with increased cancer susceptibility. These discoveries have illuminated the importance of deregulations in translational control to very specific cellular processes that contribute to cancer etiology. In addition, a growing body of evidence supports the view that deregulation of translational control is a common mechanism by which diverse oncogenic pathways promote cellular transformation and tumor development. Indeed, activation of these key oncogenic pathways induces rapid and dramatic translational reprogramming both by increasing overall protein synthesis and by modulating specific mRNA networks. These translational changes promote cellular transformation, impacting almost every phase of tumor development. This paradigm represents a new frontier in the multihit model of cancer formation and offers significant promise for innovative cancer therapies. Current research, in conjunction with cutting edge technologies, will further enable us to explore novel mechanisms of translational control, functionally identify translationally controlled mRNA groups, and unravel their impact on cellular transformation and tumorigenesis.
During tumor development, key oncogenic pathways (e.g., Myc) monopolize specific components of the translational machinery (e.g., initiation factors) to increase overall protein synthesis and modulate specific mRNA networks.
Given the fact that translation is the ultimate step for producing a functional protein, it is surprising how much the cancer biology community has historically overlooked the importance of its deregulation toward cancer development. Indeed, decades of research into the molecular programs that govern cellular transformation have mainly focused on the cancer transcriptome (van ’t Veer et al. 2002; Hawkins and Ren 2006). The microarray era enabled the research community to catalog genome-wide variations in the repertoire of transcriptional outputs downstream of specific oncogenic signaling pathways. Perhaps the reluctance in hypothesizing that changes in translational control may coordinate major events underlying cancer formation is because of the misconception that specificity in gene expression is primarily due to transcription regulation. An exciting body of research now shows that changes in mRNA translation control distinct cellular processes including metabolism, cell migration, cell adhesion, cell growth, cell-cycle control, and tumorigenesis (Silvera et al. 2010). These studies have been important in revealing not only novel general mechanisms of translational control, but also new paradigms for cellular transformation and cancer development (Fig. 1). For example, key oncogenic pathways such as Myc and PI3K monopolize the levels and activity of specific components of the translational machinery (e.g., initiation factors, ribosomal proteins) to direct specific posttranscriptional changes in gene expression directly at the level of protein production (Hannan et al. 2011). In addition, an entire class of inherited syndromes collectively referred to as “ribosomopathies” harbor mutations in distinct ribosome components and is characterized by increased cancer susceptibility (Ganapathi and Shimamura 2008). The realization that there are posttranscriptional control mechanisms in cancer development has provided a strong rationale in the design of new cancer therapies that may ultimately eradicate the abnormal translational program of cancer cells. I provide here an analysis and perspective on how the regulation of protein synthesis plays a critical and pivotal role in the etiology of cancer.
Figure 1.
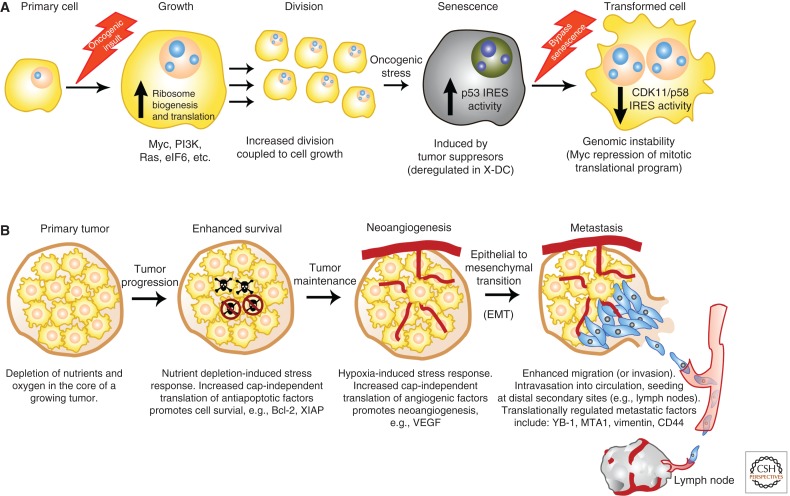
Deregulations in translational control contribute to each step of cellular transformation and tumor progression. (A) Upon receiving an oncogenic insult (lightning bolt) such as Myc overexpression or PI3K hyperactivation, ribosome biogenesis and global protein synthesis are augmented resulting in increased cell size coupled to cell division. Upon this oncogenic stress, cells initiate a tumor-suppressive response associated with increased internal ribosome entry site (IRES)-mediated translation, leading to cell-cycle arrest and senescence. To overcome the barrier of oncogene-induced senescence (OIS), cells acquire additional mutations known as secondary hits (lightning bolt). One such mechanism to overcome tumor-suppressive checkpoints such as OIS is the promotion of genome instability by decreasing IRES-dependent translation of the CDK11/p58 tumor suppressor. (B) Once established, a primary tumor will undergo unrestrained growth, which triggers a stress response including depletion of oxygen and essential nutrients from the core of the tumor. Lack of key nutrients such as growth factors often leads to increased apoptosis (skulls). Tumor cells up-regulate cap-independent translation of antiapoptotic factors (e.g., Bcl-2 and XIAP) as a mechanism to promote survival (blocked skulls). To bypass stress caused by low levels of oxygen in the tumor, cells induce cap-independent translation of neoangiogenesis promoting factors such as vascular endothelial growth factor (VEGF). Primary tumors metastasize to secondary organs. Epithelial-to-mesenchymal transition (EMT) facilitates metastatic dissemination; specifically, cancer cells lose their epithelial characteristics (yellow cells) and acquire mesenchymal traits (blue cells) including enhanced mobility and invasiveness. Multiple stages of metastatic cell dissemination include degradation of the basement membrane, intravasation into the circulatory system, and extravasation at the distal site. eIF4E regulates the translation of an invasive messenger RNA (mRNA) signature (i.e., YB-1, MTA1, vimentin, and CD44) that promotes metastasis formation. Increased translation of prometastatic factors localized at the leading edge of the tumor’s invasive front may facilitate metastasis colonization.
THE CANCEROUS TRANSLATION MACHINERY
Initiation Factors (eIFs)
Genetic studies of human cancers have made a compelling case for the role of deregulations in translational control within the multihit model of cancer etiology. Genes encoding initiation factors are aberrantly expressed during cancer progression (Table 1). Most importantly, clinical research reveals that these genes are gained or lost in human tumors, consistent with a possible causal role in cancer etiology (Table 1). However, it will be critical to determine to what extent these genetic changes in eIFs directly contribute to cancer development.
Table 1.
Genetic alterations in translational components are associated with increased cancer susceptibility
| Gene | Genetic lesion | Expression/activity | Cancer association | Reference(s) | |
|---|---|---|---|---|---|
| Ribosomopathies | |||||
| X-linked dyskeratosis congenita | DKC1 | Mutation/deletion | Decrease | Increased incidence of hematological and solid tumors | Heiss et al. 1998 |
| Diamond-Blackfan anemia | RPS 7, 10, 15, 17, 19, 24, 26, 27A RPL 5, 11, 35A, 36 |
Mutation/deletion | Decrease | Increased incidence of hematological and solid tumors | Draptchinskaia et al. 1999; Gazda et al. 2006, 2008; Cmejla et al. 2007, 2009; Farrar et al. 2008; Doherty et al. 2010 |
| 5q deletion syndrome | RPS14 | Deletion | Decrease | Increased incidence of hematological tumors | Ebert et al. 2008 |
| Shwachman-Diamond syndrome | SDBS | Mutation | Decrease | Increased incidence of hematological tumors | Boocock et al. 2003; Austin et al. 2005 |
| Cartilage-hair hypoplasia | RMRP | Mutation | Decrease | Increased incidence of hematological and solid tumors | Ridanpaa et al. 2001 |
| Translation initiation factors | |||||
| eIF2a | ND | Increase | In hematological and solid tumors | Wang et al. 1999; Rosenwald et al. 2001 | |
| eIF3a | ND | Increase | In solid tumors | Bachmann et al. 1997; Dellas et al. 1998 | |
| eIF3c | ND | Increase | In solid tumors | Rothe et al. 2000 | |
| eIF3e | ND | Increase | In solid tumors | Marchetti et al. 2001 | |
| eIF3f | LOH | Decrease | In solid tumors | Shi et al. 2006; Doldan et al. 2008a,b | |
| eIF3h | Amplification | Increase | In solid tumors | Saramaki et al. 2001 | |
| eIF3i | ND | Increase | In solid tumors | Rauch et al. 2004; Ahlemann et al. 2006 | |
| eIF4A | ND | Increase | In solid tumors | Eberle et al. 1997; Shuda et al. 2000 | |
| eIF4E | Amplification | Increase | In hematological and solid tumors | De Benedetti and Graff 2004 | |
| eIF4G | Amplification | Increase | In solid tumors | Brass et al. 1997; Bauer et al. 2002 | |
| eIF5A | ND | Increase | In hematological tumors | Balabanov et al. 2007 | |
| eIF5A2 | Amplification | Increase | In solid tumors | Guan et al. 2001 | |
| eIF6 | ND | Increase | In hematological and solid tumors | Sanvito et al. 2000; Harris et al. 2004 | |
| Translation elongation factors | |||||
| eEF2 | ND | Increase | In solid tumors | Nakamura et al. 2009 | |
| eEF1A2 | Amplification | Increase | In solid tumors | Anand et al. 2002; Lee 2003 | |
| IRES trans-acting factors | |||||
| hnRNP A1 | ND | Increase | In solid tumors | Ma et al. 2009 | |
| hnRNP E1 | ND | Decrease | In solid tumors | Wang et al. 2010; Zhang et al. 2010 | |
| hnRNP E2 | ND | Decrease | In solid tumors | Roychoudhury et al. 2007 | |
| La | ND | Increase | In solid tumors | Trotta et al. 2003 | |
| PTB | ND | Increase | In solid tumors | Jin et al. 2000 | |
| YB-1 | ND | Increase | Correlates with higher grade tumors | Stratford et al. 2007 | |
| Ribosome biogenesis factors | |||||
| Ubf1 | ND | Increase | In solid tumors | Huang et al. 2002 | |
| Bop1 | Amplification | Increase | In solid tumors | Killian et al. 2006 | |
| Npm | Mutation | Decrease | In hematological tumors | Yun et al. 2007 | |
Abbreviations: RPS, ribosomal protein small subunit; RPL, ribosomal protein large subunit; ND, not determined; PTB, polypyrimidine tract-binding protein; IRES, internal ribosome entry site; LOH, loss of heterozygosity.
Initiation is one of the most regulated steps in translation (Sonenberg and Hinnebusch 2009). During initiation, many checkpoints are coordinated by several initiation factors to control not only whether a specific mRNA is translated but also the rate at which mRNA translation occurs. This process ultimately contributes to the overall abundance of the specific protein in the cell. Genes encoding initiation factors (e.g., eIF3, eIF4G, eIF4E, eIF5A2) are amplified in a variety of human cancers and regulate specific steps of translation initiation including the ribosome–mRNA interaction and initiator Met-tRNAi binding (Table 1). For example, the eIF3 subunit eIF3h is amplified along with the Myc oncogene in breast cancer (Nupponen et al. 1999). Importantly, high-level amplification of eIF3h is found in advanced stage, androgen-independent, and poor-prognosis prostate cancer, but no copy number changes are found in nonmalignant (benign prostatic hyperplasia) or premalignant (prostatic intraepithelial neoplasia) prostates (Saramaki et al. 2001). Intriguingly, loss of heterozygosity and gene copy number analyses showed that loss of the gene encoding eIF3f, another subunit of the eIF3 initiation complex, occurs in pancreatic cancer as well as in melanoma (Doldan et al. 2008a,b). These data suggest that discrete subunits of the same initiation complex (eIF3) could differentially modulate protein synthesis (see below), acting either as oncogenes or tumor suppressors. The genetic loci of eIF4E and eIF4G, two key initiation factors involved in cap-dependent translation, were originally found amplified in breast cancer and squamous cell lung carcinoma (Sorrells et al. 1998; Bauer et al. 2001) (see section below). Furthermore, gene amplification and overexpression of eIF5A2, which enhances the formation of the first peptide bond, has been reported in ovarian cancer as well as in other solid tumors including breast cancer, hepatocellular carcinoma, and nonsmall cell lung cancer (Guan et al. 2004; Clement et al. 2006; Tang et al. 2010; He et al. 2011). The genetic lesions of the initiation factors described above have been shown to affect the overall expression levels of their mRNA and/or protein. In addition, many other components of the translation machinery involved in the initiation step are also misexpressed in a variety of hematological malignancies and solid tumors (see Table 1), although the genetic bases for some of these abnormalities are unknown.
What is the evidence that these changes exert direct and causal effects in cancer? Proving causality is challenging. One approach is to define the mechanism by which alterations in these initiation factors modulate gene expression at the translational level, ultimately contributing to cellular transformation and cancer development. The challenge in unraveling this mechanism resides in the fact that overexpression of some of these initiation factors modulates both global protein synthesis and translation efficiency of specific mRNAs. For example, in NIH 3T3 cells, overexpression of five of the 12 individual subunits that comprise the eIF3 initiation complex (eIF3a, eIF3b, eIF3c, eIF3h, and eIF3i) results in a modest increase in general protein synthesis rate (Zhang et al. 2007). In the same studies, the investigators reported that overexpression of eIF3c or eIF3h results in a shift of cyclin D1, Myc, ODC, and FGF2 mRNAs into larger polysomes, suggesting that overexpression of a transforming eIF3 subunit can also affect the translational efficiency of specific oncogenic mRNAs. Furthermore, in inflammatory breast cancer (IBC), a rare but lethal form of the disease, overexpression of eIF4G has also been shown to specifically control the internal ribosome entry site (IRES)-dependent translation (see below) of two oncogenic mRNAs (VEGF and p120 catenin) important for IBC tumor survival and dissemination (Silvera et al. 2009).
Overall, it remains uncertain whether misexpression of eIFs causes changes in overall protein synthesis (quantitative), or alters the expression of specific mRNAs (qualitative)—distinct outcomes that could both contribute to key steps in cancer formation. Based on the role attributed to these general initiation factors, it stands to reason that increases in their activity would in turn augment global protein synthesis. I will discuss below how this quantitative increase might impact cell growth during cellular transformation, representing the first “hit” for cancer formation. On the other hand, up-regulating the rate of translation initiation through overexpression of these eIFs may also qualitatively enhance the translation efficiency of the poorly translated mRNAs described above. Importantly, many of these mRNAs encode key proteins that critically impact tumorigenesis by modulating the cell cycle and survival. However, the causal contribution of quantitative and qualitative changes in translational control toward cancer development still needs to be shown in a more physiological context. To this end, future studies should use inducible tissue-specific mouse models that harbor the gain or loss of function mutation for these specific initiation factors. These in vivo approaches would be invaluable for discriminating whether the genomic alterations of these translation factors found in human cancers represent driver mutations in tumor initiation or rather contribute to progression and/or metastatic stages of the disease. It is also very important to determine whether the putative target mRNAs of eIF3, eIF4E, eIF4G, and/or eIF5A2 play a functional role during tumor development.
Although many questions still need to be addressed, it is nevertheless an exciting moment in the field of translation and cancer. It is clear that even initiation factors widely assumed to play only a housekeeping function likely show activity that would impinge on specific gene expression programs underlying distinct stages of cancer initiation and/or progression. There are now new technologies such as ribosome profiling (see below) that may shed light on important questions currently pursued in the field and facilitate the examination of the translational landscape of the cancer genome in a tissue- and cell-specific manner.
Do Mutations in the Ribosome Cause Cancer?
One of the most surprising recent discoveries in cancer biology has been the realization that mutations in ribosomal components are directly linked to tumorigenesis. Elucidating the mechanistic basis of this novel relationship has proven to be a conceptual challenge. Even a decade ago, it would have been inconceivable that impaired ribosomal activity would alter specific cellular processes, as it was a commonly held belief that perturbations in the ribosome inevitably would trigger cell death. Human genetic research has now identified ribosomopathies, which are characterized by increased cancer susceptibility. These syndromes harbor mutations in distinct ribosomal components (Table 1). One of the most interesting aspects of these ribosomopathies is the high degree of cell and tissue specificity in the presentation of their pathological features. These findings challenge the dogma that the activity and composition of ribosomes are identical in each cell and tissue in an organism—in other words, that the ribosome is just a static factory to produce proteins and not a regulatory element of the translational apparatus. The discovery of ribosomopathies as well as recent findings in the developmental biology field (see below) strongly suggests that we should reevaluate our perspective on the ribosome and its role in translational control.
One clear example of how defects in the ribosome may alter the translational landscape and contribute to specific disease pathologies and cancer susceptibility is the ribosomopathy X-linked dyskeratosis congenita (X-DC). X-DC is the most common and severe form of dyskeratosis congenita (DC), and is invariably associated with mutations of the DKC1 gene, encoding dyskerin (Table 1) (Heiss et al. 1998; Ruggero et al. 2003). Although prominent features of X-DC pathogenesis include bone marrow failure and skin abnormalities, a wide variety of tumor types including carcinomas and hematopoietic malignancies are also manifest. Dyskerin is an evolutionarily conserved enzyme responsible for the modification of approximately 100 specific uridine residues in ribosomal RNA (rRNA) into pseudouridine (Ni et al. 1997; Lafontaine et al. 1998). The role of these rRNA modifications in translational control has historically been poorly understood. The unexpected human genetic mutations associated with this key enzyme responsible for site-specific rRNA modifications were enigmatic, especially with respect to the tissue-specific phenotypes present in X-DC. The specific features of X-DC pathogenesis, including cancer susceptibility, can be explained, at least in part, by the finding that a distinct subset of mRNAs is not efficiently translated in an X-DC mouse model, X-DC patient cells, or dyskerin-knockdown cancer cells (Yoon et al. 2006; Bellodi et al. 2010a,b; Montanaro et al. 2010). On the other hand, general protein synthesis appears to be unperturbed (Yoon et al. 2006; Montanaro et al. 2010). Surprisingly, these studies have uncovered an unexpected requirement for rRNA modifications in translational control. Specifically, this subset of mRNAs shares a common regulatory motif in their 5′ untranslated region (5′ UTR): an IRES element that acts to initiate translation in a cap-independent manner. It has been proposed that IRES elements may exist in ∼10% of mRNAs, including many tumor suppressor genes and antiapoptotic factors (Fig. 2) (Graber and Holcik 2007; Komar and Hatzoglou 2011). Several studies have highlighted the importance of RNA-binding proteins known as IRES trans-acting factors (ITAFs) in modulating IRES-mediated translation of several cellular mRNAs (Spriggs et al. 2005; Lewis and Holcik 2008; Komar and Hatzoglou 2011). IRES-dependent translation acts to fine-tune gene expression at the posttranscriptional level (Holcik and Sonenberg 2005; Spriggs et al. 2010; Komar and Hatzoglou 2011). In this context, it has been shown that defects in the IRES-mediated translation of a subset of mRNAs, including important tumor suppressors such as p53 and p27, in DKC1 mutant cells contribute to cellular transformation and tumor development (Bellodi et al. 2010a,b; Montanaro et al. 2010). Recent findings also show an evolutionarily conserved role for rRNA pseudouridylation in faithfully maintaining the translational reading frame. Specifically, impaired pseudouridylation of rRNA promotes increased programmed ribosomal frameshifting (PRF) of both viral and cellular mRNAs containing a PRF signal (Jack et al. 2011). It would be interesting to assess whether rRNA pseudouridylation regulates, at the translational level, the expression of mRNAs containing a PRF signal, perhaps in a cell- and/or tissue-specific manner. An emerging role for rRNA modifications in regulating specific modes of translation has recently been highlighted by findings that methylation of rRNA is also implicated in IRES-dependent translation of specific cellular mRNAs (Chaudhuri et al. 2007; Belin et al. 2009; Basu et al. 2011). Furthermore, loss of rRNA methylation at specific sites in zebrafish leads to severe developmental defects (Higa-Nakamine et al. 2012).
Figure 2.
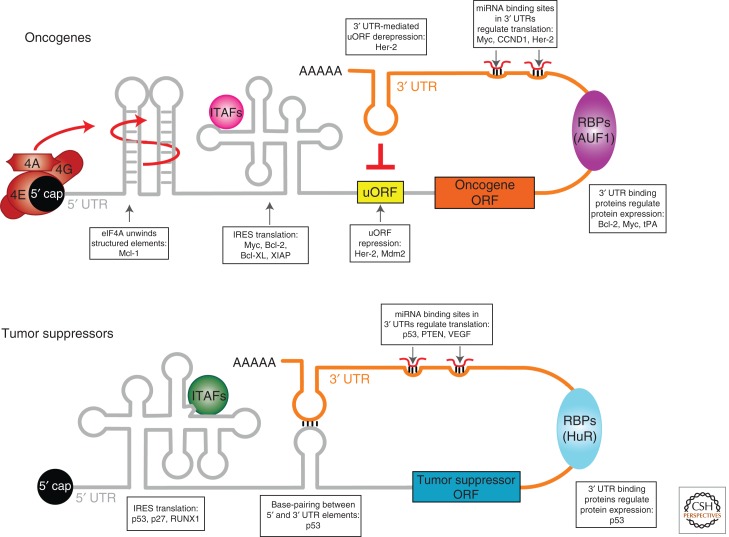
Oncogenes and tumor suppressors are exquisitely regulated at the translational level through specific regulatory elements in their mRNAs. Depictions of cis-regulatory elements present in oncogene (top) and tumor suppressor (bottom) mRNAs including key cell-cycle and survival factors. Examples of regulated mRNAs are given (boxes). The cap-binding protein eIF4E binds the 5′ cap of mRNAs. Increased eIF4E activity recruits eIF4G and the eIF4A helicase to unwind (red arrow) structured elements in the 5′ UTR of poorly translated mRNAs, increasing the expression of many growth promoting and prosurvival genes such as Mcl-1. IRES elements are structures that also direct translational regulation by promoting cap-independent protein synthesis of both tumor suppressors (p53) and prosurvival factors (XIAP) during different stages of tumor development (see Fig. 1). IRES trans-acting factors (ITAFs) modulate translational regulation directed by specific IRES elements. Upstream open reading frames (uORF) are present in select mRNAs (including growth promoting factors such as Mdm2 and Her-2) and inhibit translation initiation by preventing the ribosome from scanning to the start codon. Additionally, a region in the 3′ UTR of the Her-2 mRNA can inhibit uORF-mediated repression of translation. 3′ UTRs contain many distinct elements that interact with RNA-binding proteins (RBPs) and microRNAs (miRNAs) to promote or inhibit translation. One interesting example is the regulation of p53 translation mediated by base-pairing interactions between elements in the 5′ and 3′ UTRs of p53.
Taken as a whole, these studies raise several important questions that remain to be addressed. In particular, what regulates ribosomal RNA (rRNA) modifications? Can rRNA modifications modulate the recruitment of specific factors to the ribosome that would enhance translation specificity? What specific mRNAs are sensitive to ribosome modifications? For example, in addition to IRES elements, do rRNA modifications affect the translation of mRNAs possessing distinct cis-acting regulatory elements?
Diamond-Blackfan anemia (DBA) is another ribosomopathy in which mutations in several different ribosomal proteins (RPs) lead to bone marrow failure and an increased risk of leukemia and solid tumors (Table 1) (Flygare and Karlsson 2007; Lipton and Ellis 2010; Narla et al. 2011). This is consistent with studies in zebrafish showing that mutations in RPs can cause cancer (Amsterdam et al. 2004). In DBA, alterations in rRNA processing have been described and proposed to trigger a stress response marked by p53 activation, which would lead to cell-cycle arrest or programmed cell death (Danilova et al. 2008; McGowan et al. 2008; Dutt et al. 2011). Although the induction of p53 may explain certain pathological features of DBA, it does not provide a molecular basis for the increased cancer susceptibility associated with the disease, the cause of which remains almost completely unknown. One possibility is that RPs may exert more specialized functions in translational control, either on or off the ribosome, altering gene expression to contribute to the cancer susceptibility associated with DBA (Mazumder et al. 2003; Landry et al. 2009). Loss of function of RPS25 does not affect general protein synthesis but rather impairs IRES-dependent translation (Landry et al. 2009). Very recent research has shown that a single ribosomal protein, RPL38, regulates transcript-specific translational control of an important class of Hox mRNAs. In this study, the authors showed that RPL38+/− mice harbor homeotic transformations due to loss of function of Hox mRNA translational control (Kondrashov et al. 2011). Moreover, it appears that ribosomal protein expression may be dynamically regulated in a tissue-specific manner, suggesting that the protein composition of the ribosomal subunits may differ from cell to cell, even within the same organ (Kondrashov et al. 2011). As such, it is tempting to speculate that mutations in ribosomal proteins may have more specific roles in translational control that underlie the increased cancer susceptibility observed in DBA (Xue and Barna 2012).
It is becoming increasingly apparent that the role of ribosomal proteins in cancer development is more complex than originally envisioned, as overexpression of many RPs has been associated with cancer (Ruggero and Pandolfi 2003; Lai and Xu 2007). It is still unknown whether increases in ribosomal protein levels augment the translational capacity of a cell and whether RP overexpression is a cause or just a consequence of cellular transformation. In addition, research data suggest that extraribosomal functions exerted by a few RPs may also play a tumor-suppressive role in cancer development. For example, it has been shown that RPL11 negatively modulated c-Myc levels and activity in cultured cells (Dai et al. 2007; Challagundla et al. 2011). Other studies suggest that the L11-Mdm2-p53 pathway may act as a tumor-suppressive barrier against c-Myc-induced tumorigenesis (Macias et al. 2010).
Additional notable examples of “ribosomopathies” include cartilage–hair hypoplasia (CHH) syndrome, Shwachman-Diamond syndrome (SDS), and 5q deletion syndrome (−5q), which also show increased risk of developing certain cancers of the skin (basal cell carcinoma) as well as blood (leukemia and lymphoma) (Barlow et al. 2010; Shimamura and Alter 2010). In all of these syndromes, mutations in regulatory factors important for ribosome biogenesis and translational control have been identified (Table 1). In certain cases, these factors may also show extraribosomal functions that might contribute to disease pathogenesis (Austin et al. 2008; Dutt et al. 2011). These are intriguing lines of research that go beyond the scope of this article and are further reviewed in (Ganapathi and Shimamura 2008; Burwick et al. 2011; Fumagalli and Thomas 2011). Recent findings have uncovered a role for SBDS (the gene mutated in SDS syndrome) in ribosomal subunit joining and translational control (Finch et al. 2011; Sen et al. 2011; Wong et al. 2011). In hematopoietic stem cells from −5q patients in which RPS14 is found mutated, alterations in expression of key components of the translational machinery have been reported (Pellagatti et al. 2008). Therefore, it is likely that defects in translational control can contribute to features of these ribosomopathies including increased incidences of cancer.
Collectively, inherited ribosomopathies reveal that mutations in regulatory factors important for ribosome activity may produce mutant ribosomes lacking important constituents such as ribosomal proteins or rRNA modifications (Table 1). Can such mutant forms of the translational machinery be referred to as “cancer ribosomes”? If so, an outstanding question is the mechanism by which the “cancer ribosome” could promote cancer development via aberrant translational control. This is an important question to resolve, as at first glance it may appear counterintuitive that alterations in ribosome function could cause cancer, especially given the important connection between increased protein synthesis and cell growth (see below and Fig. 1). However, recent findings show surprising specificity in the classes of mRNAs that are specifically deregulated and underlie cancer susceptibility as a consequence of perturbations in ribosome function, as illustrated by X-DC. These advances also reflect an emerging appreciation for and increased knowledge of more specialized and dynamic regulation of translational control in vivo at an organismal level (Xue and Barna 2012).
CANCER CELLS EXPLOIT TRANSLATIONAL CONTROL FOR THEIR ONCOGENIC PROGRAM
Oncogenic Signaling Pathways Perturb Specific Translational Components
There is a growing body of evidence supporting the idea that deregulation of translational control serves as a common mechanism by which diverse oncogenic pathways (e.g., PI3K, Myc, and Ras) promote cellular transformation and tumor development (Silvera et al. 2010; van Riggelen et al. 2010; Hannan et al. 2011). In a normal cell, these pathways act as sensors of energy, stress, and nutrient availability, as well as growth factor signals, and integrate these inputs to direct control of ribosome production and gene expression at the translational level. One of the primary reasons for this cross talk is to couple these external stimuli with the execution of cell growth, division, and survival, which are directly coupled to protein synthesis (see below). Importantly, all these signals become oncogenic when hyperactivated. How do these signaling pathways alter translational control when mutated in cancer? This question has been answered, at least in part, by the discovery that the oncogenic signals directly modulate the activity and expression of specific translational components (Fig. 3). Notable examples include the PI3K-AKT-mTOR and Ras-MAPK signal transduction pathways, as well as transcriptional programs regulated by oncogenic Myc (Frederickson et al. 1992; Waskiewicz et al. 1999; Schuhmacher et al. 2001; Zeller et al. 2006; Furic et al. 2010; Silvera et al. 2010; van Riggelen et al. 2010; Hannan et al. 2011). I discuss the role of Myc in protein synthesis and cell growth in the next section.
Figure 3.
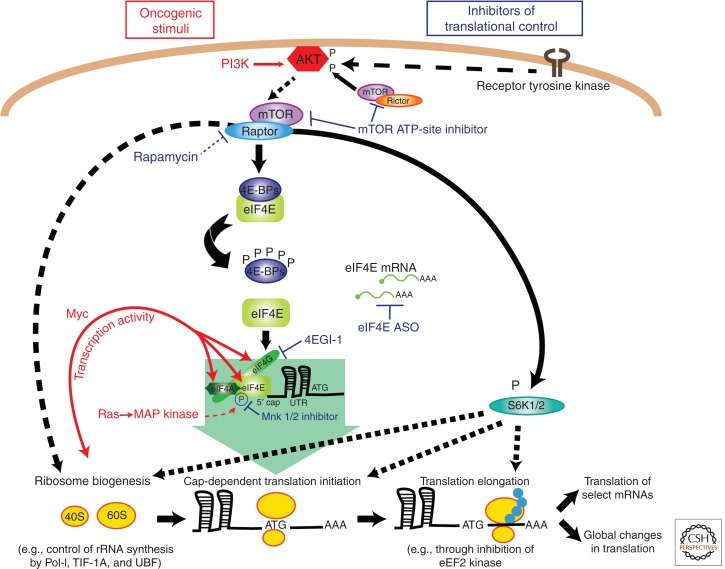
Oncogenic signals regulate each stage of translation. Oncogenic stimuli (red) such as PI3K-AKT-mTOR, Myc, and Ras promote protein synthesis by coordinating the regulation of ribosome biogenesis, translation initiation, and translation elongation. The PI3K-AKT-mTOR signaling pathway promotes ribosome biogenesis through both enhanced rRNA synthesis and enhanced ribosomal protein production (Hannan et al. 2003; Martin et al. 2004; Mayer et al. 2004). This signaling pathway stimulates translation initiation predominantly through mTORC1-dependent hyperactivation of eIF4E. In the absence of signaling, hypophosphorylated 4E-BPs bind to and inhibit eIF4E, blocking its ability to interact with eIF4G. PI3K-AKT signaling activates mTORC1, initiating a series of phosphorylations that release 4E-BPs from eIF4E. This allows for eIF4G binding to eIF4E and the subsequent recruitment of the 40S ribosomal subunit. Furthermore, S6K1/2 downstream mammalian target of rapamycin (mTOR) affects the efficiency of translation initiation and elongation (Ma and Blenis 2009). In addition, Ras-MAP kinase signaling up-regulates eIF4E activation via phosphorylation at serine 209. Myc promotes protein synthesis by increasing the transcription of multiple translational components including eIF4E mRNA. Together, these oncogenic stimuli regulate the multiple stages of translation to drive both global changes in protein synthesis as well as selective changes in the translation of specific mRNAs. Multiple approaches (blue) are used to therapeutically target the translational apparatus including rapamycin, ATP-active site inhibitors of mTOR, MNK1/2 kinase inhibitors, 4EGI-1, and eIF4E antisense oligonucleotides (ASO).
One of the best-studied examples of oncogenic signaling impinging on translational control is the PI3K-AKT-mTOR pathway, which directly modulates translation initiation largely through activation of the kinase mammalian target of rapamycin complex 1 (mTORC1) (Zoncu et al. 2011). The PI3K-AKT-mTOR pathway is one of the most commonly mutated pathways in cancer (Vogt et al. 2011). mTORC1 phosphorylates ribosomal protein S6 kinase 1/2 (S6K1/2) and the eIF4E-binding proteins (4E-BPs), which negatively regulate the major cap-binding protein eIF4E (Dennis et al. 1996; Hara et al. 1997; Gingras et al. 1999). Phosphorylation of 4E-BPs leads to a conformational change that frees eIF4E, which, with the help of other eIFs, ultimately recruits the 40S ribosomal subunit to the 5′ cap of mRNAs during translation initiation (Gingras et al. 1999; Ruggero and Sonenberg 2005). Initiation is thought to be the rate-limiting step of cap-dependent translation and eIF4E is considered the key factor in controlling this step (Duncan et al. 1987). This deduction is in part based on the fact that eIF4E activity is highly regulated at both the mRNA and protein level. Indeed, eIF4E is up-regulated at the mRNA level by a number of transcription factors including the oncogene Myc (Jones et al. 1996; De Benedetti and Graff 2004; Ruggero and Sonenberg 2005). At the protein level, eIF4E activity is also controlled through phosphorylation at serine 209 by the MAP kinase targets MNK1/2, in addition to inhibitory interactions with 4E-BPs (see above and Fig. 3) (Lachance et al. 2002; Ueda et al. 2004, 2010; Wendel et al. 2007; Furic et al. 2010). This tight regulation of eIF4E activity provides a rapid mechanism for cells to modulate translation initiation in response to numerous stimuli, including growth factor and oncogenic signaling.
The 4E-BPs/eIF4E axis is among the most well-characterized nodes in translation control and cancer. For example, eIF4E gene amplification has been reported in human breast, head, and neck cancer specimens, and eIF4E is also found overexpressed in a variety of tumors (Sorrells et al. 1998, 1999; Crew et al. 2000; Berkel et al. 2001; Rosenwald et al. 2001; Salehi and Mashayekhi 2006; Wang et al. 2009a; Yoshizawa et al. 2010). The oncogenic potential of eIF4E hyperactivity has been faithfully recapitulated both in vitro and in vivo. Overexpression of eIF4E is sufficient to induce transformation of immortalized murine fibroblasts and human epithelial cells (Lazaris-Karatzas et al. 1990; Avdulov et al. 2004). Constitutive overexpression of eIF4E in a mouse model that mimics the human oncogenic lesion leads to increased cancer susceptibility; eIF4E transgenic mice develop lymphomas, angiosarcomas, lung carcinomas, and hepatomas (Ruggero et al. 2004). Furthermore, in vivo overexpression of eIF4E cooperates with c-Myc to drive lymphomagenesis in part via a mechanism by which eIF4E overcomes Myc-induced apoptosis, a cellular barrier to tumor formation (Ruggero et al. 2004; Wendel et al. 2004). Phosphorylation of eIF4E also plays an important role in cancer formation. In fact, by using an adoptive transfer method in vivo, it has been shown that phosphorylated eIF4E contributes to Myc-induced tumorigenesis mainly by suppressing apoptosis (Wendel et al. 2007). In addition, whole body expression of a knockin mutant of eIF4E, which can no longer be phosphorylated at the serine 209 residue, was found to decrease the incidence and grade of prostatic intraepithelial neoplasia in a mouse prostate cancer model driven by PTEN loss (Furic et al. 2010).
Although these findings support the notion that eIF4E is a bona fide oncogene, they do not address the extent to which specific oncogenic signaling (i.e., PI3K-AKT-mTOR pathway) relies on eIF4E translational activity for tumor development. Some of the first evidence for such a connection came from a study showing that pharmacological inhibition of oncogenic Ras and AKT in glioblastoma cells caused a rapid and profound change in mRNA translation that far outweighed transcriptional changes. These translational changes were associated with loss of mTORC1-dependent phosphorylation of 4E-BPs (Rajasekhar et al. 2003). In addition, transfection of a 4E-BP1 phosphorylation site mutant into breast carcinoma cells suppressed their tumorigenicity by inducing apoptosis (Avdulov et al. 2004). It has recently been shown in vitro and in vivo that the 4E-BPs/eIF4E axis exerts significant control over cap-dependent translation, cell growth, cancer initiation, and progression downstream of mTOR hyperactivation (Dowling et al. 2010; Hsieh et al. 2010). Most importantly, restoring eIF4E oncogenic activity to normal levels downstream of AKT-mTOR hyperactivation results in blockage of tumor progression in a mouse model for lymphomagenesis associated with a drastic increase in overall survival (Hsieh et al. 2010).
Do these findings indicate that cells transformed by mTOR hyperactivation become “addicted” to oncogenic eIF4E-mediated translation control? Oncogenic addiction describes a tumor’s dependence on specific oncogenic lesions for its survival and growth. This represents a potential “Achilles heel” in tumor formation by which inactivation of a specific oncogene can lead to apoptosis, senescence, and tumor regression (Weinstein 2000, 2002; Weinstein and Joe 2006). The importance of this concept resides in the fact that although cancer cells harbor many oncogenic lesions, targeting the mediators of oncogenic addiction would herald a landmark improvement in cancer therapy. Although the mechanisms underlying this “Achilles heel” are poorly understood, it is tempting to speculate that the 4E-BPs/eIF4E axis may represent a novel node of oncogenic addiction to target for therapeutic intervention (see below). At the mechanistic level, targeting eIF4E activity in cancer cells can be a key to initiate a cell death program resulting in tumor regression. In support of this hypothesis, it has been shown that eIF4E hyperactivation is able to enhance the translation of select mRNAs (De Benedetti and Graff 2004), including some encoding antiapoptotic factors (Mamane et al. 2007). The 5′ UTR of these mRNAs are believed to harbor regulatory elements, such as complex secondary structures, that impart this selectivity (Fig. 2). One important example is Mcl-1, an antiapoptotic factor containing a complex 5′ UTR that is translationally up-regulated specifically upon eIF4E hyperactivation, leading to enhanced survival of cancer-initiating cells (Mills et al. 2008; Hsieh et al. 2010). Therefore, cancer cells may be addicted to eIF4E-promoted survival, which would serve as a promising target for therapeutic intervention.
Regulation of eIF4E is not the only node in which information from signaling pathways is received by the translational machinery. It is now clear that an entire repertoire of translational components may be co-opted to promote cancer initiation. For example, AKT hyperactivation also modulates translation elongation (Fig. 3; Table 1) (Wang et al. 2001). Additional regulated translational components include eIF2α, which is part of the ternary complex required to chaperone the initiator transfer RNA (tRNA) to the ribosome. Phosphorylation of eIF2α by several kinases including the double-stranded RNA-dependent protein kinase (PKR) and the endoplasmic reticulum kinase PERK leads to consequent inhibition of protein synthesis and this is a major cell growth checkpoint (Ron and Harding 2007). Disruption of the eIF2α checkpoint can lead to the transformation of immortalized rodent and human cells (Koromilas et al. 1992b; Meurs et al. 1993; Barber et al. 1995; Donze et al. 1995). However, prevention of eIF2α phosphorylation is not tumorigenic in vivo (Yang et al. 1995; Abraham et al. 1999; Scheuner et al. 2001). In addition, it has been shown that the PERK-eIF2α axis represents a survival pathway for cancer cells that need to adapt and overcome hypoxic stress during tumor progression (Fels and Koumenis 2006). Transformed cells with inactivating PERK or eIF2α mutations form smaller tumors that grow more slowly in nude mice that grow more slowly, and show higher levels of apoptosis in hypoxic areas compared to control cells with intact eIF2α phosphorylation (Bi et al. 2005). eIF2α is also commonly overexpressed in cancers and may thereby provide an uncontrolled stimulus leading to increased rates of protein synthesis (Wang et al. 1999). Interestingly, overexpression of the initiator tRNA itself is able to drive cellular transformation (Marshall et al. 2008). Another translation factor that promotes cellular transformation is eIF6, which regulates the joining of the 60S ribosomal subunit to the 48S preinitiation complex to begin translation (Gandin et al. 2008). Importantly, eIF6 has been shown to be rate limiting for translation, cell growth, and transformation (Gandin et al. 2008). This initiation factor also interacts with RACK1, a ribosome-associated scaffolding protein that coordinates signaling by PKC and src kinases (Ceci et al. 2003). Therefore, signaling through RACK1 to eIF6 may be another key node of oncogenic regulation.
Although a wealth of research (described above), including elegant genetic studies, has shown that eIF4E hyperactivation and other nodes of translational control play a key role in cancer development, the repertoire of mRNAs that are specifically sensitive to translational perturbations is not completely understood. There are now emerging technologies that may facilitate their identification. In particular, the ability to deep-sequence ribosome-protected mRNAs will enable codon-by-codon resolution of ribosome occupancy on specific mRNAs (Ingolia et al. 2009). Furthermore, through deep sequencing, it is now possible to determine the secondary structures of mRNAs by using a novel strategy termed parallel analysis of RNA structures (PARS) (Kertesz et al. 2010). The combination of these two technologies may provide a very accurate portrait of how mRNA secondary structures control the translation of the cancer genome. Furthermore, techniques such as high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) and affinity RNA purification followed by mass spectrometry have successfully identified cis-regulatory elements and specific regulatory factors associated with RNAs (Ji et al. 2004; Darnell et al. 2011; Tsai et al. 2011; Darnell and Richter 2012). Such technologies could be used to characterize the mRNAs and the translational complex directly bound to eIFs such as eIF3, eIF4G, eIF5B, and eIF4A. Undertaking these approaches in a time course manner during the different steps in tumorigenesis would be valuable in identifying novel translational regulatory networks and might help to reveal the interplay between these networks during the stages of tumor development. Fluorescence-based living cell approaches such as fluorescence resonance energy transfer (FRET) could also enable the analysis of molecular interactions of a specific translational complex in real time during cellular transformation (Huranova et al. 2009; Lorenz 2009). Simply analyzing a static complex paints an incomplete picture. By studying the dynamic assembly and disassembly of these regulatory nodes, we could gain crucial insight into their ability to promote distinct translation initiation modes, ultimately contributing to cancer development.
Why Has the Translational Control of Specific mRNAs Been Selected for Cancer Development?
To consider this question, it is perhaps important to consider mechanisms that drive cancer evolution, whereby altering specific cellular processes may lead to uncontrolled growth. In this respect, perturbing translational control could provide a rapid change in gene expression upon environmental cues that allows the cancer cell to adapt and survive even under unfavorable conditions.
But how does the cancer program control specific mRNAs to support the growth of cancer cells? A central principle emerging from detailed molecular studies is that important regulatory elements (e.g., IRES, complex 5′ and 3′ UTRs, RNA-binding proteins, and microRNA sites) may render mRNAs encoding key tumor suppressors and protooncogenes (including cell-cycle and survival factors) exquisitely vulnerable to any perturbations in translational control (Fig. 2). Many of these mRNAs possess long 5′ UTRs that contain upstream ORFs and/or stable secondary structures (Stoneley and Willis 2003; Pickering and Willis 2005; Sonenberg and Hinnebusch 2009). These specific cis features act as inhibitory elements that interfere with the activity of the eIF4F initiation complex and ribosome scanning, thereby maintaining the translational efficiency of these mRNAs at a level that does not exceed a threshold necessary for cellular homeostasis. Importantly, these mRNAs are aberrantly translated as a consequence of overexpressed eIF4E or the other components of the eIF4F complex, which overcome the translational inhibitory effects imposed by their 5′ UTR cis elements (Fig. 2) (Koromilas et al. 1992a; Stoneley and Willis 2003; Pickering and Willis 2005; Ruggero and Sonenberg 2005; Mills et al. 2008; Hsieh et al. 2010). In addition, overexpression of RNA-binding proteins such as CPEB4 has been shown to increase the translation of specific oncogenic mRNAs by binding to cis elements present in their 3′ UTRs (Ortiz-Zapater et al. 2011; for further review, please see van Kouwenhove et al. 2011). Furthermore, the sequence and structural integrity of the 5′ and 3′ UTRs in the p53 tumor suppressor mRNA are required for its translation (Fig. 2) (Chen and Kastan 2010).
Do genetic perturbations in mRNA regulatory elements directly lead to cancer formation? This is an intriguing line of research that remains to be fully explored. Alterations in the 5′ UTR of MDM2, δ-catenin, or Myc oncogenes increase the translational level of their mRNAs in multiple myeloma, Burkitt’s lymphoma, and prostate cancer (Paulin et al. 1996; Landers et al. 1997; Capoulade et al. 1998; Stoneley et al. 1998; Brown et al. 1999; Jin et al. 2003; Wang et al. 2009b). A mutation in the 5′ UTR of the CDKN2a tumor suppressor (also known as p16INK4A) is associated with increased susceptibility to melanoma. This mutation creates an aberrant upstream translation initiation codon impairing translation initiation efficiency from the wild-type AUG (Liu et al. 1999). Additionally, cancer-derived mutations in the p53 gene may alter an IRES element present in the 5′ UTR of its mRNA decreasing p53 protein expression (Grover et al. 2011).
Altogether, the remarkable repertoire of deregulated translational components, whose activity is directly controlled downstream of specific oncogenic signals, strongly supports their critical and causal role in cancer initiation and progression. What also emerges from these studies is that perturbations in translational control provide highly specific outcomes for gene expression that impinge on distinct steps along the pathway toward cancer development.
EXAMINING THE MECHANISMS BY WHICH THE TRANSLATIONAL MACHINERY PROMOTES THE MULTISTEP PROCESSES OF CELLULAR TRANSFORMATION AND TUMOR DEVELOPMENT
Cellular Transformation and Tumor Initiation
Increases in cell mass (cell growth), which occur mainly in the G1 phase of the cell cycle, is a prerequisite for accurate cell division (Fig 1) (Hall et al. 2004). Ribosome biogenesis and global protein synthesis are tightly and dynamically regulated to accommodate the growth demands of a cell (Hall et al. 2004). Therefore, an important relationship exists between the cell cycle, ribosome production, and translational control. This balance is maintained in the cell through key checkpoints. For example, down-regulation of ribosome formation and activity is required during M phase to ensure proper cytokinesis (Pyronnet et al. 2001; Boisvert et al. 2007; Barna et al. 2008; Marash et al. 2008; Sivan et al. 2011). As such, it appears that a translational program that interfaces with the cell-cycle machinery should ensure the translation of specific mRNAs at appropriate levels during each window of cell growth and division. However, we have yet to determine which portion of the genome is translationally regulated during each phase of the cell cycle.
In cancer cells the balance between growth and division is broken, leading to unrestrained increases in protein synthesis and cell size (Fig. 1). Studies on the oncogenic activity of Myc provide an excellent window for understanding this mechanism of cellular transformation and tumor initiation. The Myc transcription factor, which is commonly deregulated in human cancers, is an exquisite regulator of ribosome biogenesis, protein synthesis, and cell growth (Ruggero 2009; van Riggelen et al. 2010). Myc directly increases protein synthesis rates by controlling the expression of multiple components of the protein synthetic machinery, including ribosomal proteins, initiation factors, Pol III, and recombinant DNA (rDNA) (Fernandez et al. 2003; Gomez-Roman et al. 2003; Grandori et al. 2005; Zeller et al. 2006). Through its transcriptional regulation of the translational apparatus, Myc activity leads to increased cell size often associated with changes in nucleolar architecture (Fig. 1) (Kim et al. 2000; Arabi et al. 2003; Grandori et al. 2005; Grewal et al. 2005; Shiue et al. 2010). Interestingly, one of the initial markers for cancer cells, discovered more than 100 years ago, is an increase in the size and number of nucleoli (Pianese 1896). However, whether the increases in ribosome biogenesis and cell growth are a direct cause of cellular transformation or, as many researchers have long assumed, a consequence of the need to sustain elevated rates of proliferation in tumor development, remains a critical and challenging question. This query has been in part addressed with novel genetic strategies that restore aberrant protein synthesis to normal levels before tumor formation in Myc transgenic mice. These studies showed that the ability of Myc to increase protein synthesis directly augments cell growth (Ruggero 2009). Is it possible that, on its own, increased cell growth represents one of the oncogenic “hits” that leads to cellular transformation? For example, cell growth may provide a significant competitive advantage to a preneoplastic cell in a specific tissue type. In this situation, a larger cell may outcompete neighboring normal cells in uptake of nutrients and growth factors. Consequently, a somatic clone harboring such a cancerous lesion may possess an early competitive advantage. This advantage may result in cell death or starvation of normal cells and the destruction of the organ boundary concomitantly with the expansion of the neoplastic clone.
Aberrant cell growth due to oncogene-dependent increases in protein synthesis rates may directly accelerate cell-cycle progression. As mentioned above, the two processes are intimately linked. In this case, the oversized cells may be more susceptible to entering unscheduled cell division, as an aberrant increase in cell growth may mimic the normal G1 phase of the cell cycle (Fig. 1). This hypothesis may explain why restricting protein synthesis downstream of Myc results in restoration of normal cell growth as well as cell division (Barna et al. 2008). Importantly, Myc’s oncogenic potential is dependent on its ability to up-regulate protein synthesis (Barna et al. 2008). Other recent genetic studies have provided compelling evidence that the manipulation of the translational machinery by Myc is integral to its oncogenic activity. Inactivation of one copy of eIF6 limits Myc-driven lymphomagenesis (Miluzio et al. 2011). Furthermore, Myc also synergizes with AKT to regulate Pol I activity, another important determinant of cell growth and cancer development (Chan et al. 2011). The significance of Pol I for cancer therapy is underscored by the fact that Cylene Pharmaceuticals has developed two potent and specific inhibitors of this RNA polymerase, which show antitumor activity in xenograft models (Drygin et al. 2009, 2011).
However, we have yet to understand the exact molecular mechanisms by which oncogenic signaling, affecting ribosome biogenesis and translational control, couples cell growth with cell division during cellular transformation. For example, we do not know whether an increase in ribosome number and/or nucleolar size, as a consequence of Myc overexpression, sends a “signal” to the cell division machinery to facilitate cell-cycle progression, or whether the translational program downstream of oncogenic activation directly changes the expression of key components of cell growth and division. In this latter scenario, it stands to reason that specific mRNAs (e.g., cyclins or replication enzymes) would be more sensitive to translational perturbation, representing key translational nodes for controlling cell growth and division.
Increased cell division as a consequence of increased cell growth is not sufficient to induce cellular transformation. Indeed, even upon aberrant cell-cycle progression, a critical cellular response can counteract cellular transformation. This response is known as oncogene-induced senescence (OIS) (Fig. 1). OIS is characterized by cell-cycle arrest and induction of cell-cycle inhibitors such as p15, p16, and p53, which restrain the proliferative potential of preneoplastic clones (Serrano et al. 1997; Gil and Peters 2006). Importantly, during OIS, a switch between cap- and IRES-dependent translation occurs (Bellodi et al. 2010a). During this switch, an IRES element positioned in the 5′ UTR of p53 is engaged to promote p53 translation (Bellodi et al. 2010a). Therefore specialized translational control of mRNAs such as p53 provides a molecular barrier for cellular transformation (Takagi et al. 2005; Ray et al. 2006; Bellodi et al. 2010a; Montanaro et al. 2010). Interestingly, defects in rRNA modifications specifically perturb p53 IRES-dependent translation, resulting in defective OIS and expansion of preneoplastic clones (Bellodi et al. 2010a). This may be a critical mechanism that underlies cancer susceptibility in X-DC, discussed above (Bellodi et al. 2010a; Montanaro et al. 2010). Most recently, another translational control mechanism that modulates p53 expression in the senescence phenotype has been discovered. In this case, p53 mRNA polyadenylation/translation is hierarchically regulated by the interplay between noncanonical poly(A) polymerases, cytoplasmic polyadenylation element-binding protein (CPEB), and miR-122 (Burns and Richter 2008; Burns et al. 2011). Ultimately, alterations in translational control of p53 mRNAs would appear to be an essential step leading to transformation.
A subsequent step to overcoming tumor-suppressive barriers such as OIS, which is important to achieve full cellular transformation, is the acquisition of additional genetic lesions. These lesions are commonly referred to as secondary hits (Fig. 1). However, the mechanisms by which alterations in translational control results in the accumulation of such hits remain poorly understood. We have recently learned that deregulations in mitotic translational control play an important role at this step by promoting genomic instability (Wilker et al. 2007; Barna et al. 2008). During mitosis, only a small fraction of mRNA is translated in a cap-independent manner (Pyronnet et al. 2000; Qin and Sarnow 2004; Komar and Hatzoglou 2011). CDK11/p58 is a well-characterized endogenous mRNA that is only translated during mitosis by an IRES element (Cornelis et al. 2000; Wilker et al. 2007). Distinct ITAFs, including PTB (polypyrimidine tract-binding protein) and UNR (upstream of N-RAS), have been shown to regulate CDK11/p58 expression (Tinton et al. 2005; Ohno et al. 2011). In the context of an oncogenic lesion such as Myc hyperactivation, an aberrant increase in cap-dependent translation specifically impairs the mitotic translational switch from cap- to IRES-dependent translation (Fig. 1). This results in reduced expression of CDK11/p58, which leads to cytokinesis defects associated with increased centrosome numbers and genome instability (Barna et al. 2008). Given that deregulations in mitotic translational control directly promote genome instability, further studies will be important to illuminate the precise relationship between specific genetic lesions such as Myc and the altered “oncogenic” translational machinery.
Progression and Metastasis
Once a tumor has been established it will progress through a number of stages and may ultimately become metastatic (Fig. 1). The functional role of translational control during these stages of tumorigenesis is only now beginning to be explored. Indeed, much of the data to date are derived primarily from expression analysis of translation factors in fixed human samples. For example, the incidences of eIF5A2 overexpression increase in human colorectal tumor specimens from benign tumors to primary carcinomas to metastatic lesions (Xie et al. 2008). The expression and activity of eIF4E has also been associated with malignant progression in many human cancers including prostate, head and neck, bladder, colon, breast, lung, and lymphoma (De Benedetti and Graff 2004; Graff et al. 2009). In addition, hyperactivation of specific signal transduction pathways (see above) appears to directly increase translation factor activity during tumor progression. Nevertheless, we still have an incomplete understanding of the precise impact of translational control in the development of metastatic features in cancer cells. Unraveling the mechanistic basis of this process will shed light on new aspects of translational control and help identify novel targets for cancer therapy. In this respect, it appears that tumor cells have co-opted specific modes of translational regulation to express key survival factors for their continued development. For example, during cancer progression, tumor cells may survive stress conditions such as nutrient and oxygen deprivation through their ability to promote cap-independent translation of specific antiapoptotic factors such as Bcl-2 and XIAP (Fig. 1). In addition, IRES-dependent translation of the neoangiogenic factor VEGF promotes tumor size by enhancing blood flow to the tumor (Holcik and Korneluk 2000; Sherrill et al. 2004; Silvera et al. 2009; Silvera and Schneider 2009). Overexpression of eIF4G and 4E-BP1 is functionally linked to the promotion of IRES-dependent mRNA translation during cancer progression (Silvera et al. 2009).
Why is translational control of specific mRNAs important for the coordination of cell invasion and metastasis? To address this question, it is useful to consider the common features underlying these tumorigenic steps. For instance, cancer cells need to maintain a high capacity for migration and invasion to bypass tissue barriers, intravasate into the blood stream, and extravasate at distal secondary sites (Valastyan and Weinberg 2011). This multistep process requires rapid and specific modulation of gene expression; in this context, it is conceivable that translational control plays an important role. In support of this hypothesis, recent studies have shown that eIF4E and eIF4GI were associated with the Golgi apparatus and membrane microdomains such as the perinuclear region and lamellipodia, which are essential for cell motility. Most importantly, a fraction of these initiation factors localizes at sites of active translation near the leading edge of migrating cells (Willett et al. 2011). It is tempting to speculate that this phenomenon may represent a rapid mechanism to achieve a critical threshold of migratory proteins surrounding the leading edge of the cell. This localized translation may also facilitate the secretion of key proteins such as metalloproteases, which are necessary to degrade the basement membrane, an obligatory step before the cancer cells enter the bloodstream. Therefore, this spatial increase in translational control at the leading edge could aid the invasion of cancer cells from the primary tumor to secondary sites.
Another cellular process associated with increased metastatic potential is known as epithelial-to-mesenchymal transition (EMT), in which epithelial cancer cells decrease their epithelial properties and acquire mesenchymal features (Fig. 1B) (Floor et al. 2011; Valastyan and Weinberg 2011). This process is associated with increases in cell motility and changes in cell–cell or cell–matrix adhesion. Importantly, very recent studies using ribosome profiling have delineated at a codon-by-codon resolution that oncogenic mTOR signaling has a striking effect on the translational landscape of the cancer genome of metastatic cells (Hsieh et al. 2012). This work has functionally characterized a novel mRNA signature associated with cancer cell invasion and metastasis in vivo. This signature is comprised of vimentin, an intermediate filament protein highly up-regulated during EMT (Lahat et al. 2010); MTA1 (metastasis associated 1), a putative chromatin-remodeling protein that has been shown to drive cancer metastasis by promoting neoangiogenesis (Yoo et al. 2006); CD44, commonly overexpressed in tumor-initiating cells and is implicated in cancer metastasis (Liu et al. 2011); and YB-1 (Y-box binding protein), also called YBX1. Interestingly, it has been recently shown that the YB-1 protein promotes the cap-independent translation of specific subsets of mRNAs encoding key proteins underlying EMT (Evdokimova et al. 2009). Mechanistically, eIF4E regulates the translation of these mRNAs encoding key players in cancer metastasis at least in part through a novel regulatory element present in their 5′ UTR known as pyrimidine-rich translation element (PRTE) (Hsieh et al. 2012). Significantly, a novel clinical ATP-site inhibitor of mTOR (INK128, see below) exquisitely targets this translational signature with therapeutic benefit at all stages of prostate cancer progression, including metastasis (Hsieh et al. 2012). However, the identity and number of the key components of the translational apparatus that may regulate metastatic potential demand further investigation. Furthermore, both the repertoire of translationally regulated mRNAs that contributes to tumor invasion and metastatic dissemination, and the mechanisms that regulate their expression, remain an exciting frontier that merits further exploration.
FROM THE BENCH TO THE PATIENT: CLINICAL-“TRANSLATIONAL” ADVANCES
After key findings establishing the causal role of the deregulated translational machinery within the multihit model of cancer etiology, we are arriving at an exciting crossroad between discovery and clinical application. I will briefly summarize some of the main preclinical and clinical strategies that directly or indirectly target the protein synthesis apparatus (Hsieh and Ruggero 2010; see also Malina et al. 2012). The eIF4E oncogene, which is frequently hyperactivated in human cancer, represents an attractive target for rational drug design (Fig. 3). There are currently several approaches being pursued to therapeutically inhibit eIF4E, but perhaps the most direct of these approaches is the use of specific antisense oligonucleotides (ASOs) that bind to eIF4E mRNA and mediate its destruction by RNase H. The eIF4E-ASO is currently being tested in combination with chemotherapy in new phase I/II clinical trials of metastatic lung cancer and prostate cancer (Graff et al. 2007). Additional attempts to target eIF4E have focused on blocking its ability to interact with eIF4G. The interaction between eIF4E and eIF4G is dependent on an eIF4G Y(X)4LΦ motif, in which X is variable and Φ is hydrophobic (Marcotrigiano et al. 1999). High-throughput screens for inhibitors that could prevent eIF4E from binding to the Y(X)4LΦ motif identified 4EGI-1 as a candidate compound. 4EGI-1 is both a cytotoxic and cytostatic agent acting across multiple cell lines. Importantly, 4EGI-1 inhibits proliferation and clonogenic growth of transformed cells more effectively than untransformed cells (Moerke et al. 2007; Tamburini et al. 2009). 4EGI-1 is licensed to Eugenix, which is working toward an investigational new drug application for this compound (G Wagner, pers. comm.). Concurrently, indirect approaches to target oncogenic eIF4E activity are proving particularly effective (Hsieh et al. 2011). For example, inhibiting the mTOR kinase holds tremendous promise for the clinic. In vitro and in vivo characterizations of recently developed mTOR ATP-site inhibitors have shown greater efficacy than allosteric mTOR inhibitors (rapalogs) (see also Malina et al. 2012; Roux and Topisirovic 2012). Unlike rapalogs, mTOR ATP-site inhibitors target 4E-BP phosphorylation as well as the mTORC2 complex (Benjamin et al. 2011). PP242, the first of a series of reported ATP-site mTOR inhibitors (Feldman et al. 2009), has been used in a preclinical trial for AKT-driven lymphomagenesis (Hsieh et al. 2010). In this study, PP242 (but not rapamycin) suppressed tumor growth by inhibiting 4E-BP1 phosphorylation. In line with these studies, the antiproliferative effect of both PP242 and Torin (another ATP-site inhibitor) was attenuated in cultured 4E-BP null cells (Thoreen et al. 2009; Dowling et al. 2010). Altogether, these findings strongly suggest that the therapeutic efficacy of ATP-site inhibitors may be in large part associated with their ability to block mTORC1-dependent 4E-BP phosphorylation and eIF4E oncogenic activity (Hsieh et al. 2010). Perhaps most notably, INK128, a more potent derivative of PP242, as well as several other ATP-site inhibitors, are currently in phase I/II clinical trials in patients with advanced solid tumors and hematological malignancies (Benjamin et al. 2011; Hsieh et al. 2012).
Equally significant for the development of future cancer treatments, recent studies have shown that amplification of the eIF4E gene leads to drug resistance in immortalized human mammary epithelial cells (HMECs) that were treated with the dual PI3K/mTOR inhibitor, BEZ235 (Ilic et al. 2011). Specifically, BEZ235 elicited a cytostatic effect only in HMECs that do not display eIF4E amplification and the associated increases in cap-dependent translation (Ilic et al. 2011). Therefore, deregulation in translational control may also represent a mechanism for drug resistance. For example, increases in expression and activity of distinct translational components might evade the inhibitory activity of compounds that target key upstream regulators of the translational machinery.
It remains to be seen how the deregulation of other translation components, in addition to eIF4E, may act either as therapeutic targets or mechanisms of resistance to mTOR inhibitors. Ultimately, to understand these contributions, we need to characterize the translationally deregulated mRNAs downstream of these events.
CONCLUDING REMARKS
It is clear that the study of deregulations in the translational machinery has moved into the center stage of cancer research. Indeed, even if many questions remain to be addressed, we are now closer than ever to understanding the causal relationship between alterations in translational control and the etiology of cancer development. The critical function of translational control in cancer is underscored by the discovery of mutations in translational components underlying genetic syndromes associated with cancer susceptibility. In addition, several studies have described gene amplification of distinct key regulators of protein synthesis in somatic cancers (Table 1). Furthermore, there is a growing appreciation for the exquisite specificity in translational regulation mediated by the core components of the translational apparatus. For example, the unexpected discovery that only specific mRNAs harboring IRES elements are translationally affected by impairments in rRNA modifications has changed our perception of how the translation machinery regulates gene expression. Additionally, mutant ribosomes lacking important constituents such as ribosomal proteins may be largely functionally active, but preferentially defective in specific aspects of translational control, contributing to cancer development.
Altering the translational landscape of the cancer genome appears to be a key driver in tumor formation associated with hyperactivation of signal transduction pathways. Indeed, distinct oncogenic signals appear to monopolize translational control at almost every stage of cancer initiation and development, resulting in specific and distinct cellular outcomes (Fig. 1). Important studies have also uncovered a surprising level of specificity for controlling gene expression at the posttranscriptional level downstream of the activation of these oncogenic signals. This translational specificity is also dictated by distinct regulatory elements within key mRNAs, such as IRES elements or structured 5′ UTRs, which control the expression of these mRNAs during specific steps of cellular transformation and tumor development (Fig. 2). These translational control mechanisms would provide cancer cells ample opportunity to grow, survive, and expand indefinitely even under unfavorable conditions. Future research can build on these recent significant findings to elucidate the dynamic relationship between the regulatory components of the translational machinery and oncogenic signaling. Taken as a whole, alterations in translational control may represent an “oncogenic addiction” node. Such a node could serve as a potential “Achilles heel” for tumor suppression (Fig. 3). Ultimately, the significance of this research is further reflected by a number of novel and promising therapeutic approaches to target specific translational components in cancer that are presently in clinical trials.
ACKNOWLEDGMENTS
I am indebted to Kimhouy Tong for all her help with this article. I am grateful to Maria Barna and Craig Stumpf and the other members of the Ruggero Laboratory for many helpful discussions and for critically reviewing this manuscript. I apologize to those whose work I was unable to cite. This work is supported by National Institutes of Health grants R01 HL085572, R01 CA140456, and R01 CA154916, and Phi Beta Psi Sorority. Davide Ruggero is a Leukemia and Lymphoma Society Research Scholar.
Footnotes
Editors: John W.B. Hershey, Nahum Sonenberg, and Michael B. Mathews
Additional Perspectives on Protein Synthesis and Translational Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abraham N, Stojdl DF, Duncan PI, Methot N, Ishii T, Dube M, Vanderhyden BC, Atkins HL, Gray DA, McBurney MW, et al. 1999. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem 274: 5953–5962 [DOI] [PubMed] [Google Scholar]
- Ahlemann M, Zeidler R, Lang S, Mack B, Munz M, Gires O 2006. Carcinoma-associated eIF3i overexpression facilitates mTOR-dependent growth transformation. Mol Carcinog 45: 957–967 [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N 2004. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol 2: E139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand N, Murthy S, Amann G, Wernick M, Porter LA, Cukier IH, Collins C, Gray JW, Diebold J, Demetrick DJ, et al. 2002. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat Genet 31: 301–305 [DOI] [PubMed] [Google Scholar]
- Arabi A, Rustum C, Hallberg E, Wright AP 2003. Accumulation of c-Myc and proteasomes at the nucleoli of cells containing elevated c-Myc protein levels. J Cell Sci 116: 1707–1717 [DOI] [PubMed] [Google Scholar]
- Austin KM, Leary RJ, Shimamura A 2005. The Shwachman-Diamond SBDS protein localizes to the nucleolus. Blood 106: 1253–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin KM, Gupta ML, Coats SA, Tulpule A, Mostoslavsky G, Balazs AB, Mulligan RC, Daley G, Pellman D, Shimamura A 2008. Mitotic spindle destabilization and genomic instability in Shwachman-Diamond syndrome. J Clin Invest 118: 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman PB, et al. 2004. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell 5: 553–563 [DOI] [PubMed] [Google Scholar]
- Bachmann F, Banziger R, Burger MM 1997. Cloning of a novel protein overexpressed in human mammary carcinoma. Cancer Res 57: 988–994 [PubMed] [Google Scholar]
- Balabanov S, Gontarewicz A, Ziegler P, Hartmann U, Kammer W, Copland M, Brassat U, Priemer M, Hauber I, Wilhelm T, et al. 2007. Hypusination of eukaryotic initiation factor 5A (eIF5A): A novel therapeutic target in BCR-ABL-positive leukemias identified by a proteomics approach. Blood 109: 1701–1711 [DOI] [PubMed] [Google Scholar]
- Barber GN, Wambach M, Thompson S, Jagus R, Katze MG 1995. Mutants of the RNA-dependent protein kinase (PKR) lacking double-stranded RNA binding domain I can act as transdominant inhibitors and induce malignant transformation. Mol Cell Biol 15: 3138–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JL, Drynan LF, Trim NL, Erber WN, Warren AJ, McKenzie AN 2010. New insights into 5q- syndrome as a ribosomopathy. Cell Cycle 9: 4286–4293 [DOI] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D 2008. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456: 971–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Das P, Chaudhuri S, Bevilacqua E, Andrews J, Barik S, Hatzoglou M, Komar AA, Mazumder B 2011. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular internal ribosome entry sites. Mol Cell Biol 31: 4482–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C, Diesinger I, Brass N, Steinhart H, Iro H, Meese EU 2001. Translation initiation factor eIF-4G is immunogenic, overexpressed, and amplified in patients with squamous cell lung carcinoma. Cancer 92: 822–829 [DOI] [PubMed] [Google Scholar]
- Bauer C, Brass N, Diesinger I, Kayser K, Grasser FA, Meese E 2002. Overexpression of the eukaryotic translation initiation factor 4G (eIF4G-1) in squamous cell lung carcinoma. Int J Cancer 98: 181–185 [DOI] [PubMed] [Google Scholar]
- Belin S, Beghin A, Solano-Gonzalez E, Bezin L, Brunet-Manquat S, Textoris J, Prats AC, Mertani HC, Dumontet C, Diaz JJ 2009. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS ONE 4: e7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, Kopmar N, Ruggero D 2010a. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J 29: 1865–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L, Ruggero D 2010b. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res 70: 6026–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin D, Colombi M, Moroni C, Hall MN 2011. Rapamycin passes the torch: A new generation of mTOR inhibitors. Nat Rev Drug Discov 10: 868–880 [DOI] [PubMed] [Google Scholar]
- Berkel HJ, Turbat-Herrera EA, Shi R, de Benedetti A 2001. Expression of the translation initiation factor eIF4E in the polyp-cancer sequence in the colon. Cancer Epidemiol Biomarkers Prev 10: 663–666 [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, et al. 2005. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 24: 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI 2007. The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM 2003. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet 33: 97–101 [DOI] [PubMed] [Google Scholar]
- Brass N, Heckel D, Sahin U, Pfreundschuh M, Sybrecht GW, Meese E 1997. Translation initiation factor eIF-4γ is encoded by an amplified gene and induces an immune response in squamous cell lung carcinoma. Hum Mol Genet 6: 33–39 [DOI] [PubMed] [Google Scholar]
- Brown CY, Mize GJ, Pineda M, George DL, Morris DR 1999. Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene 18: 5631–5637 [DOI] [PubMed] [Google Scholar]
- Burns DM, Richter JD 2008. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev 22: 3449–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns DM, D’Ambrogio A, Nottrott S, Richter JD 2011. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature 473: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwick N, Shimamura A, Liu JM 2011. Non-Diamond Blackfan anemia disorders of ribosome function: Shwachman Diamond syndrome and 5q- syndrome. Semin Hematol 48: 136–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoulade C, Bressac-de Paillerets B, Lefrere I, Ronsin M, Feunteun J, Tursz T, Wiels J 1998. Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt’s lymphoma cells. Oncogene 16: 1603–1610 [DOI] [PubMed] [Google Scholar]
- Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, Biffo S 2003. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426: 579–584 [DOI] [PubMed] [Google Scholar]
- Challagundla KB, Sun XX, Zhang X, DeVine T, Zhang Q, Sears RC, Dai MS 2011. Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol Cell Biol 31: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JC, Hannan KM, Riddell K, Ng PY, Peck A, Lee RS, Hung S, Astle MV, Bywater M, Wall M, et al. 2011. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Sci Signal 4: ra56. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S, Vyas K, Kapasi P, Komar AA, Dinman JD, Barik S, Mazumder B 2007. Human ribosomal protein L13a is dispensable for canonical ribosome function but indispensable for efficient rRNA methylation. RNA 13: 2224–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kastan MB 2010. 5′-3′-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev 24: 2146–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement PM, Johansson HE, Wolff EC, Park MH 2006. Differential expression of eIF5A-1 and eIF5A-2 in human cancer cells. FEBS J 273: 1102–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D 2007. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat 28: 1178–1182 [DOI] [PubMed] [Google Scholar]
- Cmejla R, Cmejlova J, Handrkova H, Petrak J, Petrtylova K, Mihal V, Stary J, Cerna Z, Jabali Y, Pospisilova D 2009. Identification of mutations in the ribosomal protein L5 (RPL5) and ribosomal protein L11 (RPL11) genes in Czech patients with Diamond-Blackfan anemia. Hum Mutat 30: 321–327 [DOI] [PubMed] [Google Scholar]
- Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R 2000. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell 5: 597–605 [DOI] [PubMed] [Google Scholar]
- Crew JP, Fuggle S, Bicknell R, Cranston DW, de Benedetti A, Harris AL 2000. Eukaryotic initiation factor-4E in superficial and muscle invasive bladder cancer and its correlation with vascular endothelial growth factor expression and tumour progression. Br J Cancer 82: 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Arnold H, Sun XX, Sears R, Lu H 2007. Inhibition of c-Myc activity by ribosomal protein L11. EMBO J 26: 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, Lin S 2008. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood 112: 5228–5237 [DOI] [PubMed] [Google Scholar]
- *.Darnell JC, Richter JD 2012. Cytoplasmic RNA binding proteins and the control of complex brain function. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146: 247–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti A, Graff JR 2004. eIF-4E expression and its role in malignancies and metastases. Oncogene 23: 3189–3199 [DOI] [PubMed] [Google Scholar]
- Dellas A, Torhorst J, Bachmann F, Banziger R, Schultheiss E, Burger MM 1998. Expression of p150 in cervical neoplasia and its potential value in predicting survival. Cancer 83: 1376–1383 [DOI] [PubMed] [Google Scholar]
- Dennis PB, Pullen N, Kozma SC, Thomas G 1996. The principal rapamycin-sensitive p70(s6k) phosphorylation sites, T-229 and T-389, are differentially regulated by rapamycin-insensitive kinase kinases. Mol Cell Biol 16: 6242–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty L, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Clinton C, Schneider HE, Sieff CA, Newburger PE, Ball SE, et al. 2010. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet 86: 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doldan A, Chandramouli A, Shanas R, Bhattacharyya A, Cunningham JT, Nelson MA, Shi J 2008a. Loss of the eukaryotic initiation factor 3f in pancreatic cancer. Mol Carcinog 47: 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doldan A, Chandramouli A, Shanas R, Bhattacharyya A, Leong SP, Nelson MA, Shi J 2008b. Loss of the eukaryotic initiation factor 3f in melanoma. Mol Carcinog 47: 806–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze O, Jagus R, Koromilas AE, Hershey JW, Sonenberg N 1995. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J 14: 3828–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. 2010. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328: 1172–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. 1999. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet 21: 169–175 [DOI] [PubMed] [Google Scholar]
- Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, et al. 2009. Anticancer activity of CX-3543: A direct inhibitor of rRNA biogenesis. Cancer Res 69: 7653–7661 [DOI] [PubMed] [Google Scholar]
- Drygin D, Lin A, Bliesath J, Ho CB, O’Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, et al. 2011. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res 71: 1418–1430 [DOI] [PubMed] [Google Scholar]
- Duncan R, Milburn SC, Hershey JW 1987. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem 262: 380–388 [PubMed] [Google Scholar]
- Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, Wilson FH, Currie T, Khanna-Gupta A, Berliner N, et al. 2011. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 117: 2567–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle J, Krasagakis K, Orfanos CE 1997. Translation initiation factor eIF-4A1 mRNA is consistently overexpressed in human melanoma cells in vitro. Int J Cancer 71: 396–401 [DOI] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, et al. 2008. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 451: 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, Sorokin A, Ovchinnikov LP, Davicioni E, Triche TJ, et al. 2009. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 15: 402–415 [DOI] [PubMed] [Google Scholar]
- Farrar JE, Nater M, Caywood E, McDevitt MA, Kowalski J, Takemoto CM, Talbot CC Jr, Meltzer P, Esposito D, Beggs AH, et al. 2008. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood 112: 1582–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM 2009. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol 7: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fels DR, Koumenis C 2006. The PERK/eIF2α/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther 5: 723–728 [DOI] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B 2003. Genomic targets of the human c-Myc protein. Genes Dev 17: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, Gonzalez Fernandez A, Simpson P, D’Santos CS, Arends MJ, et al. 2011. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev 25: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floor S, van Staveren WC, Larsimont D, Dumont JE, Maenhaut C 2011. Cancer cells in epithelial-to-mesenchymal transition and tumor-propagating-cancer stem cells: Distinct, overlapping or same populations. Oncogene 30: 4609–4621 [DOI] [PubMed] [Google Scholar]
- Flygare J, Karlsson S 2007. Diamond-Blackfan anemia: Erythropoiesis lost in translation. Blood 109: 3152–3154 [DOI] [PubMed] [Google Scholar]
- Frederickson RM, Mushynski WE, Sonenberg N 1992. Phosphorylation of translation initiation factor eIF-4E is induced in a ras-dependent manner during nerve growth factor-mediated PC12 cell differentiation. Mol Cell Biol 12: 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Thomas G 2011. The role of p53 in ribosomopathies. Semin Hematol 48: 97–105 [DOI] [PubMed] [Google Scholar]
- Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, et al. 2010. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci 107: 14134–14139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi KA, Shimamura A 2008. Ribosomal dysfunction and inherited marrow failure. Br J Haematol 141: 376–387 [DOI] [PubMed] [Google Scholar]
- Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S 2008. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 455: 684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball SE, Orfali KA, et al. 2006. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet 79: 1110–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda HT, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, et al. 2008. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet 83: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Peters G 2006. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: All for one or one for all. Nat Rev Mol Cell Biol 7: 667–677 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N 1999. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev 13: 1422–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman N, Grandori C, Eisenman RN, White RJ 2003. Direct activation of RNA polymerase III transcription by c-Myc. Nature 421: 290–294 [DOI] [PubMed] [Google Scholar]
- Graber TE, Holcik M 2007. Cap-independent regulation of gene expression in apoptosis. Mol Biosyst 3: 825–834 [DOI] [PubMed] [Google Scholar]
- Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, Dowless MS, et al. 2007. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest 117: 2638–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, Parsons SH, Brail LH, Colligan BM, Koop JW, et al. 2009. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res 69: 3866–3873 [DOI] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318 [DOI] [PubMed] [Google Scholar]
- Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA 2005. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 7: 295–302 [DOI] [PubMed] [Google Scholar]
- Grover R, Sharathchandra A, Ponnuswamy A, Khan D, Das S 2011. Effect of mutations on the p53 IRES RNA structure: Implications for de-regulation of the synthesis of p53 isoforms. RNA Biol 8: 132–142 [DOI] [PubMed] [Google Scholar]
- Guan XY, Sham JS, Tang TC, Fang Y, Huo KK, Yang JM 2001. Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer Res 61: 3806–3809 [PubMed] [Google Scholar]
- Guan XY, Fung JM, Ma NF, Lau SH, Tai LS, Xie D, Zhang Y, Hu L, Wu QL, Fang Y, et al. 2004. Oncogenic role of eIF-5A2 in the development of ovarian cancer. Cancer Res 64: 4197–4200 [DOI] [PubMed] [Google Scholar]
- Hall MN, Raff M, Thomas G, eds. 2004. Cell growth: Control of cell size. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, et al. 2003. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23: 8862–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan KM, Sanij E, Hein N, Hannan RD, Pearson RB 2011. Signaling to the ribosome in cancer—It is more than just mTORC1. IUBMB Life 63: 79–85 [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J 1997. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem 272: 26457–26463 [DOI] [PubMed] [Google Scholar]
- Harris MN, Ozpolat B, Abdi F, Gu S, Legler A, Mawuenyega KG, Tirado-Gomez M, Lopez-Berestein G, Chen X 2004. Comparative proteomic analysis of all-trans-retinoic acid treatment reveals systematic posttranscriptional control mechanisms in acute promyelocytic leukemia. Blood 104: 1314–1323 [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Ren B 2006. Genome-wide location analysis: Insights on transcriptional regulation. Hum Mol Genet 15 (Spec No 1): R1–R7 [DOI] [PubMed] [Google Scholar]
- He LR, Zhao HY, Li BK, Liu YH, Liu MZ, Guan XY, Bian XW, Zeng YX, Xie D 2011. Overexpression of eIF5A-2 is an adverse prognostic marker of survival in stage I non-small cell lung cancer patients. Int J Cancer 129: 143–150 [DOI] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I 1998. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 19: 32–38 [DOI] [PubMed] [Google Scholar]
- Higa-Nakamine S, Suzuki T, Uechi T, Chakraborty A, Nakajima Y, Nakamura M, Hirano N, Kenmochi N 2012. Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res 40: 391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Korneluk RG 2000. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: Role of La autoantigen in XIAP translation. Mol Cell Biol 20: 4648–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N 2005. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327 [DOI] [PubMed] [Google Scholar]
- Hsieh AC, Ruggero D 2010. Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin Cancer Res 16: 4914–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D 2010. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell 17: 249–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Truitt ML, Ruggero D 2011. Oncogenic AKTivation of translation as a therapeutic target. Br J Cancer 105: 329–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. 2012. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485: 55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Wu T, Xu L, Liu A, Ji Y, Hu G 2002. Upstream binding factor up-regulated in hepatocellular carcinoma is related to the survival and cisplatin-sensitivity of cancer cells. FASEB J 16: 293–301 [DOI] [PubMed] [Google Scholar]
- Huranova M, Jablonski JA, Benda A, Hof M, Stanek D, Caputi M 2009. In vivo detection of RNA-binding protein interactions with cognate RNA sequences by fluorescence resonance energy transfer. RNA 15: 2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic N, Utermark T, Widlund HR, Roberts TM 2011. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci 108: E699–E708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR, et al. 2011. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell 44: 660–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Fraser CS, Yu Y, Leary J, Doudna JA 2004. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci 101: 16990–16995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, McCutcheon IE, Fuller GN, Huang ES, Cote GJ 2000. Fibroblast growth factor receptor-1 α-exon exclusion and polypyrimidine tract-binding protein in glioblastoma multiforme tumors. Cancer Res 60: 1221–1224 [PubMed] [Google Scholar]
- Jin X, Turcott E, Englehardt S, Mize GJ, Morris DR 2003. The two upstream open reading frames of oncogene mdm2 have different translational regulatory properties. J Biol Chem 278: 25716–25721 [DOI] [PubMed] [Google Scholar]
- Jones RM, Branda J, Johnston KA, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt EV 1996. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol 16: 4754–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E 2010. Genome-wide measurement of RNA secondary structure in yeast. Nature 467: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian A, Sarafan-Vasseur N, Sesboue R, Le Pessot F, Blanchard F, Lamy A, Laurent M, Flaman JM, Frebourg T 2006. Contribution of the BOP1 gene, located on 8q24, to colorectal tumorigenesis. Genes Chromosomes Cancer 45: 874–881 [DOI] [PubMed] [Google Scholar]
- Kim S, Li Q, Dang CV, Lee LA 2000. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc Natl Acad Sci 97: 11198–11202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M 2011. Cellular IRES-mediated translation: The war of ITAFs in pathophysiological states. Cell Cycle 10: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas AE, Lazaris-Karatzas A, Sonenberg N 1992a. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J 11: 4153–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N 1992b. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science 257: 1685–1689 [DOI] [PubMed] [Google Scholar]
- Lachance PE, Miron M, Raught B, Sonenberg N, Lasko P 2002. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol Cell Biol 22: 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev 12: 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahat G, Zhu QS, Huang KL, Wang S, Bolshakov S, Liu J, Torres K, Langley RR, Lazar AJ, Hung MC, et al. 2010. Vimentin is a novel anti-cancer therapeutic target; insights from in vitro and in vivo mice xenograft studies. PLoS ONE 5: e10105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lai MD, Xu J 2007. Ribosomal proteins and colorectal cancer. Curr Genomics 8: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers JE, Cassel SL, George DL 1997. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res 57: 3562–3568 [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR 2009. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 23: 2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345: 544–547 [DOI] [PubMed] [Google Scholar]
- Lee JM 2003. The role of protein elongation factor eEF1A2 in ovarian cancer. Reprod Biol Endocrinol 1: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SM, Holcik M 2008. For IRES trans-acting factors, it is all about location. Oncogene 27: 1033–1035 [DOI] [PubMed] [Google Scholar]
- Lipton JM, Ellis SR 2010. Diamond Blackfan anemia 2008–2009: Broadening the scope of ribosome biogenesis disorders. Curr Opin Pediatr 22: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Dilworth D, Gao L, Monzon J, Summers A, Lassam N, Hogg D 1999. Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat Genet 21: 128–132 [DOI] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al. 2011. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 17: 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M 2009. Visualizing protein-RNA interactions inside cells by fluorescence resonance energy transfer. RNA 15: 97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J 2009. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Ma YL, Peng JY, Zhang P, Huang L, Liu WJ, Shen TY, Chen HQ, Zhou YK, Zhang M, Chu ZX, et al. 2009. Heterogeneous nuclear ribonucleoprotein A1 is identified as a potential biomarker for colorectal cancer based on differential proteomics technology. J Proteome Res 8: 4525–4535 [DOI] [PubMed] [Google Scholar]
- Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, Zhang Y 2010. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell 18: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Malina A, Mills JR, Pelletier J 2012. Emerging therapeutics targeting mRNA translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, Sonenberg N 2007. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE 2: e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O, Kimchi A 2008. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell 30: 447–459 [DOI] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Pellegrini S, Bertacca G, Callahan R 2001. Reduced expression of INT-6/eIF3-p48 in human tumors. Int J Oncol 18: 175–179 [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 3: 707–716 [DOI] [PubMed] [Google Scholar]
- Marshall L, Kenneth NS, White RJ 2008. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell 133: 78–89 [DOI] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119: 969–979 [DOI] [PubMed] [Google Scholar]
- Mayer C, Zhao J, Yuan X, Grummt I 2004. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev 18: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL 2003. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115: 187–198 [DOI] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, et al. 2008. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet 40: 963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs EF, Galabru J, Barber GN, Katze MG, Hovanessian AG 1993. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci 90: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, Trojahn U, Wendel HG, Charest A, Bronson RT, et al. 2008. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci 105: 10853–10858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miluzio A, Beugnet A, Grosso S, Brina D, Mancino M, Campaner S, Amati B, de Marco A, Biffo S 2011. Impairment of cytoplasmic eIF6 activity restricts lymphomagenesis and tumor progression without affecting normal growth. Cancer Cell 19: 765–775 [DOI] [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, et al. 2007. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128: 257–267 [DOI] [PubMed] [Google Scholar]
- Montanaro L, Calienni M, Bertoni S, Rocchi L, Sansone P, Storci G, Santini D, Ceccarelli C, Taffurelli M, Carnicelli D, et al. 2010. Novel dyskerin-mediated mechanism of p53 inactivation through defective mRNA translation. Cancer Res 70: 4767–4777 [DOI] [PubMed] [Google Scholar]
- Nakamura J, Aoyagi S, Nanchi I, Nakatsuka S, Hirata E, Shibata S, Fukuda M, Yamamoto Y, Fukuda I, Tatsumi N, et al. 2009. Overexpression of eukaryotic elongation factor eEF2 in gastrointestinal cancers and its involvement in G2/M progression in the cell cycle. Int J Oncol 34: 1181–1189 [PubMed] [Google Scholar]
- Narla A, Hurst SN, Ebert BL 2011. Ribosome defects in disorders of erythropoiesis. Int J Hematol 93: 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573 [DOI] [PubMed] [Google Scholar]
- Nupponen NN, Porkka K, Kakkola L, Tanner M, Persson K, Borg A, Isola J, Visakorpi T 1999. Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am J Pathol 154: 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Shibayama M, Sato M, Tokunaga A, Yoshida N 2011. Polypyrimidine tract-binding protein regulates the cell cycle through IRES-dependent translation of CDK11 (p58) in mouse embryonic stem cells. Cell Cycle 10: 3706–3713 [DOI] [PubMed] [Google Scholar]
- Ortiz-Zapater E, Pineda D, Martinez-Bosch N, Fernandez-Miranda G, Iglesias M, Alameda F, Moreno M, Eliscovich C, Eyras E, Real FX, et al. 2011. Key contribution of CPEB4-mediated translational control to cancer progression. Nat Med 18: 83–90 [DOI] [PubMed] [Google Scholar]
- Paulin FE, West MJ, Sullivan NF, Whitney RL, Lyne L, Willis AE 1996. Aberrant translational control of the c-myc gene in multiple myeloma. Oncogene 13: 505–513 [PubMed] [Google Scholar]
- Pellagatti A, Hellstrom-Lindberg E, Giagounidis A, Perry J, Malcovati L, Della Porta MG, Jadersten M, Killick S, Fidler C, Cazzola M, et al. 2008. Haploinsufficiency of RPS14 in 5q- syndrome is associated with deregulation of ribosomal- and translation-related genes. Br J Haematol 142: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianese G 1896. Beitrag zur histologie und aetiologie der carcinoma. Histologische und experimentelle untersuchungen. Beitr Pathol Anat Allg Pathol: 1–193 [Google Scholar]
- Pickering BM, Willis AE 2005. The implications of structured 5′ untranslated regions on translation and disease. Semin Cell Dev Biol 16: 39–47 [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Pradayrol L, Sonenberg N 2000. A cell cycle-dependent internal ribosome entry site. Mol Cell 5: 607–616 [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Dostie J, Sonenberg N 2001. Suppression of cap-dependent translation in mitosis. Genes Dev 15: 2083–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Sarnow P 2004. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J Biol Chem 279: 13721–13728 [DOI] [PubMed] [Google Scholar]
- Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC 2003. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell 12: 889–901 [DOI] [PubMed] [Google Scholar]
- Rauch J, Ahlemann M, Schaffrik M, Mack B, Ertongur S, Andratschke M, Zeidler R, Lang S, Gires O 2004. Allogenic antibody-mediated identification of head and neck cancer antigens. Biochem Biophys Res Commun 323: 156–162 [DOI] [PubMed] [Google Scholar]
- Ray PS, Grover R, Das S 2006. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep 7: 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, et al. 2001. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 104: 195–203 [DOI] [PubMed] [Google Scholar]
- Ron D, Harding HP 2007. eIF2α phosphorylation in cellular stress-responses and disease. In Translational control in biology and medicine (ed. Matthews MB, Sonenberg N, Hershey JW), pp. 345–368 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Rosenwald IB, Hutzler MJ, Wang S, Savas L, Fraire AE 2001. Expression of eukaryotic translation initiation factors 4E and 2α is increased frequently in bronchioloalveolar but not in squamous cell carcinomas of the lung. Cancer 92: 2164–2171 [DOI] [PubMed] [Google Scholar]
- Rothe M, Ko Y, Albers P, Wernert N 2000. Eukaryotic initiation factor 3 p110 mRNA is overexpressed in testicular seminomas. Am J Pathol 157: 1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Roux PP, Topisirovic I 2012. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury P, Paul RR, Chowdhury R, Chaudhuri K 2007. HnRNP E2 is downregulated in human oral cancer cells and the overexpression of hnRNP E2 induces apoptosis. Mol Carcinog 46: 198–207 [DOI] [PubMed] [Google Scholar]
- Ruggero D 2009. The role of Myc-induced protein synthesis in cancer. Cancer Res 69: 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP 2003. Does the ribosome translate cancer? Nat Rev Cancer 3: 179–192 [DOI] [PubMed] [Google Scholar]
- Ruggero D, Sonenberg N 2005. The Akt of translational control. Oncogene 24: 7426–7434 [DOI] [PubMed] [Google Scholar]
- Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP 2003. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299: 259–262 [DOI] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP 2004. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med 10: 484–486 [DOI] [PubMed] [Google Scholar]
- Salehi Z, Mashayekhi F 2006. Expression of the eukaryotic translation initiation factor 4E (eIF4E) and 4E-BP1 in esophageal cancer. Clin Biochem 39: 404–409 [DOI] [PubMed] [Google Scholar]
- Sanvito F, Vivoli F, Gambini S, Santambrogio G, Catena M, Viale E, Veglia F, Donadini A, Biffo S, Marchisio PC 2000. Expression of a highly conserved protein, p27BBP, during the progression of human colorectal cancer. Cancer Res 60: 510–516 [PubMed] [Google Scholar]
- Saramaki O, Willi N, Bratt O, Gasser TC, Koivisto P, Nupponen NN, Bubendorf L, Visakorpi T 2001. Amplification of EIF3S3 gene is associated with advanced stage in prostate cancer. Am J Pathol 159: 2089–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 7: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al. 2001. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res 29: 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Wang H, Nghiem CL, Zhou K, Yau J, Tailor CS, Irwin MS, Dror Y 2011. The ribosome-related protein, SBDS, is critical for normal erythropoiesis. Blood 118: 6407–6417 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE 2004. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem 279: 29066–29074 [DOI] [PubMed] [Google Scholar]
- Shi J, Kahle A, Hershey JW, Honchak BM, Warneke JA, Leong SP, Nelson MA 2006. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene 25: 4923–4936 [DOI] [PubMed] [Google Scholar]
- Shimamura A, Alter BP 2010. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev 24: 101–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiue CN, Arabi A, Wright AP 2010. Nucleolar organization, growth control and cancer. Epigenetics 5: 200–205 [DOI] [PubMed] [Google Scholar]
- Shuda M, Kondoh N, Tanaka K, Ryo A, Wakatsuki T, Hada A, Goseki N, Igari T, Hatsuse K, Aihara T, et al. 2000. Enhanced expression of translation factor mRNAs in hepatocellular carcinoma. Anticancer Res 20: 2489–2494 [PubMed] [Google Scholar]
- Silvera D, Schneider RJ 2009. Inflammatory breast cancer cells are constitutively adapted to hypoxia. Cell Cycle 8: 3091–3096 [DOI] [PubMed] [Google Scholar]
- Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, Hochman T, Formenti SC, Schneider RJ 2009. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol 11: 903–908 [DOI] [PubMed] [Google Scholar]
- Silvera D, Formenti SC, Schneider RJ 2010. Translational control in cancer. Nat Rev Cancer 10: 254–266 [DOI] [PubMed] [Google Scholar]
- Sivan G, Aviner R, Elroy-Stein O 2011. Mitotic modulation of translation elongation factor 1 leads to hindered tRNA delivery to ribosomes. J Biol Chem 286: 27927–27935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells DL, Black DR, Meschonat C, Rhoads R, De Benedetti A, Gao M, Williams BJ, Li BD 1998. Detection of eIF4E gene amplification in breast cancer by competitive PCR. Ann Surg Oncol 5: 232–237 [DOI] [PubMed] [Google Scholar]
- Sorrells DL, Ghali GE, Meschonat C, DeFatta RJ, Black D, Liu L, De Benedetti A, Nathan CO, Li BD 1999. Competitive PCR to detect eIF4E gene amplification in head and neck cancer. Head Neck 21: 60–65 [DOI] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Mitchell SA, Willis AE 2005. Internal ribosome entry segment-mediated translation during apoptosis: The role of IRES-trans-acting factors. Cell Death Differ 12: 585–591 [DOI] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE 2010. Translational regulation of gene expression during conditions of cell stress. Mol Cell 40: 228–237 [DOI] [PubMed] [Google Scholar]
- Stoneley M, Willis AE 2003. Aberrant regulation of translation initiation in tumorigenesis. Curr Mol Med 3: 597–603 [DOI] [PubMed] [Google Scholar]
- Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE 1998. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 16: 423–428 [DOI] [PubMed] [Google Scholar]
- Stratford AL, Habibi G, Astanehe A, Jiang H, Hu K, Park E, Shadeo A, Buys TP, Lam W, Pugh T, et al. 2007. Epidermal growth factor receptor (EGFR) is transcriptionally induced by the Y-box binding protein-1 (YB-1) and can be inhibited with Iressa in basal-like breast cancer, providing a potential target for therapy. Breast Cancer Res 9: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB 2005. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123: 49–63 [DOI] [PubMed] [Google Scholar]
- Tamburini J, Green AS, Bardet V, Chapuis N, Park S, Willems L, Uzunov M, Ifrah N, Dreyfus F, Lacombe C, et al. 2009. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood 114: 1618–1627 [DOI] [PubMed] [Google Scholar]
- Tang DJ, Dong SS, Ma NF, Xie D, Chen L, Fu L, Lau SH, Li Y, Guan XY 2010. Overexpression of eukaryotic initiation factor 5A2 enhances cell motility and promotes tumor metastasis in hepatocellular carcinoma. Hepatology 51: 1255–1263 [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinton SA, Schepens B, Bruynooghe Y, Beyaert R, Cornelis S 2005. Regulation of the cell-cycle-dependent internal ribosome entry site of the PITSLRE protein kinase: Roles of Unr (upstream of N-ras) protein and phosphorylated translation initiation factor eIF-2α. Biochem J 385: 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta R, Vignudelli T, Candini O, Intine RV, Pecorari L, Guerzoni C, Santilli G, Byrom MW, Goldoni S, Ford LP, et al. 2003. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell 3: 145–160 [DOI] [PubMed] [Google Scholar]
- Tsai BP, Wang X, Huang L, Waterman ML 2011. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol Cell Proteomics 10: M110 007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R 2004. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol 24: 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Sasaki M, Elia AJ, Chio II, Hamada K, Fukunaga R, Mak TW 2010. Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proc Natl Acad Sci 107: 13984–13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA 2011. Tumor metastasis: Molecular insights and evolving paradigms. Cell 147: 275–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R 2011. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 11: 644–656 [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309 [DOI] [PubMed] [Google Scholar]
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536 [DOI] [PubMed] [Google Scholar]
- Vogt PK, Rommel C, Vanhaesebroeck B, eds. 2011. Phosphoinositide 3-kinase in health and disease. Springer-Verlag, Berlin/Heidelberg [Google Scholar]
- Wang S, Rosenwald IB, Hutzler MJ, Pihan GA, Savas L, Chen JJ, Woda BA 1999. Expression of the eukaryotic translation initiation factors 4E and 2α in non-Hodgkin’s lymphomas. Am J Pathol 155: 247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG 2001. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J 20: 4370–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Geng J, Wang JH, Chu XY, Geng HC, Chen LB 2009a. Overexpression of eukaryotic initiation factor 4E (eIF4E) and its clinical significance in lung adenocarcinoma. Lung Cancer 66: 237–244 [DOI] [PubMed] [Google Scholar]
- Wang T, Chen YH, Hong H, Zeng Y, Zhang J, Lu JP, Jeansonne B, Lu Q 2009b. Increased nucleotide polymorphic changes in the 5′-untranslated region of δ-catenin (CTNND2) gene in prostate cancer. Oncogene 28: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Vardy LA, Tan CP, Loo JM, Guo K, Li J, Lim SG, Zhou J, Chng WJ, Ng SB, et al. 2010. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell 18: 52–62 [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA 1999. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol 19: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB 2000. Disorders in cell circuitry during multistage carcinogenesis: The role of homeostasis. Carcinogenesis 21: 857–864 [DOI] [PubMed] [Google Scholar]
- Weinstein IB 2002. Cancer. Addiction to oncogenes—The Achilles heal of cancer. Science 297: 63–64 [DOI] [PubMed] [Google Scholar]
- Weinstein IB, Joe AK 2006. Mechanisms of disease: Oncogene addiction—A rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol 3: 448–457 [DOI] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW 2004. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 428: 332–337 [DOI] [PubMed] [Google Scholar]
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, et al. 2007. Dissecting eIF4E action in tumorigenesis. Genes Dev 21: 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker EW, van Vugt MA, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, et al. 2007. 14–3-3sigma controls mitotic translation to facilitate cytokinesis. Nature 446: 329–332 [DOI] [PubMed] [Google Scholar]
- Willett M, Brocard M, Davide A, Morley SJ 2011. Translation initiation factors and active sites of protein synthesis co-localize at the leading edge of migrating fibroblasts. Biochem J 438: 217–227 [DOI] [PubMed] [Google Scholar]
- Wong CC, Traynor D, Basse N, Kay RR, Warren AJ 2011. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood 118: 4305–4312 [DOI] [PubMed] [Google Scholar]
- Xie D, Ma NF, Pan ZZ, Wu HX, Liu YD, Wu GQ, Kung HF, Guan XY 2008. Overexpression of EIF-5A2 is associated with metastasis of human colorectal carcinoma. Hum Pathol 39: 80–86 [DOI] [PubMed] [Google Scholar]
- Xue S, Barna M 2012. Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 13: 357–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams BR, Aguet M, Weissmann C 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J 14: 6095–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo YG, Kong G, Lee MO 2006. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1α protein by recruiting histone deacetylase 1. EMBO J 25: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D 2006. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 312: 902–906 [DOI] [PubMed] [Google Scholar]
- Yoshizawa A, Fukuoka J, Shimizu S, Shilo K, Franks TJ, Hewitt SM, Fujii T, Cordon-Cardo C, Jen J, Travis WD 2010. Overexpression of phospho-eIF4E is associated with survival through AKT pathway in non-small cell lung cancer. Clin Cancer Res 16: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun JP, Miao J, Chen GG, Tian QH, Zhang CQ, Xiang J, Fu J, Lai PB 2007. Increased expression of nucleophosmin/B23 in hepatocellular carcinoma and correlation with clinicopathological parameters. Br J Cancer 96: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. 2006. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci 103: 17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Pan X, Hershey JW 2007. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem 282: 5790–5800 [DOI] [PubMed] [Google Scholar]
- Zhang T, Huang XH, Dong L, Hu D, Ge C, Zhan YQ, Xu WX, Yu M, Li W, Wang X, et al. 2010. PCBP-1 regulates alternative splicing of the CD44 gene and inhibits invasion in human hepatoma cell line HepG2 cells. Mol Cancer 9: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM 2011. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]