Summary
The switch between an inactive and active conformation is an important transition for signaling proteins, yet the mechanisms underlying such switches are not clearly understood. Escherichia coli CheY, a response regulator protein from the two-component signal transduction system that regulates bacterial chemotaxis, is an ideal protein for the study of allosteric mechanisms. By utilizing 15N CPMG relaxation dispersion experiments, we monitored the inherent dynamic switching of unphosphorylated CheY. We show that CheY does not undergo a two-state concerted switch between the inactive and active conformations. Interestingly, partial saturation of Mg2+ enhances the intrinsic allosteric motions. Taken together with chemical shift perturbations, these data indicate that the µs-ms timescale motions underlying CheY allostery are segmental in nature. We propose an expanded allosteric network of residues, including W58, that undergo asynchronous, local switching between inactive and active-like conformations as the primary basis for the allosteric mechanism.
Introduction
Allosteric conformational change is critical for the function of many proteins. At the present time, it is not generally understood how allosteric conformational changes are executed or how many different execution strategies exist. As a simpler alternative to defining precise conformational change trajectories, there has been intense focus directed at the “selected fit” versus “induced fit” paradigms (Okazaki and Takada, 2008; Hammes et al., 2009; Wlodarski and Zagrovic, 2009; Zhou, 2010; Csermely et al., 2010), as well as at concerted vs. propagated conformational changes (Cui and Karplus, 2008; Tsai et al., 2009). The pairing of selected fit with a concerted conformational change – that is, a simple “switch” – has been particularly popular. It is widely recognized that dynamics are central to these processes, yet there are few experiments that have been carried out to directly assess some of the basic assumptions of allostery. Hence there is a need for experimental dynamics data on the timescale of conformational equilibria in allosteric proteins.
Receiver domains from response regulator (RR) proteins have been studied extensively and, because of their small size, have become favored as models for understanding conformational allostery. RRs, along with sensor kinases, comprise the two-component system ubiquitous in prokaryotes. RRs usually consist of an input receiver domain that is activated by phosphorylation, and an output domain that transmits the signal into various activities such as DNA binding (Bourret, 2010; Galperin, 2010). Accordingly, the ability of the receiver domain to undergo a well-defined conformational change is a vital component of RR function (Lee et al., 2001; Gao et al., 2007). The chemotaxis protein Y (CheY) from Escherichia coli is a RR that regulates chemotactic flagellar rotation (Clegg and Koshland, 1984; Matsumura et al., 1984). Because it lacks an output domain, the CheY receiver domain must both accept a phosphoryl group and directly activate its downstream effector; in response to phosphorylation at D57 (Sanders et al., 1989), which requires the presence of Mg2+ (Lukat et al., 1990), CheY undergoes a conformational change that enables it to directly bind the flagellar motor switch protein FliM at a surface distal to D57. CheY binding to FliM promotes a change in the flagellar rotation from counterclockwise to clockwise (Baker et al., 2006). Unphosphorylated CheY also interacts with FliM, though with considerably reduced affinity (Barak and Eisenbach, 1992; Welch et al., 1993).
As is common in allosteric signaling molecules, CheY samples an active conformation (“A”, FliM binding competent) and an inactive conformation (“I”, FliM binding incompetent). In the I conformation, FliM binding is sterically hindered by the location of Y106 in a solvent exposed or “out” position (Figure 1A). Upon activation by phosphorylation, Y106 (~11 Å away from the phosphoaspartate) rotates to the “in”, buried position, relieving the hindrance. T87 hydrogen bonds with the phosphoryl group and has been shown to be an important link for Y106 rotation (Appleby and Bourret, 1998; Lee et al., 2001a). Motion of the β4-α4 loop, consisting of residues 88–91, facilitates burial of Y106 (Ma and Cui, 2007; Simonovic and Volz, 2001), leading to the possibility that residues in the loop (in addition to T87) may be involved in the signaling. Together with small changes in the β5-α5 loop and side-chain motions of K109 and F14, the I and A conformations differ mainly in the Y106 rotation and the location of the β4-α4 loop (Lee et al., 2001a).
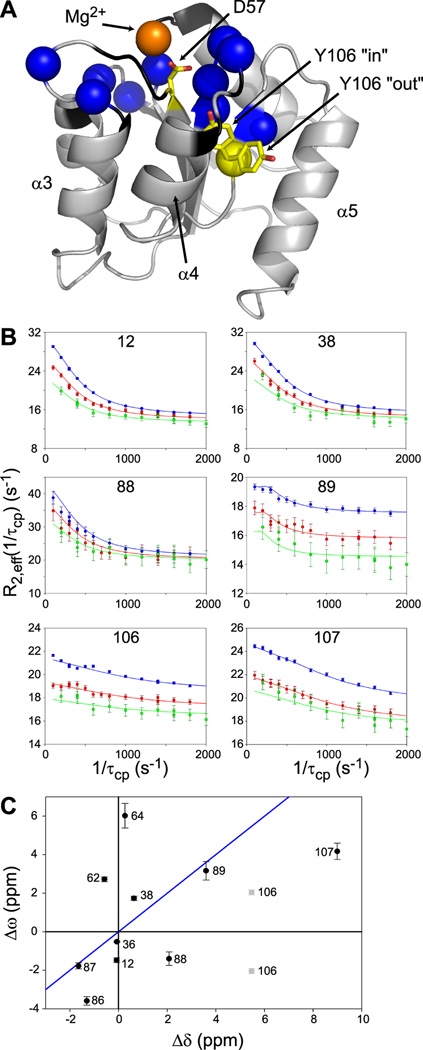
Figure 1. 15N CPMG Relaxation dispersion data from unphosphorylated CheY in the presence of 10 mM Mg2+.
(A) Structure of unphosphorylated CheY using 3CHY with the side-chain orientation of D57 and Mg2+ location from 2CHE. Residues with non-zero Rex values at 10 mM Mg2+ are displayed as blue spheres. Black indicates residues that are too broad to be measured. Highlighted in yellow are Y106 and the site of phosphorylation, D57. For Y106, the “in” rotamer indicates active and “out” indicates inactive CheY.
(B) Raw CPMG dispersion curves are displayed for 6 example residues at 700 MHz (blue), 600 MHz (red) and 500 MHz (green). Lines are local fits to the Carver-Richards equation with parameters given in Table 1.
(C) Δω values from local fits of relaxation dispersion are plotted against Δδ using the difference between unphosphorylated CheY and BeFx-bound CheY both with 10 mM Mg2+. Δδ was determined using Δδ′ = N10mM Mg2+ − NBeFx, where to get the final Δδ value we made an additional adjustment. Since unphosphorylated CheY is not completely inactive and BeFx-bound CheY is not completely active, we used Δδ = 1.05Δδ′. The blue line has a slope of 1. See also Figures S1 and S2.
The prevailing conceptual framework for receiver domain activation/allostery has been that the protein exists in a dynamic equilibrium that accesses both the I and A conformations, in line with the Monod-Wyman-Changeux (MWC) model of allostery. The following findings provide evidence of an I-to-A state conformational equilibrium: (1) in the absence of phosphorylation, CheY still has the ability to stimulate clockwise flagellar rotation (Barak and Eisenbach, 1992); (2) in the crystal structure of unphosphorylated, Mg2+-free CheY, both “out” and “in” conformations of the Y106 side chain were observed (Volz and Matsumura, 1991); (3) binding of FliM, CheA, and CheZ peptides to CheY affect its ability to phosphorylate and can be explained by a ligand-induced shift of the I-to-A equilibrium (Schuster et al., 2001); (4) NMR studies of the receiver domains of Spo0F and NtrC showed that in regions where structural changes upon phosphorylation were observed, there was enhanced transverse relaxation due to conformational exchange on the μs-ms time scale (Volkman et al., 2001; Feher and Cavanagh, 1999); and (5) mutant NtrC proteins revealed a correlation between activity/inactivity and a two-state equilibrium between active and inactive conformations (Volkman et al., 2001; Gardino et al., 2009). While these data are consistent with a two-state switch of RRs, direct detection of a two-state dynamic process between active and inactive conformations has been elusive.
Complicating the simple idea of a pre-existing equilibrium between I and A conformations are several crystal structures of CheY that show intermediate conformations (Simonovic and Volz, 2001; Dyer and Dahlquist, 2006; Guhaniyogi et al., 2006). Additionally, molecular dynamics simulations indicated that Y106 rotation and the formation of a hydrogen bond between T87 and the phosphoryl group are independent of one another (Ma and Cui, 2007). These studies showed that CheY is not restricted to the two end states and taken together suggest that it can be trapped in metastable states that presumably are sampled along the allosteric conformational change trajectory. It is unknown, however, whether such trapped states are functionally relevant or are merely artifacts from crystallization; it also remains unknown what the relevant timescales are for conformational switching in CheY.
In order to further understand the allosteric switch CheY undergoes upon phosphorylation, we investigated the conformational equilibrium that occurs in the unphosphorylated protein. NMR relaxation dispersion was used to measure the dynamics of CheY switching to test for consistency with a two-state model. We found that a physiological level of Mg2+ likely plays a critical role in promoting allosteric conformational changes. Nevertheless, whether Mg2+ is present or absent, unphosphorylated CheY appears not to undergo two-state concerted switching between I and A conformations. Rather, the data are more suggestive of a model in which CheY switches in a non-concerted, segmental fashion. Local sites may occupy their active conformations at different times utilizing a previously undescribed signaling network consisting of A88 with T87 and the quartet of W58, M85, E89, and Y106.
Results
Microsecond-millisecond motions are along the allosteric path
Large scale conformational changes in proteins frequently occur on the slow, or μs-ms, timescale (Henzler-Wildman and Kern, 2007; Kleckner and Foster, 2011). In an attempt to measure the dynamic switching between the I and A conformations of unphosphorylated CheY, we used 15N Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion experiments to measure motions on this timescale (Palmer et al., 2001). These experiments elucidate processes on the µs-ms timescale that contribute to broadening (or “width”) of NMR peaks. Specifically, the NMR line width is proportional to the transverse relaxation rate, R2, which is comprised of an “intrinsic” rate, R2°, and a rate that arises from conformational exchange processes on the µs-ms timescale, Rex:
| (1) |
The CPMG relaxation dispersion experiment measures the suppression of Rex contributions to R2 as a function of spacing between 180° pulses in the CPMG train, τcp (Loria et al., 1999). For a two-state exchange process in the limit of fast exchange on the NMR timescale,
| (2) |
where pI and pA are populations of the major and minor states, Δω is the difference in chemical shift between the two states, and kex is the rate of exchange between I and A (= k1 + k−1). A longer expression (the Carver-Richards equation) exists for the general case (Palmer et al., 2001). A particular residue’s Rex value is considered to be non-zero (i.e. affected by conformational exchange) if Rex > 2 s−1, as described in the Experimental Procedures.
Pilot relaxation dispersion experiments indicated that the dispersion curves were more pronounced at lower temperatures, and 15 °C was determined to be a good compromise between pronounced dispersion curves and signal-to-noise. Under the standard conditions of 10 mM Mg2+, 15N CPMG relaxation dispersion data were collected at 500, 600, and 700 MHz for CheY. Residues with non-zero Rex values localized to the active site and FliM binding interface, which includes residues T87, A88, and E89 (in the β4-α4 loop), as well as Y106 and V107 (Figure 1A). This was not surprising since all substantial conformational differences between the I and A states are limited to this region (Lee et al., 2001a). There were no non-zero Rex values elsewhere in the protein, detected either by CPMG relaxation dispersion or by model-free analysis of T1, T2, and {1H}-15N NOE data. Therefore, at a qualitative level, these data are consistent with motion corresponding to the I-to-A transition.
Motions measured by relaxation dispersion do not result from a single conformational switch event
The relaxation dispersion curves (Figure 1B) were quantitatively analyzed using the general Carver-Richards equation (Palmer et al., 2001) to fit each residue individually (i.e., “local fits”, see Experimental Procedures). A total of 11 residues could be fit with reasonable precision. If CheY switches concertedly in a simple two-state manner between I and A conformations, we would expect to measure the same exchange rate and populations for all residues. However, the fits yielded a range of kinetic and thermodynamic parameters (Table 1). Exchange rates varied from slow (~1300 s−1 for residues 12 and 38) to fast (~3000 s−1 for residues 106 and 107) and populations varied from 90% – 99% of I. The different exchange rates were also evident in the raw dispersion curves (compare rows of Figure 1B). In addition, two-state or three-state concerted switching could not be rationalized based on attempts to group fit the data (see Experimental Procedures for details).
Table 1.
Local fits of 15N CPMG relaxation dispersion for CheY in the presence of 10 mM Mg2+.
| Residue | kex (s−1) | Δω (ppm) | pI |
R2° (s−1) 500 MHz |
R2° (s−1) 600 MHz |
R2° (s−1) 700 MHz |
χ2 |
|---|---|---|---|---|---|---|---|
| 12 | 1220 ± 50 | 1.5 ± 0.1 | 0.94 ± 0.01 | 13.3 ± 0.3 | 13.9 ± 0.2 | 14.6 ± 0.1 | 4.0 |
| 36 | 1910 ± 300 | 0.52 ± 0.04 | 0.91 ± 0.02 | 13.1 ± 0.2 | 13.2 ± 0.1 | 14.1 ± 0.1 | 2.6 |
| 38 | 1370 ± 70 | 1.7 ± 0.1 | 0.95 ± 0.01 | 14.0 ± 0.4 | 14.3 ± 0.2 | 15.1 ± 0.2 | 46 |
| 62 | 2060 ± 180 | 2.7 ± 0.1 | 0.99 ± 0.01 | 12.0 ± 0.1 | 12.2 ± 0.1 | 13.1 ± 0.1 | 0.75 |
| 64 | 3620 ± 460 | 6.0 ± 0.6 | 0.99 ± 0.01 | 13.7 ± 0.5 | 13.0 ± 0.6 | 13.8 ± 0.7 | 5.3 |
| 86 | 2100 ± 330 | 3.6 ± 0.2 | 0.99 ± 0.01 | 15.1 ± 0.3 | 15.5 ± 0.2 | 16.9 ± 0.2 | 13 |
| 87 | 1370 ± 180 | 1.8 ± 0.2 | 0.98 ± 0.01 | 15.3 ± 0.4 | 14.8 ± 0.3 | 16.6 ± 0.2 | 3.6 |
| 88 | 1230 ± 280 | 1.4 ± 0.4 | 0.91 ± 0.04 | 20.1 ± 1 | 20.2 ± 0.9 | 21.0 ± 0.8 | 14 |
| 89 | 246 ± 31 | 3.2 ± 0.5 | 0.99 ± 0.01 | 14.5 ± 0.3 | 15.8 ± 0.2 | 17.5 ± 0.1 | 2.9 |
| 106 | 3100 ± 410 | 2.0 ± 0.1 | 0.99 ± 0.01 | 16.4 ± 0.2 | 17.1 ± 0.2 | 18.5 ± 0.2 | 4.5 |
| 107 | 2840 ± 380 | 4.2 ± 0.4 | 0.99 ± 0.01 | 17.6 ± 0.3 | 17.7 ± 0.3 | 19.3 ± 0.4 | 5.5 |
Yet another test for two-state behavior is to compare the dispersion-based differences in chemical shifts to known differences in chemical shifts for two defined structural states. Accordingly, we compared Δω from the local fits of relaxation dispersions to Δδ from the chemical shift perturbations between unphosphorylated and BeFx-bound (phosphoryl mimic (Yan et al., 1999)) CheY. A poor correlation between Δω and Δδ was obtained (Figure 1C), further indicating that the I-to-A transition cannot be described by a simple two-state transition.
Taken together, the non-uniform values of the individual exchange parameters, the inability to group fit, and poor correlation of Δω and Δδ indicates that CheY does not undergo concerted, two-state switching in the presence of 10 mM Mg2+ at 15 °C.
Removal of Mg2+ alters dynamics of unphosphorylated CheY
Another possible source of non-zero Rex values in CheY and other RRs is the reversible binding of Mg2+. A divalent metal ion is necessary for all RR phosphorylation and dephosphorylation (Lukat et al., 1990). In the experiments described in the previous section, near physiological concentrations of Mg2+ (10 mM) were used to characterize CheY in an environment similar to inside of cells and to have consistency with previous biochemical and NMR work on CheY (Moy et al., 1994; Hubbard et al., 2003). Because the binding affinity of Mg2+ to CheY is 1.5 ± 0.3 mM under our conditions (determined by NMR 1H-15N HSQC peak shifts, Figure S1), at 10 mM Mg2+ and 1 mM CheY, 86% of CheY is bound by the ion. Thus, if the Mg2+ binding kinetics are on the appropriate timescale, the binding of Mg2+ and associated side-chain rearrangements could be detected by relaxation dispersion and this could complicate the interpretation in terms of conformational exchange.
To separate motions associated with Mg2+ binding and release from those intrinsic to CheY, we carried out relaxation dispersion experiments without Mg2+ present (i.e., with 1 mM ethylenediaminetetraacetic acid, EDTA). In the absence of Mg2+, 13 residues displayed non-zero Rex values for CheY, compared to 11 when Mg2+ is present at 10 mM. These residues are localized to the FliM binding interface or the active site (Figure 2A) just as when Mg2+ is present (Figure 1A). The locally fit parameters, when assuming a two-state mechanism, are diverse (Figure 2B and Table 2). In comparison with dynamics in the presence of Mg2+, many of the same residues display significant dispersion, including T87 and V107. The locally fit parameters of the common residues differ significantly between the Mg2+-free and Mg2+-present conditions (Tables 1 and 2). Additionally, Δω and Δδ do not correlate (Figure 2C), as was the case with Mg2+ (Figure 1C). Therefore, even without the potentially complicating effects of Mg2+, CheY appears not to undergo two-state switching.
Figure 2. 15N CPMG Relaxation dispersion data from unphosphorylated CheY in the absence of Mg2+ and presence of 1 mM EDTA.
(A) Similar to Figure 1, residues with non-zero Rex values at 1 mM EDTA are displayed as green spheres.
(B) At 700 MHz (blue) and 600 MHz (red), data are displayed with local fits using the Carver-Richards equation with parameters given in Table 2.
(C) Plot of Δω vs. Δδ using Δδ between CheY with 1 mM EDTA and CheY with saturating BeFx and 10 mM Mg2+.
See also Figures S1 and S2.
Table 2.
Local fits of 15N CPMG relaxation dispersion for CheY in the presence of 1 mM EDTA.
| Residue | kex (s−1) | Δω (ppm) | pI |
R2° (s−1) 600 MHz |
R2° (s−1) 700 MHz |
χ2 |
|---|---|---|---|---|---|---|
| 18 | 288 ± 57 | 3.9 ± 0.7 | 0.99 ± 0.01 | 15.7 ± 0.3 | 17.0 ± 0.3 | 2.9 |
| 19 | 180 ± 30 | 5.7 ± 1 | 0.96 ± 0.01 | 16.1 ± 0.7 | 17.4 ± 0.9 | 7.6 |
| 36 | 2400 ± 500 | 0.6 ± 0.1 | 0.86 ± 0.04 | 13.3 ± 0.2 | 14.1 ± 0.3 | 2.6 |
| 38 | 2360 ± 640 | 3.6 ± 0.4 | 0.99 ± 0.01 | 14.0 ± 0.5 | 15.8 ± 0.7 | 4.3 |
| 62 | 1820 ± 520 | 3.7 ± 0.4 | 0.99 ± 0.01 | 13.4 ± 0.4 | 13.8 ± 0.7 | 11 |
| 64 | 96 ± 16 | 9.4 ± 1 | 0.80 ± 0.04 | 20.0 ± 1 | 22.2 ± 2 | 13 |
| 67 | 1860 ± 920 | 0.4 ± 0.1 | 0.73 ± 0.1 | 15.2 ± 0.4 | 16.3 ± 0.6 | 3.5 |
| 68 | 1400 ± 790 | 3.5 ± 0.8 | 0.99 ± 0.01 | 16.1 ± 0.4 | 15.9 ± 0.6 | 1.5 |
| 69 | 2400 ± 1100 | 2.3 ± 0.5 | 0.99 ± 0.01 | 15.0 ± 0.5 | 15.0 ± 0.8 | 1.1 |
| 87 | 2130 ± 540 | 0.53 ± 0.1 | 0.81 ± 0.05 | 14.2 ± 0.3 | 15.9 ± 0.4 | 2.3 |
| 94 | 1330 ± 800 | 0.27 ± 0.1 | 0.67 ± 0.2 | 15.4 ± 0.2 | 16.1 ± 0.3 | 3.3 |
| 107 | 2440 ± 570 | 0.52 ± 0.1 | 0.80 ± 0.08 | 14.8 ± 0.2 | 16.1 ± 0.4 | 2.6 |
| 127 | 1980 ± 550 | 1.6 ± 0.2 | 0.98 ± 0.01 | 15.8 ± 0.3 | 16.5 ± 0.5 | 2.5 |
It is possible that bound Mg2+ induces a conformational change in CheY. The crystal structure of Mg2+-bound E. coli CheY reveals a conformational difference from Mg2+-free CheY that localizes to α4 and the β4-α4 loop (Bellsolell et al., 1994). However, chemical shift perturbations of CheY upon the addition of Mg2+ are strongly correlated with closeness to the ion and in general are small in regions of allosteric conformational change (Figure S2), suggesting no significant structural rearrangements. In addition, the crystal structure of Salmonella typhimurium CheY (Stock et al., 1993) (which differs by 3 amino acids from E. coli) and the NMR structure of E. coli CheY (Moy et al., 1994), both with Mg2+ present, have no indication of any large structural rearrangement. For these reasons, the different CheY dynamics observed with or without Mg2+ do not appear to be the direct result of Mg2+-induced conformational change.
Physiological Mg2+ concentrations enhance allosteric dynamics
To gain greater insight into the effect of Mg2+ on CheY dynamics, we obtained estimates of Rex at additional Mg2+ concentrations of 1 mM and 75 mM (Figure 3). Thus, assuming a single binding site (see Supplemental Discussion), 1 mM CheY was calculated (based on the Kd of 1.5 mM) to be bound by Mg2+ at a level of 0%, 31%, 86%, and 98% for 0 mM, 1 mM, 10 mM, and 75 mM concentrations of Mg2+, respectively. For all concentrations of Mg2+, estimates of Rex were obtained as the difference of R2,eff values at the lowest and highest values of τcp used for dispersion measurements. Overall, the dependence of Rex on the concentration of Mg2+ is immediately apparent: Rex in the presence of no Mg2+ or very high concentration of Mg2+ (75 mM) is relatively low, and Rex at intermediate levels of Mg2+ (1 mM) is very high (Figure 3). Furthermore, peak broadening was evident in many residues at intermediate Mg2+ concentrations, implying increased motion. While high Rex levels at intermediate concentrations of Mg2+ are expected for residues in close proximity to the bound ion (assuming appropriate line-broadening kinetics), high Rex levels are not expected at residues distal to Mg2+ that are not structurally affected by the ion, such as on the FliM binding interface. Yet, Rex at V107 is quite pronounced (28 s−1) at 1 mM Mg2+. We note that Mg2+ binding induces a small chemical shift change of 0.52 ppm for the amide nitrogen of V107 (Figure S2), especially compared to the large chemical shift change of 8.6 ppm brought about by binding of BeFx. Furthermore, A88 experiences extreme line broadening at intermediate concentrations of Mg2+. The dispersion experienced at this position is larger than expected when compared to neighboring residues. Thus, there is something particular about partial Mg2+ saturation that increases conformational dynamics at A88 and even further at the FliM binding interface more than 10 Å away. The difference in Rex far from Mg2+ is due to a change in the I-to-A transition (see Supplemental Discussion and Figures S3 and S4). Importantly, this occurs without significant changes to the structure of the ground state (see above), even though phosphorylation – which occurs at nearly the same location as Mg2+ binding – induces a large structural change. The Mg2+ concentration’s effect on the I-to-A transition is likely biologically relevant since Rex is largest at intermediate concentrations of Mg2+ (1 mM); this concentration is close to the Kd for Mg2+ and CheY (1.5 mM), and also close to the concentration of Mg2+ in E. coli (~1–2 mM Mg2+ (Alatossava et al., 1985)).
Figure 3. Effect of Mg2+ concentration on Rex.

Rex is approximated by the change in R2,eff of two CPMG relaxation dispersion planes. Approximate Rex is shown for CheY in the presence of 1 mM EDTA (blue), 1 mM Mg2+ (red), 10 mM Mg2+ (green), and 75 mM Mg2+ (black) for all residues with non-zero Rex values at 1 mM Mg2+. Residues G39 and A88 are broadened away at 1 mM Mg2+ presumably from exchange, indicated by *. Also, there are no data for Y106 at 1 mM Mg2+ and V108 at 1 mM EDTA because of peak overlap.
See also Figures S1, S2, S3 and S4.
Key residues display linear chemical shifts in response to Mg2+ binding
We observed that, for Y106, V107, and E89, the NH chemical shifts move along a line for various states of CheY, which include apo, BeFx-bound, inactive mutant T87I CheY (Ganguli et al., 1995), and activated mutant A113P CheY (Smith et al., 2004) (Figures 4A, 4B, and 4C). Linear chemical shifts are indicative of a classic, two-state, I-to-A switch, as was described for the receiver domain of the NtrC RR (Volkman et al., 2001). To test whether linear shifts were observed throughout CheY, we plotted all Mg2+-induced CSPs on a normalized vector scale (Figure 4D). If Mg2+ shifts this equilibrium, all chemical shifts should fall along the dotted line from inactive mutant T87I to BeFx-bound wild-type CheY. However, with a few exceptions, the CSPs distribute essentially randomly, with no apparent preferred “direction”. Thus, it appears that, overall, Mg2+ binding elicits either a new conformational state(s) that has only subtle differences from the I state and bears no resemblance to A-like chemical shifts, or Mg2+ has essentially no significant effect on CheY conformation. This is further supported by no significant change in the affinity of binding of FliM to unphosphorylated CheY with the addition of Mg2+ (Figure S5). Nevertheless, there are a few key residues (e.g. Y106, E89) that display the linear chemical shifts suggestive of an equilibrium shift (Figure 4). Interestingly, additional residues that fall near the line (Figure 4D, M85, A97 and A98) lie in the pocket surrounding W58. These residues, along with Y106 and E89, may experience a shift in conformation due to the presence of Mg2+ (discussed below).
Figure 4. Chemical shift perturbations (CSP) from inactive to BeFx activated CheY.

Overlaid 1H-15N HSQCs are shown with peaks highlighted for (A) E89, (B) Y106, and (C) V107 from various conditions and mutants of CheY: inactivating mutant T87I with 10 mM Mg2+ (green), wild-type with 1 mM EDTA (blue), wild-type with 10 mM Mg2+ (red), activating mutant A113P with 10 mM Mg2+ (orange), BeFx-bound wild-type with 10 mM Mg2+ (purple) and BeFx-bound A113P with 10 mM Mg2+ (pink).
(D) To display all residues, CSP is normalized according to inactivating mutant T87I with 10mM Mg2+ (green) and BeFx-bound wild-type CheY (purple). Each circle represents the normalized CSP of each residue of wild-type CheY in the presence of 10 mM Mg2+ (red) and 1mM EDTA (blue). Residues are only shown if was greater than 0.20 where ΔHN and ΔN are between T87I and wild-type BeFx-bound CheY. If ΔHN < 0.06 ppm or ΔN < 0.25 ppm, the chemical shifts are considered within error in that dimension and the residues were removed. Therefore, while V107 appears linear, it is not shown in (D) since the proton shift is < 0.06 ppm.
See also Figure S5.
Discussion
For half a century now, allosteric conformational change has been revealed to be an extremely successful mechanism for regulating protein function. Yet, key specific questions regarding allostery have yet to be answered. Is the conformational change concerted? Are there structural intermediates? What are those intermediates? What is the dynamic process that allows conversion between the end conformations and how is it triggered? In summary, we still have a poor understanding of how allosteric conformational changes actually happen. By conducting in-depth NMR experiments on the small allosteric protein CheY, we hoped to gain new insights into the mechanism(s) of allostery. CheY is an excellent system for probing allostery because numerous crystal structures have been solved in a variety of liganded states and it is highly amenable to NMR relaxation experiments. It also exhibits a rich complement of residues with μs-ms motion at residues that connect the physically separated upstream and downstream effector sites.
Allostery in CheY does not operate by a simple shift of a two-state equilibrium
From previous crystallographic, NMR, and functional data (Birck et al., 1999; Feher and Cavanagh, 1999; Volkman et al., 2001; Gardino et al., 2009), the evidence suggests that receiver domains primarily adopt two conformations and hence allostery is achieved by shifting a pre-existing dynamic equilibrium between these two states, consistent with the MWC model of allostery. Based on this previous work on NtrC, FixJ, and Spo0F, and our initial observation here of Rex in the allosteric network of residues, the simplest expectation would be that CheY undergoes a concerted switch between I (inactive) and A (active) conformations. Here, we show using NMR relaxation dispersion that intrinsic allostery in CheY does not appear to operate by a simple shift of a two-state equilibrium. This conclusion rests, in part, on the ability of relaxation dispersion experiments to accurately identify conformational exchange rates and populations.
It is possible that unphosphorylated CheY does not completely switch to the A state but rather samples a conformational intermediate between the I and A states, and this may account for the lack of correlation in Δδ and Δω values. However, this cannot be confirmed experimentally without having accurate chemical shifts for the intermediate state, which has yet to be isolated in solution. Alternatively, if a structural model for the intermediate exists, the chemical shifts can be calculated using SHIFTX (Neal et al., 2003); then Δδ can be calculated between the inactive and intermediate states and compared with Δω from relaxation dispersion fits. This was carried out using the partially switched structure of CheY bound to FliM without phosphorylation (2B1J (Dyer and Dahlquist, 2006)) as the intermediate, but the Δω values did not correlate. In any case, if the observed Rex values result only from unphosphorylated CheY switching concertedly to an intermediate state, one would still expect kex and populations to be uniform, yet they are not.
Distal quartet of residues directs allosteric conformational change
In order for CheY to undergo its allosteric transition it must primarily do two things: (1) reposition the β4-α4 loop, and (2) swing Y106 into the pocket just under the β4-α4 loop. We measured three residues, Y106, V107, and E89, that appeared to approach two-state behavior, although this is apparent only in the chemical shift data (Figure 4) and not in the relaxation dispersion data (Figures 1 and 2). One possible explanation for the linear chemical shifts is that because Y106 is at the solvent interface and presumably under less structural constraint, its position is to a reasonable approximation either “out” or “in”, yielding a bimodal distribution of possible chemical shift values for the amide of Y106. The strong ring currents dominate the chemical shifts of both Y106 and V107 amides. The amide chemical shift of E89, located in the β4-α4 loop, likely monitors the remodeling of this loop. We utilized SHIFTX (Neal et al., 2003) to calculate the effect on chemical shift from a conformational change. Y106 and V107 amide chemical shifts are very sensitive to the rotation of Y106, with minimal effects from the location of the β4-α4 loop. By contrast, the amide shift of E89 is very sensitive to movement of the β4-α4 loop, but not to Y106. Therefore, E89 monitors the β4-α4 loop motion, and Y106 and V107 likely monitor the rotation of Y106. Taken together, the chemical shift data reveal a localized shift to active-like states.
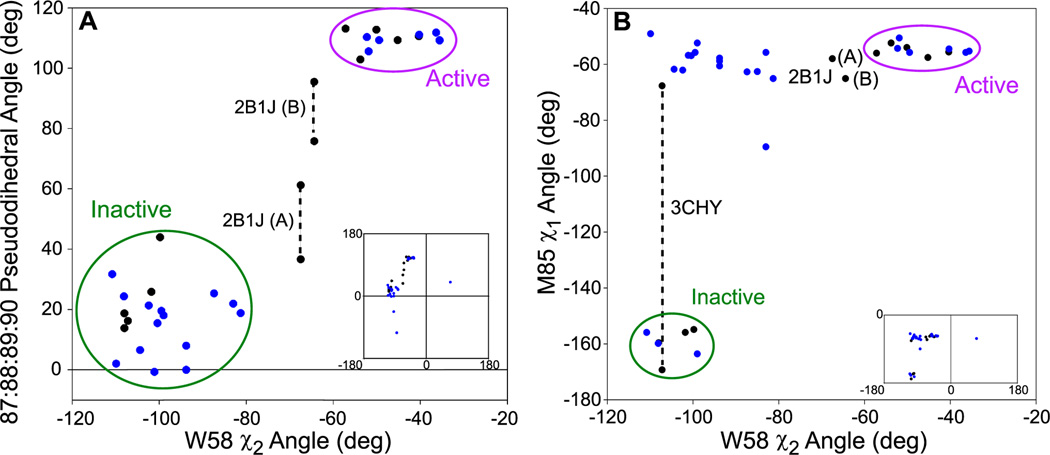
While E89, Y106, and V107 have the most linear chemical shifts, a few residues are nearly linear (M85, A97, and A98) while all others show no semblance of linearity. M85, A97, and A98 have not been discussed previously as part of the allosteric mechanism. Yet, these residues line the pocket occupied by W58 and are proximal to the β4-α4 loop and Y106. Examination of CheY in its inactive (3CHY (Volz and Matsumura, 1991)), active (1FQW (Lee et al., 2001a)), and intermediate conformations (2B1J (Dyer and Dahlquist, 2006)) reveals an extended set of conformational interactions. Upon activation, the hydroxyl of Y106 hydrogen bonds to the backbone carbonyl of E89, which is allowed by movement of the β4-α4 loop. What appears to further stabilize this interaction is a large movement of the E89 side chain towards the side chain of W58, resulting in hydrophobic contacts between the methylene groups of E89 and the tryptophan indole. Interestingly, superimposition of inactive and active CheY structures shows that the tryptophan indole group reorients by ~55° and the intermediate state (when CheY is bound only to FliM peptide) shows the indole at an intermediate angle. Although tryptophan at position 58 is not conserved amongst RRs, the receiver domain from PhoB displays a similar rotation of ~60° upon binding BeFx (Sola et al., 1999; Bachhawat et al., 2005). When analyzing a range of crystal structures, we find that the angle of the W58 indole correlates with the pseudodihedral angle 87:88:89:90 (Figure 5A). This pseudodihedral angle has been used to indicate the location of the β4-α4 loop (Dyer and Dahlquist, 2006). In order to contact E89, W58 increases its distance from M85, which is associated with W58 in the inactive conformation. Thus, M85 and E89 appear to be in competition for W58, with E89 making contact only in the active conformation. In further support of this extended mechanism, we note that in 3CHY, M85 was found to occupy two conformations (similar to Y106), of which only one is adopted in the intermediate and active conformations and the other is adopted in other crystal structures of the inactive state (Figure 5B). Additional crystal structures reveal that M85 often occupies the “active” rotamer even though W58 has not rotated to the active state (Figure 5B). This indicates that W58 is not as robustly coupled to M85 as the β4-α4 loop. However, one should be cautioned not to overinterpret the lack of coupling between M85 and W58; since the two M85 side-chain conformations have similar electron density, this leads to the possibility that the crystal structures did not accurately account for multiple conformations. We also note that the amide of W58 could not be assigned, which is consistent with its indole sampling at least two conformations prior to full activation. We propose here that the quartet of W58, M85, E89, and Y106 are coupled, as a key component of the allosteric signaling process from phosphorylation.
Figure 5. Comparison of dihedral angles in crystal structures of CheY.
(A) Comparison of the 87:88:89:90 pseudodihedral angle with the rotation of W58 and (B) comparison of M85 χ1 angle with the rotation of W58. The pseudodihedral angle was measured using Cα coordinates. In black are wild-type CheY crystal structures (1FQW, 1F4V, 2B1J, 1JBE, 3CHY, 2CHE, and 1C4W) and in blue are mutant or CheA-bound CheY crystal structures (only single site mutations and >2.5 Å resolution, full list is provided in Table S1). Insets are the full range of angles occupied while large plots are zoomed on the majority of crystal structures. See Table S1.
A88 links Mg2+ binding to the allosteric quartet
Upon Mg2+ binding to CheY, intrinsic fluorescence of W58 is quenched (Lukat et al., 1990), indicating that Mg2+ may play an additional role in this signaling cascade. T87 is considered to be the initial “sensor” of phosphorylation and triggers the allosteric response (Appleby and Bourret, 1998). However, it is unclear whether additional residues play a crucial role and which residues may sense Mg2+. A88, in addition to T87, also hydrogen bonds with the phosphoryl group via its backbone amide. Perhaps most importantly, from the relaxation dispersion data in 10 mM Mg2+, A88 fit to parameters unlike any other residue. While most residues fit to populations (pI) of 0.98–0.99, A88 fit to 0.91. Furthermore, its dispersion curve visually resembles residues 12 and 38, which are near to the bound Mg2+ ion, more than the allosteric residues 87, 89, 106, and 107 (Figure 1B). Careful inspection of the fit to A88 indicates that its R2° values are anomalously high and the quality of the fits is significantly lower than the other residues (Table 1). Thus, we suspect that in addition to the motion that gives rise to the main part of A88’s dispersion curve, there is a separate, faster motion on the μs timescale. Only A88 appears to have this more complex combination of motions. Furthermore, A88 experiences line broadening severely affected by the concentration of Mg2+. At 1 mM Mg2+, A88 is completely broadened away in the 1H-15N HSQC. Given its behavior at different Mg2+ concentrations, A88 may be especially sensitive to Mg2+ binding and release.
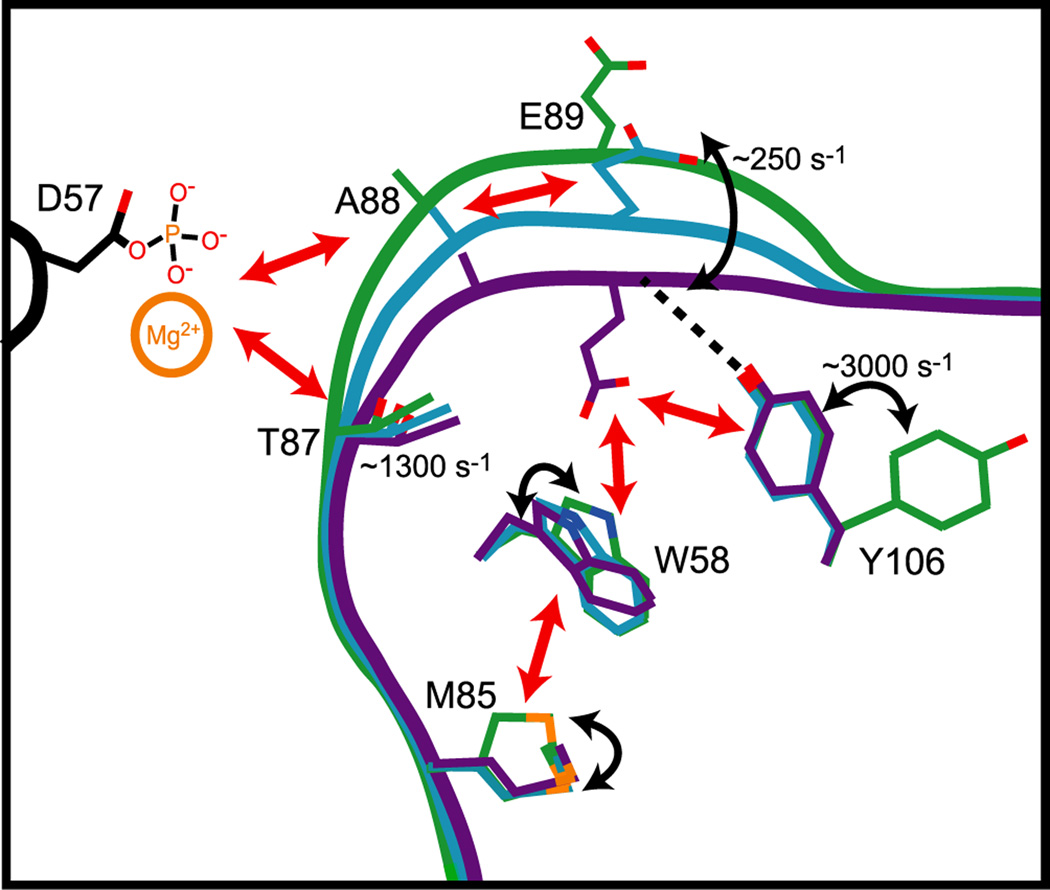
We therefore hypothesize that A88 acts as a crucial monitor of ion binding and/or phosphorylation at the active site. It is important to note that, even though they are adjacent, residues 86–89 have different exchange parameters. Therefore, at face value these residues appear largely independent. Nevertheless, because these residues lie within the allosteric pathway and all show motion on the μs-ms timescale, we suggest that in addition to T87, A88 provides a linkage to E89, and that the recruitment of W58 by E89 represents a key step in switching the β4-α4 loop. Given these considerations, we propose that the allosteric network in CheY is facilitated by several distinct behaviors that extend from D57, to T87 and A88, and ultimately to the distal quartet of W58, M85, E89, and Y106 (Figure 6).
Figure 6. Cartoon model of the allosteric signaling mechanism in CheY.
Representations of inactive (green), intermediate (blue) and active (purple) CheY with important signaling residues displayed.
Evidence for segmental allosteric dynamics
Considering the chemical shift and relaxation dispersion data together leads inexorably to a paradox. How can there be linear chemical shifts in key allosteric residues when switching appears not to be concerted by relaxation dispersion? A distribution of 2D chemical shifts along a line is typically considered to indicate a rapid exchange between two states (Volkman et al., 2001), with the spread of positions resulting from a shifting of the equilibrium. While this is indeed highly likely to be the case at the spin probes’ “site” (amide H and N could be considered as one site), if two probes show this behavior at significant distances from one another, they need not result from the same conformational exchange process. Thus, we consider if the observed linearity can result from local two-state switching. Specifically, the switching of W58 can be sensed by residues 85, 97 and 98, but the switching of Y106 appears independent of W58. It is possible that some groups of residues switch concertedly, yet with some independence from other switching events. Even though such a group appears to switch independently, there could be coupling with a time lag or interference effects. This situation could give rise to linear chemical shifts, yet show dispersion curves that cannot be fitted together, as was the case for CheY here. Thus, we propose that CheY’s allosteric switching is segmental in nature. It is not truly two-state, but there are several groupings of coupled residues that move with some independence and perhaps in a preferred sequential order. This rough model also suggests that CheY shares features of both conformational selection and induced fit. It does not appear that the A-state is fully formed in the absence of phosphorylation, since there is a poor correlation of Δω and Δδ. Yet, some residues may sample “active-like” local conformations. We speculate that binding and release of Mg2+ actuates conformational transitions along the allosteric pathway, from T87 and A88 to the W58-M85-E89-Y106 quartet (Figure 6). This is highlighted by maximal Rex at Y106 occurring at the intermediate Mg2+ concentration of 1 mM.
The segmental nature of CheY’s allosteric dynamics is perhaps best supported by the behavior of Y106. The rate of dynamic switching as observed by CPMG relaxation dispersion is clearly faster than the other switching residues, as evidenced by visual inspection of the dispersion curve (Figure 1B), as well as the fitted kex of 3100 s−1 (Table 1). Assuming that this observed motion corresponds to the allosteric conformational change, Y106 must be moving with at least partial independence from the other residues. That Y106 switches rapidly and with relative ease is supported by the significantly populated active-like inward position in 3CHY (Volz and Matsumura, 1991), as well as its low activation barrier for switching, as determined from a computational study (Ma and Cui, 2007). Even from the linear chemical shift data (Figure 4), it is seen that in 10 mM Mg2+, Y106 is only ~20% toward the active state chemical shift, whereas the other linear shift data appear to be in a range of ~20–60% toward the active state (Figure 4D). Thus, neither relaxation dispersion nor chemical shift data fully support a model for pre-existing, concerted, two-state switching dynamics.
Comparison to other systems
Most of our knowledge of allosteric dynamics in RRs derives from NMR work on the nitrogen regulatory protein C (NtrC) receiver domain. A combination of chemical shift analysis (particularly of mutants) and transverse relaxation measurements led to the proposal of a two-state dynamic equilibrium between I and A conformations in NtrC (Volkman et al., 2001). The recent CPMG relaxation dispersion study on NtrC reported that those data are consistent with a two-state pre-existing equilibrium and that unphosphorylated NtrC populates the active state at the ~14% level (Gardino et al., 2009); interestingly, it appears that that study was carried out in the absence of Mg2+. By contrast, because CheY does not appear two-state (even in the absence of Mg2+) we cannot currently estimate the population of the active state. Even when considering residues locally, the populations of “excited states” from relaxation dispersion (Table 1) are typically much lower than those deduced from chemical shift plots (Figure 4), assuming that T87I populates only the inactive state. Unfortunately, relaxation dispersion data have not been reported for any other RR to the best of our knowledge. CPMG relaxation dispersion is an excellent approach to studying allosteric mechanisms if the conformational switching occurs on the timescale between a few hundred s−1 to ~3000 s−1. Application of this method to other allosteric proteins holds the promise of revealing allosteric mechanisms. This has been done successfully for the KIX domain of the CREB binding protein (Bruschweiler et al., 2009), catabolite activator protein (CAP) (Tzeng and Kalodimos, 2009), and the PBX1 homeodomain (Farber and Mittermaier, 2011), each of which exhibited two-state allosteric switching.
Additionally, a study on the trp RNA-binding attenuation protein (TRAP) revealed allosteric motions as a series of localized conformational changes (Kleckner et al., 2012). TRAP’s behavior is similar to CheY’s segmental dynamics and shed light on the diverse mechanisms proteins may employ to achieve allostery. These results imply that a protein may not solely employ a MWC or Koshland-Nemethy-Filmer (KNF) allosteric mechanism, but may combine them to form a complicated switching event.
Experimental Procedures
Protein expression and purification
E. coli CheY DNA (provided by Dr. Bob Bourret, University of North Carolina at Chapel Hill) was subcloned into the pET28a plasmid (Novagen). Mutants were made by site-directed mutagenesis PCR. The CheY vector was transformed into BL21 Star (DE3) cells (Invitrogen) and grown in minimal media with the appropriate isotope(s): 15NH4Cl (99%) and/or D-glucose (U-13C6-99%) as the sole nitrogen and carbon sources, respectively). Samples for relaxation dispersion were grown to yield high (>80%) 2H incorporation. Cells were grown at 37 °C until A600 reached 0.6. Isopropyl 1-thio-β-D-galactopyranoside was added to a final concentration of 1 mM and cells were grown for an additional 22–26 hours (32–36 hours if 2H2O used) at 20 °C. The cells were harvested by centrifugation, resuspended in buffer A (25 mM Tris, 10 mM MgCl2, pH 8.0), and stored at −20 °C until needed.
Resuspended cells underwent three freeze/thaw cycles and sonication. The lysate was then centrifuged at 6000 rpm and dialyzed overnight into buffer A at 4 °C. The protein was purified on a Q Sepharose Fast Flow column equilibrated with buffer A. CheY eluted approximately halfway along a gradient of buffer B (buffer A with the addition of 1.5 M NaCl) from 10 to 55% buffer B over 135 mL. The eluent was passed over either a G-50 or G-75 gel-filtration column equilibrated with NMR buffer (50 mM NaPi, 0.02% NaN3, pH 7.0 and an appropriate amount of MgCl2 and/or EDTA). Pure CheY was concentrated to between 1.2 and 3 mM and stored at 4 °C. Concentrations were determined using the extinction coefficient of 10.3 cm−1/mM (Silversmith et al., 2001) and 14.3 cm−1/mM for wild-type and Y106W CheY, respectively.
NMR Spectroscopy
All NMR spectra were collected on 1 mM CheY with NMR buffer and 10% 2H2O. The appropriate amount of MgCl2 or EDTA was present in the NMR buffer, depending on the experiment. NMR experiments were collected at 15 °C on Varian INOVA spectrometers equipped with room-temperature (500 and 600 MHz) or cryogenic (700 MHz) probes. Assignments for wild-type CheY in the presence of 10 mM Mg2+ were made from triple resonance experiments, supplemented by previously published assignments (Moy et al., 1994). We made use of both triple resonance experiments and assignments of wild-type CheY to assign mutant and BeFx-bound wild-type CheY (both in the presence of 10 mM Mg2+). Assignments of CheY in the presence of 1 mM EDTA were made based on following peak shifts during titration with Mg2+. All NMR data were processed using NMRPipe (Delaglio et al., 1995) and visualized with NMRDraw and NMRView (Johnson, 2004).
Preparation of BeFx-bound CheY
NMR samples of BeFx-bound CheY were prepared with high enough concentrations of BeCl2 and NaF to ensure saturation of CheY and in an ideal ratio for a BeF3− complex. The samples were prepared by first combining all components of a typical unphosphorylated CheY sample (e.g. 1 mM CheY protein, 10 mM MgCl2, 10% D2O). Then BeCl2 and lastly NaF were added to a final concentration of 4 mM and 25 mM, respectively. The samples were then left at room temperature overnight and centrifuged briefly before transferring to an NMR tube.
15N CPMG Relaxation Dispersion
Relaxation dispersion experiments were carried out using relaxation-compensated CPMG experiments (Loria et al., 1999). Most experiments were collected with a 40 ms total CPMG period and one with 20 ms. 12 or 13 τcp values with 2 duplicates and a reference experiment were collected interleaved. R2,eff was calculated from peak intensities as previously described (Mulder et al., 2001).
NMR relaxation dispersion experiments measuring motions on the μs-ms timescale were fit assuming a two-state model. Residues were considered to have significant motions and worth analyzing if Rex > 2 s−1. These residues were analyzed as previously described (Mauldin et al., 2009). Briefly, using an F-test (αcritical = 0.01), the data were fit to both a model that assumes no exchange and a simple two-state model to determine which residues exhibited a significant dependence of R2,eff on 1/τcp. These residues were then fit to the Carver-Richards equation (Palmer et al., 2001). By fitting the dependence of R2,eff on 1/τcp, the exchange rate (kex), populations (pI, pA), and chemical shifts (Δω) were determined. Data from two and three field strengths were fit simultaneously for 1 mM EDTA and 10 mM Mg2+, respectively, using the in-house program exrate2.0. Errors were estimated by Monte Carlo simulation.
In the limit of fast exchange, eq. (2) can be used to fit dispersion curves (Palmer et al., 2001). Residues were also fit to this equation to assess appropriateness of the Carver-Richards equation. For many of the slower residues, the simplified fast equation did not give the same fit as the Carver-Richards equation and therefore we decided the latter was needed to distinguish the best parameters.
Fitting all of the residues together to the same kex and pI or “group fitting” was done in the same manner as previously described (Mauldin et al., 2009; Mulder et al., 2001). In order to justify group fitting, χ2group/ χ2local ≤ 2 where χ2local is the χ2 when a residue is locally fit and χ2group is the χ2 of the same residue when group fit. If necessary, the residue with the highest ratio is removed and the group fitting is attempted again. For CheY, the attempt to group fit resulted in elimination of all residues partly due to the high quality of data.
After all fitting was complete, two residues (G39 in 10 mM Mg2+ and D13 in 1 mM EDTA) were determined to have errors that were too large to be accurately fit. Visual inspection revealed both of the peaks to be extremely broad in the 1H-15N HSQC. Therefore, these residues undergo motions but are unable to be fit due to high error in the intensity measurements. These residues were removed from any further quantitative interpretation of the data.
Utilizing cpmg_fit (provided by Dmitry Korzhnev and Lewis Kay), curves from each residue were fit to a three-state model; however, the fits were not robust due to the limited data. These preliminary fits yielded individual three-state parameters that were diverse. Nevertheless, using an F-test (αcritical = 0.05), three residues (A36, T87, and E89) in 10 mM Mg2+ fit better to a three-state model than a two-state model. In 1mM EDTA, no residues showed improvement with a three-state fit. Additionally, group fitting to a three-state model did not provide significant statistical improvement. This suggests that CheY is not functioning as a three-state concerted switch.
The sign of Δω was determined from comparison of peak positions in the HSQC and HMQC spectra (Skrynnikov et al., 2002).
Determination of approximate Rex by relaxation dispersion
An estimate of Rex can be obtained by using a very low and a very high 1/τcp, as long as the rate of exchange is less than ~5000 s−1. We used the same experimental set-up as used for measuring full relaxation dispersion curves (as described above) except only two τcp values, 10.0 ms and 0.556 ms were employed. A 40 ms total relaxation time (T) was used and all Rex estimates were made at 15 °C. Rex was calculated from the peak intensities for each plane using
| (2) |
in which I is the peak intensity for the given τcp value.
Supplementary Material
Highlights.
NMR dynamics data reveal CheY does not undergo concerted, two-state switching
Asynchronous, local switching of a network of residues facilitates CheY activation
Mg2+ enhances the dynamics associated with the inactive-to-active transition
Acknowledgements
We thank Bob Bourret, Ruth Silversmith and Bob Immormino for kindly providing the CheY DNA, helpful discussions, and critical reading of the manuscript. We would like to thank Matthew Whitley for his help in the assignments and preparation of BeFx-bound CheY. We also thank Peter Thompson and Austin Smith for their help with analysis of A113P and Y106W mutants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alatossava T, Jutte H, Kuhn A, Kellenberger E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 1985;162:413–419. doi: 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleby JL, Bourret RB. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active-site quintet. J. Bacteriol. 1998;180:3563–3569. doi: 10.1128/jb.180.14.3563-3569.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachhawat P, Swapna GV, Montelione GT, Stock AM. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure. 2005;13:1353–1363. doi: 10.1016/j.str.2005.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MD, Wolanin PM, Stock JB. Signal transduction in bacterial chemotaxis. Bioessays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- Barak R, Eisenbach M. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry. 1992;31:1821–1826. doi: 10.1021/bi00121a034. [DOI] [PubMed] [Google Scholar]
- Bellsolell L, Prieto J, Serrano L, Coll M. Magnesium binding to the bacterial chemotaxis protein CheY results in large conformational changes involving its functional surface. J. Mol. Biol. 1994;238:489–495. doi: 10.1006/jmbi.1994.1308. [DOI] [PubMed] [Google Scholar]
- Birck C, Mourey L, Gouet P, Fabry B, Schumacher J, Rousseau P, Kahn D, Samama JP. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure. 1999;7:1505–1515. doi: 10.1016/s0969-2126(00)88341-0. [DOI] [PubMed] [Google Scholar]
- Bourret RB. Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 2010;13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruschweiler S, Schanda P, Kloiber K, Brutscher B, Kontaxis G, Konrat R, Tollinger M. Direct observation of the dynamic process underlying allosteric signal transmission. J. Am. Chem. Soc. 2009;131:3063–3068. doi: 10.1021/ja809947w. [DOI] [PubMed] [Google Scholar]
- Clegg DO, Koshland DE. The role of a signaling protein in bacterial sensing: behavioral effects of increased gene expression. P. Natl. Acad. Sci.-Biol. 1984;81:5056–5060. doi: 10.1073/pnas.81.16.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends in Biochem. Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Karplus M. Allostery and cooperativity revisited. Protein Sci. 2008;17:1295–1307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dyer CM, Dahlquist FW. Switched or not?: the structure of unphosphorylated CheY bound to the N terminus of FliM. J. Bacteriol. 2006;188:7354–7363. doi: 10.1128/JB.00637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber PJ, Mittermaier A. Concerted dynamics link allosteric sites in the PBX homeodomain. J. Mol. Biol. 2011;405:819–830. doi: 10.1016/j.jmb.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Feher VA, Cavanagh J. Millisecond-timescale motions contribute to the function of the bacterial response regulator protein Spo0F. Nature. 1999;400:289–293. doi: 10.1038/22357. [DOI] [PubMed] [Google Scholar]
- Galperin MY. Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 2010;13:150–159. doi: 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli S, Wang H, Matsumura P, Volz K. Uncoupled phosphorylation and activation in bacterial chemotaxis. The 2.1-A structure of a threonine to isoleucine mutant at position 87 of CheY. J. Biol. Chem. 1995;270:17386–17393. [PubMed] [Google Scholar]
- Gao R, Mack TR, Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends in Biochem. Sci. 2007;32:225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardino AK, Villali J, Kivenson A, Lei M, Liu CF, Steindel P, Eisenmesser EZ, Labeikovsky W, Wolf-Watz M, Clarkson MW, et al. Transient non-native hydrogen bonds promote activation of a signaling protein. Cell. 2009;139:1109–1118. doi: 10.1016/j.cell.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhaniyogi J, Robinson VL, Stock AM. Crystal structures of beryllium fluoride-free and beryllium fluoride-bound CheY in complex with the conserved C-terminal peptide of CheZ reveal dual binding modes specific to CheY conformation. J. Mol. Biol. 2006;359:624–645. doi: 10.1016/j.jmb.2006.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: a flux description of reaction mechanism. Proc. Natl. Acad. Sci. USA. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- Hubbard JA, MacLachlan LK, King GW, Jones JJ, Fosberry AP. Nuclear magnetic resonance spectroscopy reveals the functional state of the signalling protein CheY in vivo in Escherichia coli. Mol. Microbiol. 2003;49:1191–1200. doi: 10.1046/j.1365-2958.2003.03628.x. [DOI] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta. 2011;1814:942–968. doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner IR, Gollnick P, Foster MP. Mechanisms of allosteric gene regulation by NMR quantification of microsecond-millisecond protein dynamics. J. Mol. Biol. 2012;415:372–381. doi: 10.1016/j.jmb.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Cho HS, Pelton JG, Yan D, Berry EA, Wemmer DE. Crystal structure of activated CheY. Comparison with other activated receiver domains. J. Biol. Chem. 2001;276:16425–16431. doi: 10.1074/jbc.M101002200. [DOI] [PubMed] [Google Scholar]
- Loria JP, Rance M, Palmer AG. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 1999;121:2331–2332. [Google Scholar]
- Lukat GS, Stock AM, Stock JB. Divalent metal ion binding to the CheY protein and its significance to phosphotransfer in bacterial chemotaxis. Biochemistry. 1990;29:5436–5442. doi: 10.1021/bi00475a004. [DOI] [PubMed] [Google Scholar]
- Ma L, Cui Q. Activation mechanism of a signaling protein at atomic resolution from advanced computations. J. Am. Chem. Soc. 2007;129:10261–10268. doi: 10.1021/ja073059f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura P, Rydel JJ, Linzmeier R, Vacante D. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J. Bacteriol. 1984;160:36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauldin RV, Carroll MJ, Lee AL. Dynamic dysfunction in dihydrofolate reductase results from antifolate drug binding: modulation of dynamics within a structural state. Structure. 2009;17:386–394. doi: 10.1016/j.str.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy FJ, Lowry DF, Matsumura P, Dahlquist FW, Krywko JE, Domaille PJ. Assignments, secondary structure, global fold, and dynamics of chemotaxis Y protein using three- and four-dimensional heteronuclear (13C,15N) NMR spectroscopy. Biochemistry. 1994;33:10731–10742. [Google Scholar]
- Mulder FA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 2001;8:932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- Neal S, Nip AM, Zhang HY, Wishart DS. Rapid and accurate calculation of protein H-1, C-13 and N-15 chemical shifts. J. Biomol. NMR. 2003;26:215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Takada S. Dynamic energy landscape view of coupled binding and protein conformational change: induced-fit versus population-shift mechanisms. Proc. Natl. Acad. Sci. USA. 2008;105:11182–11187. doi: 10.1073/pnas.0802524105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- Sanders DA, Gillece-Castro BL, Stock AM, Burlingame AL, Koshland DE., Jr. Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J. Biol. Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- Schuster M, Silversmith RE, Bourret RB. Conformational coupling in the chemotaxis response regulator CheY. Proc. Natl. Acad. Sci. USA. 2001;98:6003–6008. doi: 10.1073/pnas.101571298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silversmith RE, Smith JG, Guanga GP, Les JT, Bourret RB. Alteration of a nonconserved active site residue in the chemotaxis response regulator CheY affects phosphorylation and interaction with CheZ. J. Biol. Chem. 2001;276:18478–18484. doi: 10.1074/jbc.M011418200. [DOI] [PubMed] [Google Scholar]
- Simonovic M, Volz K. A distinct meta-active conformation in the 1.1-A resolution structure of wild-type apoCheY. J. Biol. Chem. 2001;276:28637–28640. doi: 10.1074/jbc.C100295200. [DOI] [PubMed] [Google Scholar]
- Skrynnikov NR, Dahlquist FW, Kay LE. Reconstructing NMR spectra of "invisible" excited protein states using HSQC and HMQC experiments. J. Am. Chem. Soc. 2002;124:12352–12360. doi: 10.1021/ja0207089. [DOI] [PubMed] [Google Scholar]
- Smith JG, Latiolais JA, Guanga GP, Pennington JD, Silversmith RE, Bourret RB. A search for amino acid substitutions that universally activate response regulators. Mol. Microbiol. 2004;51:887–901. doi: 10.1046/j.1365-2958.2003.03882.x. [DOI] [PubMed] [Google Scholar]
- Sola M, Gomis-Ruth FX, Serrano L, Gonzalez A, Coll M. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J. Mol. Biol. 1999;285:675–687. doi: 10.1006/jmbi.1998.2326. [DOI] [PubMed] [Google Scholar]
- Stock AM, Martinez-Hackert E, Rasmussen BF, West AH, Stock JB, Ringe D, Petsko GA. Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol. Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462 doi: 10.1038/nature08560. 368-U139. [DOI] [PubMed] [Google Scholar]
- Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J. Biol. Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- Welch M, Oosawa K, Aizawa S, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarski T, Zagrovic B. Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin. Proc. Natl. Acad. Sci. USA. 2009;106:19346–19351. doi: 10.1073/pnas.0906966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Cho HS, Hastings CA, Igo MM, Lee SY, Pelton JG, Stewart V, Wemmer DE, Kustu S. Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc. Natl. Acad. Sci. USA. 1999;96:14789–14794. doi: 10.1073/pnas.96.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX. From induced fit to conformational selection: a continuum of binding mechanism controlled by the timescale of conformational transitions. Biophys. J. 2010;98:L15–L17. doi: 10.1016/j.bpj.2009.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.