Abstract
Background
In HBV-infected patients, different genotypes of the hepatitis B virus influence liver disease progression and response to antiviral therapy. Moreover, long-term antiviral therapy will eventually select for drug-resistant mutants. Detection of mutations associated to antiviral therapy and HBV genotyping are essential for monitoring treatment of chronic hepatitis B patients.
Results
In this study, a simple method of partial-S gene sequencing using a common PCR amplification was established for genotyping clinical HBV isolates sensitively, which could detect the drug-resistant mutations successfully at the same time.
Conclusions
The partial S gene sequencing assay developed in this study has potential for application in HBV genotyping and drug resistant mutation detection. It is simpler and more convenient than traditional S gene sequencing, but has nearly the same sensitivity and specificity when compared to S gene sequencing.
Background
Eight distinct genotypes (A to H) of hepatitis B virus (HBV) have been identified, and this classification is based on the distance of the nucleotide sequence from the viral genome of 8% or greater [1,2]. These genotypes also have a distinct geographical distribution, while genotypes B and C are more common in China. Since genotypes of HBV influence liver disease progression and response to antiviral therapy in HBV-infected patients, several methods have been developed for genotyping of HBV strains [3], these include sequence analysis; microarray (DNA-Chip) [4,5]; reverse hybridization [6]; restriction fragment length polymorphism (RFLP) [7]; serological assays and genotype-specific PCR assays [8,9]. These techniques have the disadvantage that they are based on specific hybridization of HBV DNA, and nucleotide changes can interfere with this process and subsequent sequence analysis. For example, RFLP and multiple PCR methods might give wrong results even for a single base mutation. Serological assay has a low cost and does not rely on PCR amplification, but it is still subjected to the effects of specific base mutation. INNO-LiPA HBV genotyping assay has the limitation of high cost, and also its likelihood to be affected by the specific binding site mutant gene [10,14]. Sequence analysis is definitely the most accurate method and not subject to these constraints, but it is also the most labor intensive technique and needs nested PCR to increase the sensitivity.
As we all know, there are several antiviral therapies--such as interferon; pegylated interferon or nucleotide/nucleoside analogs--widely used to treat HBV infection. None of these therapies can eradicate HBV infection and all often induce drug-resistant mutants. As a result, HBV genotyping and the detection of mutations that confer drug resistance help select an appropriate treatment strategy and monitor the treatment. However, there are a limited number of methods that enable simultaneous genotyping and mutation detection. In this study, we used the partial S-gene sequencing using common PCR to genotype HBV, which is simpler and more sensitive compared with the S-gene sequencing.
Partial S-gene sequencing means sequencing part of the S gene from 370 nt to 861 nt. This part of the S gene we chose is relatively conserved and has many drug-resistant mutant sites, so it could be used in both HBV genotyping and in analysis of HBV drug resistant mutation.
Results and discussion
Phylogenetic tree analysis
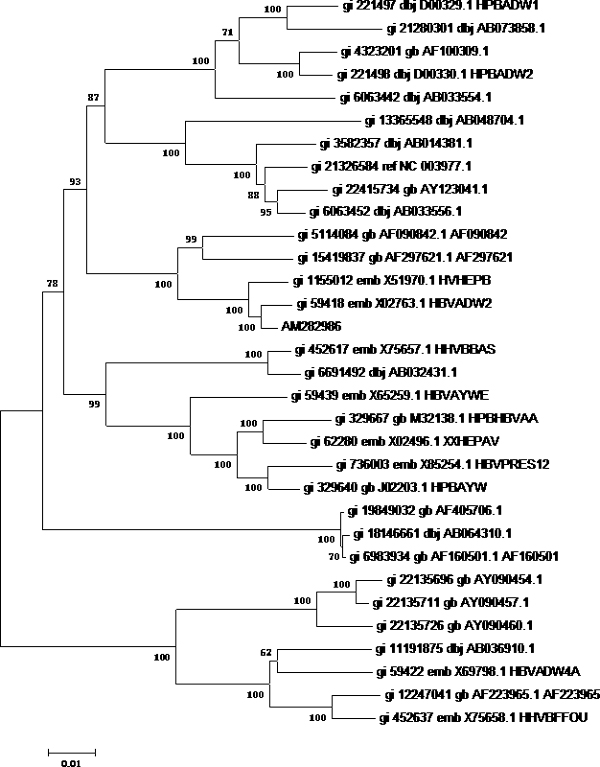
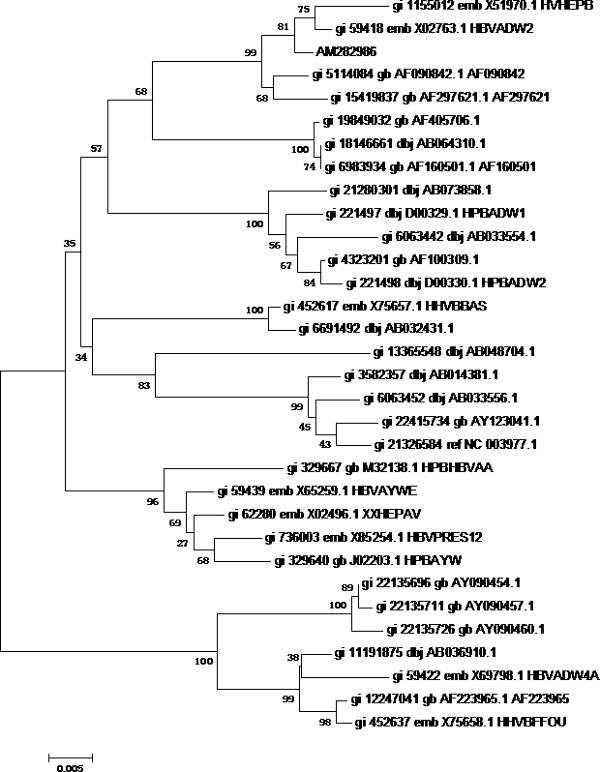
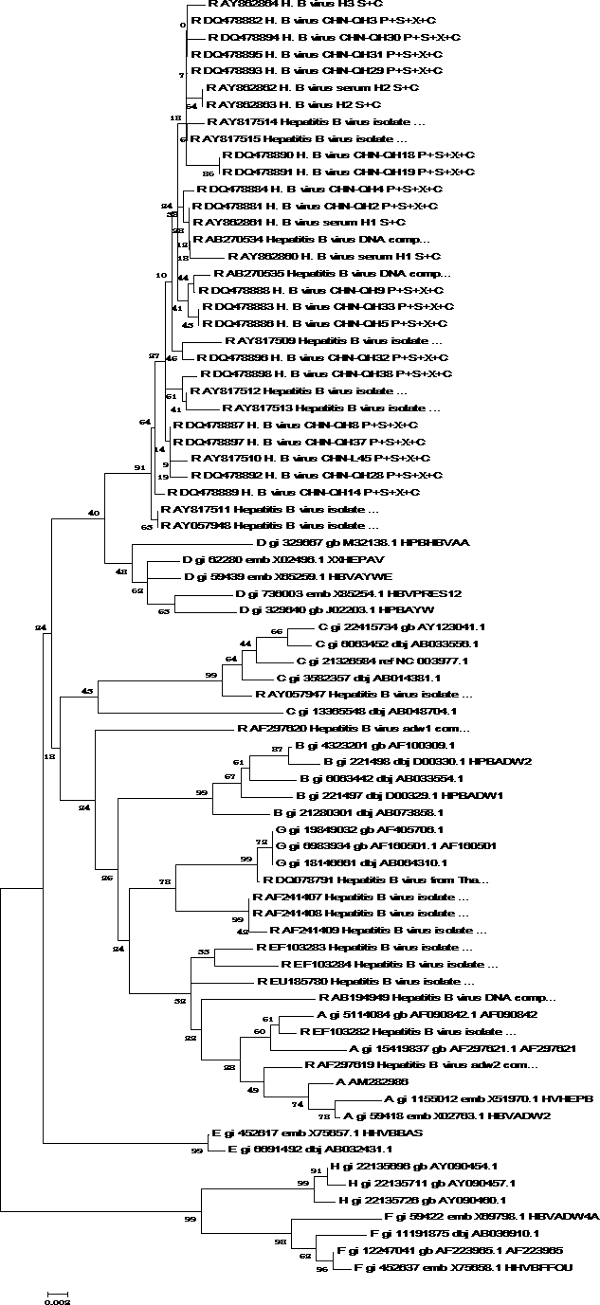
First, phylogenetic tree analysis was used for testing the possibility of HBV genotyping using partial S gene sequencing. Reference sequences from 32 HBV genomes of eight different genotypes were used (shown as Table 1). Software MEGA4 was used to analyze these genomes to get the phylogenetic trees of genome sequencing; S gene sequencing and partial S gene sequencing respectively. All these sequences could be genotyped successfully by partial S gene sequencing. The results of phylogenetic tree analysis indicated that the partial S gene sequencing had nearly the same phylogenetic tree as that of the S gene sequencing (shown as Figure 1, 2, 3).
Table 1.
Reference sequences for genotyping
| Genotype A | AM282986 | gi_59418 | gi_1155012 | gi_15419837 | gi_5114084 |
| Genotype B | gi_21280301 | gi_221497 | gi_221498 | gi_4323201 | gi_6063442 |
| Genotype C | gi_13365548 | gi_22415734 | gi_6063452 | NC_003977 | gi_3582357 |
| Genotype D | gi_329640 | gi_736003 | gi_329667 | gi_62280 | gi_59439 |
| Genotype E | gi_452617 | gi_6691492 | |||
| Genotype F | gi_11191875 | gi_59422 | gi_12247041 | gi_452637 | |
| Genotype G | gi_18146661 | gi_6983934 | gi_19849032 | ||
| Genotype H | gi_22135696 | gi_22135711 | gi_22135726 |
Figure 1.
Phylogenetic tree map of HBV genome sequencing. 32 HBV genomes of eight different genotypes were sequenced by HBV genome sequencing, and software MEGA4 was used to analyze these genomes to get the phylogenetic trees of genome sequencing. Tree Inference: [Method: Neighbor-Joining; Phylogeny Test and options: Bootstrap (1000 replicates; seed = 100000)]; Include Sites: [Gaps/Missing Data: Complete Deletion]; Substitution Model: [Model: Nucleotide: Maximum Composite Likelihood; Substitutions to Include: d: Transitions + Transversions; Pattern among Lineages: Same (Homogeneous); Rates among sites: Uniform rates]
Figure 2.
Phylogenetic tree map of HBV S gene sequencing. 32 HBV genomes of eight different genotypes were sequenced by HBV S gene sequencing, and software MEGA4 was used to analyze these genomes to get the phylogenetic trees of HBV S gene sequencing. Tree Inference: [Method: Neighbor-Joining; Phylogeny Test and options: Bootstrap (1000 replicates; seed = 100000)]; Include Sites: [Gaps/Missing Data: Complete Deletion; Codon Positions: 1st+2nd+3rd]; Substitution Model: [Model: Nucleotide: Maximum Composite Likelihood; Substitutions to Include: d: Transitions + Transversions; Pattern among Lineages: Same (Homogeneous); Rates among sites: Uniform rates]
Figure 3.
Phylogenetic tree map of HBV partial S gene sequencing. 32 HBV genomes of eight different genotypes were sequenced by HBV partial S gene sequencing, and software MEGA4 was used to analyze these genomes to get the phylogenetic trees of HBV partial S gene sequencing. Tree Inference: [Method: Neighbor-Joining; Phylogeny Test and options: Bootstrap (1000 replicates; seed = 100000)]; Include Sites: [Gaps/Missing Data: Complete Deletion; Codon Positions: 1st+2nd+3rd]; Substitution Model: [Model: Nucleotide: Maximum Composite Likelihood; Substitutions to Include: d: Transitions + Transversions; Pattern among Lineages: Same (Homogeneous); Rates among sites: Uniform rates]
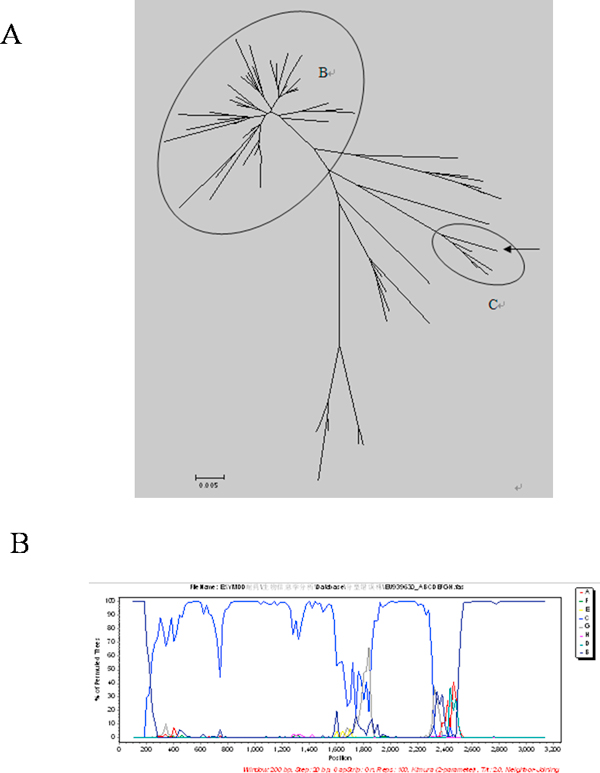
To further evaluate the effect of partial S gene sequencing, 53 HBV genotype A samples; 43 HBV genotype B samples; 50 HBV genotype C samples and 51 HBV genotype D samples were genotyped using partial S gene sequencing and S gene sequencing respectively (shown as Table 2). Compared the results of these two methods, we found that the results of partial S gene sequencing are consistent with S gene sequencing, except one sample, EU939630. From further recombinant analysis, we found that EU939630 was a C/B recombinant strain (shown as Figure 4).
Table 2.
Sequences for genotyping verification
Figure 4.
Genotype and recombinant analysis results of EU939630. A: Genotype results of 43 HBV genotype B samples using partial S gene sequencing, 42 samples of which had been genotyped correctly except one, EU939630. The arrow indicates EU939630 was genotyped as HBV genotype C by partial S gene sequencing. B: Recombinant analysis result of EU939630. From the recombinant analysis, we found that EU939630 was a C/B recombinant strain.
Sensitivity of partial S gene sequencing
147 HBV-positive (HBV copies were more than 500 copies/ml) serum samples were sequenced by partial S gene sequencing, 2 of which could not to be sequenced, so the sensitivity of our partial S gene sequencing was 98.64%.
Application of partial S gene sequencing in recombinant HBV isolates
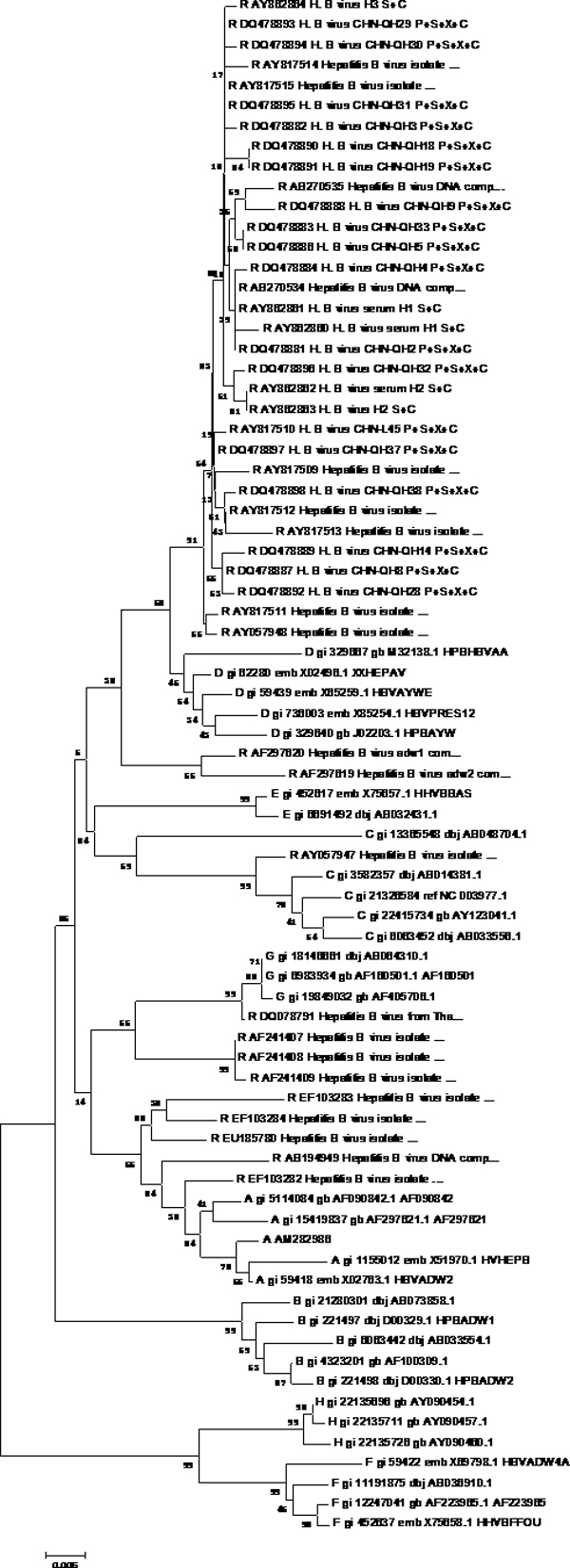
Next, we evaluated the application of partial S gene sequencing in HBV recombinant isolates (shown as Figure 5 and 6). 44 recombinant HBV isolates were collected as shown in Table 3. The genotyping results indicated that there were 38 samples with the same results using partial S gene sequencing and S gene sequencing. Other 6 samples had different genotyping results by these two methods, which were isolated from South Africa (2/6); Thailand (1/6) and Vietnam (3/6) respectively. One sample from South Africa (AF297620) failed to genotype by partial S gene sequencing, as there was a recombinant site in its S region (shown as Figure 7) which was analyzed by Simplot software and NCBI Viral genotyping tool.
Figure 5.
Phylogenetic tree map of HBV recombinant strains using S gene sequencing. 44 recombinant HBV isolates were collected to draw the phylogenetic tree map of HBV S gene sequencing by the method described above. Tree Inference: [Method: Neighbor-Joining; Phylogeny Test and options: Bootstrap (1000 replicates; seed = 100000)]; Include Sites: [Gaps/Missing Data: Complete Deletion]; Substitution Model: [Model: Nucleotide: Maximum Composite Likelihood; Substitutions to Include: d: Transitions + Transversions; Pattern among Lineages: Same (Homogeneous); Rates among sites: Uniform rates]
Figure 6.
Phylogenetic tree map of HBV recombinant strains using partial S gene sequencing. 44 recombinant HBV isolates were collected to draw the phylogenetic tree map of HBV partial S gene sequencing by the method described above. Tree Inference: [Method: Neighbor-Joining; Phylogeny Test and options: Bootstrap (1000 replicates; seed = 100000)]; Include Sites: [Gaps/Missing Data: Complete Deletion]; Substitution Model: [Model: Nucleotide: Maximum Composite Likelihood; Substitutions to Include: d: Transitions + Transversions; Pattern among Lineages: Same (Homogeneous); Rates among sites: Uniform rates]
Table 3.
Genotype results of 44 HBV recombinant strains
| Genotyping method | Source | |||
|---|---|---|---|---|
| S | partial S | genome | ||
| EU185780 | A | A | D3/A2 | Argentina |
| AB194949 | A | A | A (A3/Acmr)/E | Cameroon |
| AY817509 | D | D | C/D | China |
| AY817510 | D | D | C/D | China |
| AY817511 | D | D | C/D | China |
| AY817512 | D | D | C/D | China |
| AY817513 | D | D | C/D | China |
| AY817514 | D | D | C/D | China |
| AY817515 | D | D | C/D | China |
| AY862860 | D | D | C/D | China |
| AY862861 | D | D | C/D | China |
| AY862862 | D | D | C/D | China |
| AY862863 | D | D | C/D | China |
| AY862864 | D | D | C/D | China |
| DQ478881 | D | D | C/D | China |
| DQ478882 | D | D | C/D | China |
| DQ478883 | D | D | C/D | China |
| DQ478884 | D | D | C/D | China |
| DQ478886 | D | D | C/D | China |
| DQ478887 | D | D | C/D | China |
| DQ478888 | D | D | C/D | China |
| DQ478889 | D | D | C/D | China |
| DQ478890 | D | D | C/D | China |
| DQ478891 | D | D | C/D | China |
| DQ478892 | D | D | C/D | China |
| DQ478893 | D | D | C/D | China |
| DQ478894 | D | D | C/D | China |
| DQ478895 | D | D | C/D | China |
| DQ478896 | D | D | C/D | China |
| DQ478897 | D | D | C/D | China |
| DQ478898 | D | D | C/D | China |
| AY057947 | C | C | A/C | China |
| AY057948 | D | D | C/D | China |
| EF103282 | A | A | A/D | India |
| EF103283 | A | A | A/D | India |
| EF103284 | A | A | A/D | India |
| AB270534 | D | D | C/D | Mongolia:Ulaanbaatar |
| AB270535 | D | D | C/D | Mongolia:Ulaanbaatar |
| AF297619 | D | A | A/D | South Africa |
| AF297620 | D | - | A/D | South Africa |
| DQ078791 | C | G | G/C | Thailand |
| AF241407 | D | G | C/A/G/B | Vietnam |
| AF241408 | D | G | C/A/G/B | Vietnam |
| AF241409 | D | G | C/A/G/B | Vietnam |
Figure 7.
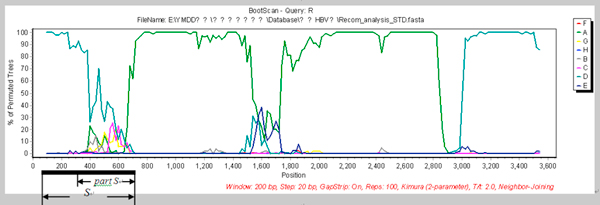
Recombinant analysis results of AF297620. AF297620 was failed to be genotyped by partial S gene sequencing because it had recombinant sites in partial S gene region.
Drug-resistant mutations analysis
The part of the S gene we chose for sequencing has many drug-resistant mutant sites, which means it had potential for use in analysis of HBV drug resistant mutation. To evaluate this, we analyzed all possible sites of resistance mutations (V521L; A529V; A529T; T532A; S550I; rtL180M; rtM204V/I; N584T and K589E) of 147 HBV-positive serum samples [16]. Through partial S gene sequencing we detected rtM204V mutation in one sample successfully (shown in Figure 8), meaning the partial S gene sequencing could be used in both HBV genotyping and in analysis of HBV drug resistant mutation.
Figure 8.
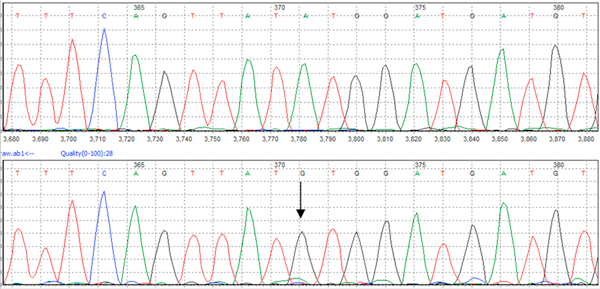
Drug resistant mutation analysis by partial S gene sequencing. Through drug resistant mutation analysis of 147 HBV positive serum samples using partial S gene sequencing, one sample was found to be rtM204V mutation. The arrow indicates the A is replaced by G.
Conclusions
HBV genotyping and the detection of drug resistance mutations are important for monitoring the treatment of chronic hepatitis B, but there are a limited number of methods for the simultaneous detection of HBV genotypes and drug resistance mutations [11,13]. We have established a partial S gene sequencing method to genotype HBV isolates as well as detect drug resistance mutations at the same time. To testify the sensitivity of our partial S gene sequencing, 147 clinical serum samples were used, and 145 samples could be sequenced successfully by this assay, with a sensitivity of 98.64%.
The part of S gene we chose for sequencing has many drug-resistant mutant sites, and we detected all possible mutant sites in this region of 145 samples and found the rtM204V mutation in one sample. That means the partial S gene sequencing could be used in analysis of HBV drug resistant mutation.
To evaluate the potential of partial S gene sequencing for use in HBV genotyping, the phylogenetic tree analysis was used. From the phylogenetic tree mapping, we found that the partial S gene sequencing had nearly the same phylogenetic tree map to that of the S gene sequencing (shown as Figure 2 and 3). This means partial S gene sequencing has the possibility to be used as a promising method in HBV genotyping. To further demonstrate this, 197 HBV positive serum samples of four different genotypes (A, B, C, and D) were genotyped using partial S gene sequencing and S gene sequencing respectively (Table 2). Although at least eight HBV genotypes have been reported, the major HBV genotypes in China are B and C [12,15]. Genotypes A and D are found in a very small proportion of Chinese patients, and genotypes E, F, G, and H have not been reported in China. As a result, we only evaluated the performance of partial S gene sequencing assay for genotypes A, B, C, and D in our study. We found the results of partial S gene sequencing were consistent with S gene sequencing, except one sample, EU939630, which was proved to be a C/B recombinant strain. From this, we have proved that this assay could specifically detect mutant and wild-type HBV in clinical serum samples.
Considering that the recombinant strain might disturb genotyping effect, we evaluated the application of partial S gene sequencing in 44 HBV recombinant isolates (Table 3). 43 samples could be genotyped correctly by partial S gene sequencing method, meanwhile, S gene sequencing genotyped 41 samples successfully. That means partial S gene sequencing had the potential to take the place of S gene sequencing in the field of HBV genotyping.
Based on these findings, we could draw the conclusion that the partial S gene sequencing assay developed in this study could be applied in HBV genotyping and drug resistant mutation detection. It might be an ideal choice for HBV genotyping for it is simpler and more convenient than traditional S gene sequencing while it has nearly the same sensitivity and specificity as S gene sequencing.
Methods
Serum samples
Serum samples are collected from four hospitals: the Second People Hospital of Guangdong Province, the First Affiliated Hospital of Guangzhou Medical College, Guangzhou Overseas Chinese Hospital, and Guangzhou Huadu Ren-Ai Hospital. All these serum samples were collected in compliance with the Helsinki Declaration, and all the patients who provided serum samples were voluntary. This study was approved by the Institutional Review Board of Wuhan University. All specimens were sampled from sterile blood vessels (excluding anticoagulant) and stored at -20°C.
Sequencing
HBV DNA was isolated from serum (QIAamp DNA Blood Mini Kit, QIAGEN, Hilden, Germany) according to the kit instructions. A product of 491 base pairs of partial S gene was amplified with the primers F (sense, 5'- TCGCTGGATGTGTCTGCGGCGTTTTAT-3') and R (antisense, 5'- ACCCCATCTTTTTGTTTTGTTAGG-3') using a PCR protocol as follows: 12 min at 95°C, 35 cycles of 1 min at 94°C, 1 min at 52°C, 1 min at 72°C, and a final elongation step of 7 min at 72°C, using the AmpliTaq Gold amplification system.
Phylogenetic tree analysis
To testify the feasibility of the partial S gene sequencing used for HBV genotyping, 32 different genotypes of the HBV (http://lancelot.otago.ac.nz) genome sequence were chosen as the reference sequences (shown in Table 1). Three different DNA sequencing methods were used to genotype serum samples for phylogenetic tree mapping: whole genome sequencing, S gene sequencing and partial S gene sequencing.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Fanjun Wang designed this study; Lili Lu and Youping Deng drafted and revised the manuscript; Changshun Yu, Zhanwu Lv participated in the collection of clinical samples and gene sequencing; Xuelian Luo, Chao Wan participated in the phylogenetic tree analysis; Zhaohui Hu, Qinyi Zhu participated in acquisition of data and data analysis; Youping Deng gave final approval of the version to be published; Chuyu Zhang participated in the design of the study and the general supervision of the research group.
Contributor Information
Fanjun Wang, Email: labwfj@kingmed.com.cn.
Lili Lu, Email: lili_lu813@yahoo.com.cn.
Youping Deng, Email: youpingd@gmail.com.
Chuyu Zhang, Email: labzhang@yahoo.cn.
Acknowledgements
This article has been published as part of BMC Medical Genomics Volume 6 Supplement 1, 2013: Proceedings of the 2011 International Conference on Bioinformatics and Computational Biology (BIOCOMP'11). The full contents of the supplement are available online at http://www.biomedcentral.com/bmcmedgenomics/supplements/6/S1. Publication of this supplement has been supported by the International Society of Intelligent Biological Medicine.
References
- Norder H, Courouce AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- Norder H, Hammas B, Lee SD, Bile K, Courouce AM, ushahwar IK, Magnius LO. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J Gen Virol. 1993;74:1341–1348. doi: 10.1099/0022-1317-74-7-1341. [DOI] [PubMed] [Google Scholar]
- Bartholomeusz A, Schaefer S. Hepatitis B virus genotypes: comparison of genotyping methods. Rev Med Virol. 2004;14:3–16. doi: 10.1002/rmv.400. [DOI] [PubMed] [Google Scholar]
- Tang XR, Zhang JS, Zhao H, Gong YH, Wang YZ, Zhao JL. Detection of hepatitis B virus genotypes using oligonucleotide chip among hepatitis B virus carriers in Eastern China. World J Gastroenterol. 2007;13:1975–1979. doi: 10.3748/wjg.v13.i13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet G, Tran N. The DNA-Chip technology as a new molecular tool for the detection of HBV mutants. J Clin Virol. 2005;34(Suppl 1):S49–S53. doi: 10.1016/s1386-6532(05)80010-1. [DOI] [PubMed] [Google Scholar]
- Osiowy C, Giles E. Evaluation of the INNO-LiPA HBV genotyping assay for determination of hepatitis B virus genotype. J Clin Microbiol. 2003;41:5473–5477. doi: 10.1128/JCM.41.12.5473-5477.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Das BC, Hussain Z, Kar P. Hepatitis B virus genotypes in acute and fulminant hepatitis patients from north India using two different molecular genotyping approaches. Hepatol Res. 2006;35:79–82. doi: 10.1016/j.hepres.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Chen J, Yin J, Tan X, Zhang H, Zhang H, Chen B, Chang W, Schaefer S, Cao G. Improved multiplex-PCR to identify hepatitis B virus genotypes A-F and subgenotypes B1, B2, C1 and C2. J Clin Virol. 2007;38:238–243. doi: 10.1016/j.jcv.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Liu WC, Mizokami M, Buti M, Lindh M, Young KC, Sun KT, Chi YC, Li HH, Chang TT. Simultaneous quantification and genotyping of hepatitis B virus for genotypes A to G by real-time PCR and two-step melting curve analysis. J Clin Microbiol. 2006;44:4491–4497. doi: 10.1128/JCM.01375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G, Wang Z, Wen S, Jiang J, Wang L, Cheng J, Tan D, Xiao F, Ma S, Li W, Luo K, Naoumov NV, Hou J. Geographic distribution, virologic and clinical characteristics of hepatitis B virus genotypes in China. J Viral Hepat. 2005;12(6):609–17. doi: 10.1111/j.1365-2893.2005.00657.x. [DOI] [PubMed] [Google Scholar]
- Degertekin B, Hussain M, Tan J, Oberhelman K, Lok AS. Sensitivity and accuracy of an updated line probe assay (HBV DR v.3) in detecting mutations associated with hepatitis B antiviral resistance. J Hepatol. 2009;49:695–701. doi: 10.1002/hep.22785. [DOI] [PubMed] [Google Scholar]
- Wang Z, Huang Y, Wen S, Zhou B, Hou J. Hepatitis B virus genotypes and subgenotypes in China. Hepatol Res. 2007;37(Suppl 1):S36–41. doi: 10.1111/j.1872-034X.2007.00102.x. [DOI] [PubMed] [Google Scholar]
- Wang YZ, Xiao JH, Liu LG, Ye CY, Shen HY, Xu TM, Zhu KZ. Simultaneous detection of hepatitis B virus genotypes and mutations associated with resistance to lamivudine, adefovir, and telbivudine by the polymerase chain reaction-ligase detection reaction. Braz J Infect Dis. 2011;15(6):560–6. doi: 10.1016/S1413-8670(11)70251-3. [DOI] [PubMed] [Google Scholar]
- Ali MM, Hasan F, Ahmad S, Al-Nakib W. Comparative evaluation of INNO-LiPA HBV assay, direct DNA sequencing and subtractive PCR-RFLP for genotyping of clinical HBV isolates. Virol J. 2010;7:111. doi: 10.1186/1743-422X-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pas SD, Tran N, de Man RA, Burghoorn-Maas C, Vernet G, Niesters HG. Comparison of reverse hybridization, microarray, and sequence analysis for genotyping hepatitis B virus. J Clin Microbiol. 2008;46(4):1268–73. doi: 10.1128/JCM.01519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuyver L, Locarnini SA, Lok A, Richman DD, Carman WF, Dienstag JL, Shinazi RF. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology. 2001;33:751–757. doi: 10.1053/jhep.2001.22166. [DOI] [PubMed] [Google Scholar]