Abstract
Equine arteritis virus (EAV) is an enveloped, positive-strand RNA virus. Genome replication of EAV has been associated with modified intracellular membranes that are shaped into double-membrane vesicles (DMVs). We showed by immuno-electron microscopy that the DMVs induced in EAV-infected cells contain double-strand (ds)RNA molecules, presumed RNA replication intermediates, and are decorated with the autophagy marker protein microtubule-associated protein 1 light chain 3 (LC3). Replication of EAV, however, was not affected in autophagy-deficient cells lacking autophagy-related protein 7 (ATG7). Nevertheless, colocalization of DMVs and LC3 was still observed in these knockout cells, which only contain the nonlipidated form of LC3. Although autophagy is not required, depletion of LC3 markedly reduced the replication of EAV. EAV replication could be fully restored in these cells by expression of a nonlipidated form of LC3. These findings demonstrate an autophagy-independent role for LC3 in EAV replication. Together with the observation that EAV-induced DMVs are also positive for ER degradation-enhancing α-mannosidase-like 1 (EDEM1), our data suggested that this virus, similarly to the distantly-related mouse hepatitis coronavirus, hijacks the ER-derived membranes of EDEMosomes to ensure its efficient replication.
Keywords: arterivirus, autophagy, double-membrane vesicles, LC3, nidoviruses
Introduction
Viruses depend on the cellular machinery of their host for their multiplication. Therefore they have developed sophisticated mechanisms to exploit these cellular machineries to their own benefit. Positive-strand RNA viruses are the largest group of viruses, to which many notorious pathogens belong. A common trait of these viruses is the cytoplasmic replication of their genomes in association with modified cellular membranes. Membranes of different cellular compartments, including endoplasmic reticulum (ER), mitochondria and endosomes, are hijacked by positive-strand RNA viruses to assemble their replication/transcription complexes and to replicate their genomes.1-3
Arteriviruses are enveloped, positive-strand RNA viruses of veterinary importance as exemplified by the equine arterivirus (EAV) and the porcine respiratory and reproductive syndrome virus. The Arteriviridae, together with the Coronaviridae and Roniviridae, have been classified into the order Nidovirales, because of similarities in both their polycistronic genome organization and replication strategy.4,5 Members from the different virus families, however, differ considerably in virion architecture, genome size and complexity. Arteriviruses contain two large open-reading frames (ORF), ORF1a and ORF1b, at the 5′ end of their genomic RNA, which occupy approximately two-thirds of the viral genome and encode for the subunits of the replicase machinery. The ORF1a- and 1b-encoded polyproteins contain an array of conserved domains, including proteinases, which process the two polyproteins into nonstructural proteins (NSPs), and clusters of hydrophobic transmembrane segments, which anchor the nascent polyproteins into the host cell membranes.
Arteriviruses induce the formation of double-membrane vesicles (DMVs) in infected cells, which are derived from the ER and have been linked to RNA synthesis.6-8 It has been shown that coexpression of two transmembrane NSPs of EAV, NSP2 and NSP3, is sufficient for the induction of membrane rearrangements, including DMVs.9 The mechanism and the host factors essential for the generation of these structures remain, however, largely unknown. The induction of DMVs by arteriviruses raises the question whether these viruses hijack the cellular pathway of autophagy, which is also characterized by DMVs, for their replication. Several pathogens have been shown to exploit autophagy for their efficient propagation in host cells.10-13 Picornaviruses in particular appear to subvert this pathway to generate DMVs that are involved in RNA synthesis.14-21
Autophagy is a conserved degradative pathway that allows the cell to eliminate large damaged and superfluous intracellular structures.22,23 Components targeted to destruction are sequestered into DMVs called autophagosomes, which then fuse with lysosomes thereby exposing their content to the interior of these lytic organelles. The autophagy protein LC3 plays an essential role in autophagosome formation and is commonly used as a marker protein for autophagosomes. LC3 is predominantly present in cells in its nonlipidated form, i.e., LC3-I.24 When autophagy is induced, a series of conjugation reactions mediated by a specific set of autophagy-related (ATG) proteins leads to the covalent linkage of LC3 to the phosphatidylethanolamine present on autophagosomal membranes.24 The lipidated form of LC3 is called LC3-II.
As little is known about how arteriviruses induce the formation of their DMVs, we have investigated the involvement of host cellular pathways in arterivirus replication and the generation of DMVs using EAV as the prototype for this virus family. Our data showed that the nonlipidated form of LC3 is associated to the EAV-induced DMVs. Although EAV needs LC3 for its efficient replication, an intact autophagy machinery is not required. These observations and the colocalization of the DMVs with EDEM1 suggest that EAV, similarly to the distantly-related mouse hepatitis coronavirus,25,26 hijacks EDEMosomes,27 ER-derived vesicles that deliver short-lived ER chaperones to the endolysosomal system, to ensure its efficient replication.
Results
dsRNA produced by EAV colocalize with LC3 and EDEM1
To get more insight into the mechanism and host factors involved in the generation of EAV-induced DMVs, we decided to explore whether they colocalize with marker proteins for specific cellular organelles by immunofluorescence. As a marker for the EAV-induced DMVs, we used a monoclonal antibody recognizing dsRNA. The same antibody has previously successfully been used to detect dsRNA generated by other positive-strand RNA viruses.28-32 In the cells infected by these viruses, the dsRNA molecules have been shown to localize to the virus-induced membrane rearrangements involved in RNA synthesis.
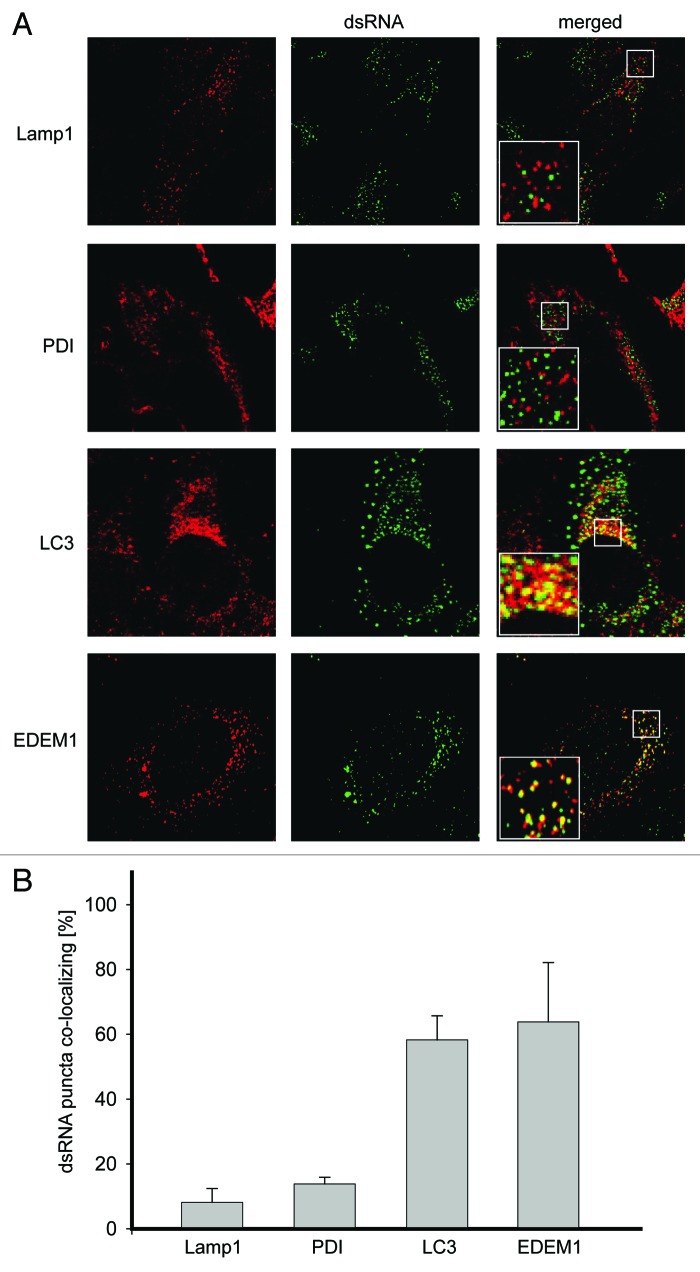
Vero E6 cells were thus infected with EAV and fixed at 16 h post infection (p.i.) before being processed for immunofluorescence analysis. As expected, no dsRNA was detected in noninfected cells (Fig. S1 and S2). In contrast, cells infected with EAV displayed the presence of numerous cytoplasmic puncta in keeping with the idea that dsRNA could represent a specific marker for EAV-induced DMVs (Fig. 1A).8 First, we analyzed whether these structures were associated with marker proteins of different subcellular compartments, including the ER, lysosomes, autophagosomes and EDEMosomes, by using antibodies recognizing the proteins disulfide isomerase (PDI), lysosomal-associated membrane protein 1 (LAMP1), LC3 or EDEM1, respectively. While we found that PDI and LAMP1 were not colocalizing with the dsRNA, LC3 and EDEM1 were distributed in numerous punctate structures, several of which were colocalizing with dsRNA-positive puncta induced by EAV (Fig. 1A). Statistical analysis of the immunofluorescence images confirmed these observations demonstrating that indeed a much higher number of dsRNA puncta are positive for LC3 or EDEM1 than for PDI or LAMP1 (Fig. 1B). A similar degree of colocalization was detected during the entire course of the EAV infection (data not shown).
Figure 1. LC3 and EDEM1 colocalize with EAV-induced dsRNA foci. (A) Vero E6 cells were infected with EAV at a multiplicity of infection of 1 TCID50/cell and fixed at 16 h p.i before being processed for immunofluorescence analysis using antibodies against dsRNA and either PDI, LAMP1, LC3 or EDEM1. Insets show enlargements of the boxed areas. (B) Summary statistics of the samples shown in (A) expressed as the percentage of dsRNA puncta colocalizing with the indicated marker protein signals. Error bars represent the standard deviations from counting 50 cells in 3 independent experiments.
EAV-induced DMVs contain dsRNA and are positive for LC3
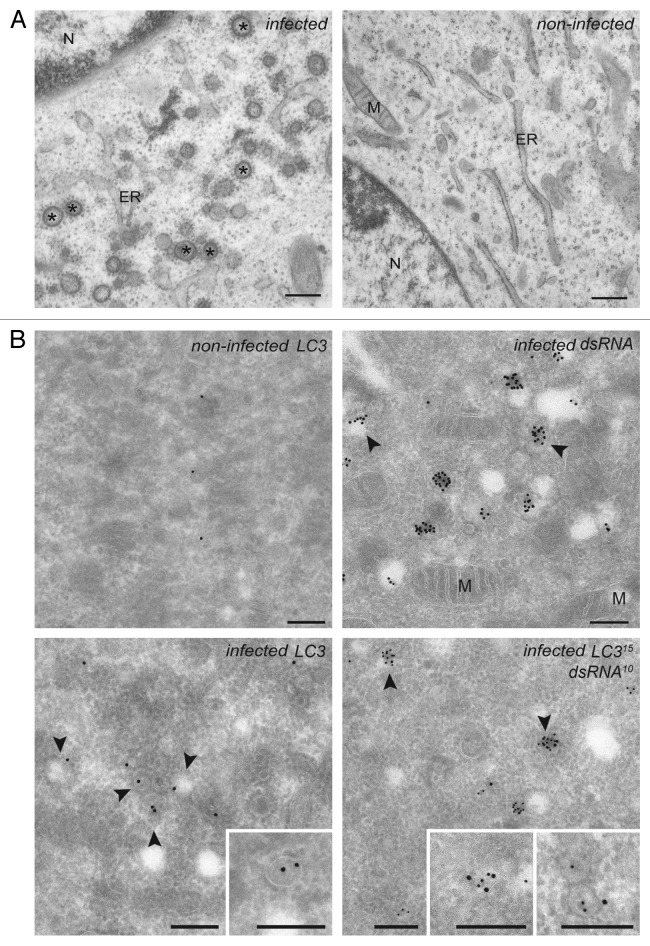
Subsequently, we explored whether the puncta positive for both dsRNA and LC3 corresponded to the EAV-induced DMVs. First, we confirmed the formation of DMVs in cells infected with EAV at the ultrastructural level. To this end, BHK-21 cells were infected with EAV and incubated at 37°C for 16 h before being processed for conventional electron microscopy (EM).33 Numerous DMVs were readily observed in the cytoplasm of cells exposed to EAV but not in the control sample (Fig. 2A). In agreement with previous studies,6,7 ribosomes were associated onto the surface of the EAV-induced DMVs, which were often found in close proximity of the ER and occasionally in continuous association with it. Essentially the same results were obtained using Vero E6 cells as well (data not shown).
Figure 2. EAV infection induces the formation of dsRNA- and LC3-positive DMVs. (A) BHK-21 cells infected with EAV or mock treated were chemically fixed at 16 h p.i. and embedded with Epon resin. Numerous DMVs associated with ribosomes were found mostly in the perinuclear region of the EAV-infected cells (left panel). Asterisks indicate some of the DMVs. (B) BHK-21 cells (mock)-infected with EAV and fixed at 16 h p.i. were processed for IEM. Cryo-sections were labeled with antibodies against dsRNA (top right panel), LC3 (top or bottom left panel) or both (bottom right panel). Arrowheads indicate some of the DMVs. Insets show examples of labeled vesicles at high magnification. The size of the gold particles is indicated in the image double labeled sample. ER, endoplasmic reticulum; M, mitochondria; N, nucleus. Scale bars: 200 nm.
To localize proteins to the EAV-induced DMVs, we next performed immuno-EM (IEM) on ultrathin cryo-preparations of noninfected and infected cells (Fig. 2B). The general cellular architecture as well as many EAV-induced DMVs were well preserved. Labeling of cryo-sections with the antibodies recognizing dsRNA revealed that dsRNA is principally localized into the interior of the DMVs (Fig. 2B), thereby confirming the dsRNA-positive puncta observed by immunofluorescence are DMVs. Importantly, these structures were also labeled by antibodies against LC3 and double labeling for LC3 and dsRNA revealed that these two components are on DMVs when colocalizing. These results proved that dsRNA is localized to the DMVs interior and this molecule is consequently a marker for these structures. In addition, they show that LC3 is associated to the EAV-induced DMVs.
EAV replication does not require an intact autophagy machinery
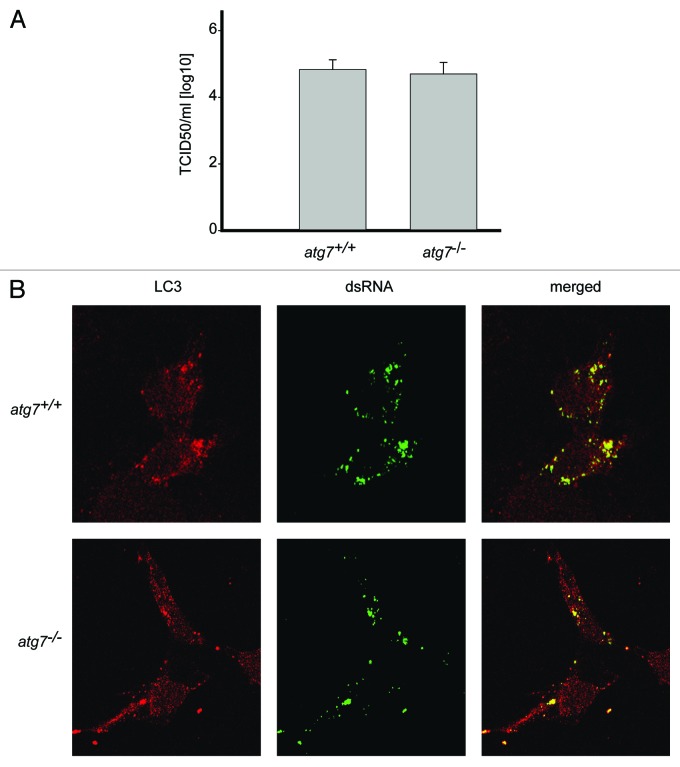
Association of LC3 with EAV-induced DMVs indicated a possible role of autophagy in EAV replication as already shown for several other viruses,10-13 including poliovirus, which also triggers the formation of DMVs.34 Furthermore, infection of cells with EAV resulted in both the redistribution of the autophagy marker protein GFP-LC3 into puncta and increase of the cellular levels of LC3-II similarly to starved cells, indicating an induction of autophagy (Fig. S2). Therefore we investigated whether deletion of atg7, a gene essential for autophagy,35 affected the replication of EAV. As expected, GFP-LC3 positive foci were observed upon starvation of wild-type (atg7+/+) murine embryonic fibroblasts (MEFs) but not in ATG7-deficient (atg7−/−) MEFs (Fig. S3).35 In parallel, we analyzed the complete life cycle of EAV in atg7+/+ and atg7−/− MEFs by determining the 50% tissue culture infectious dose (TCID50) of an EAV stock on these cells. Our results show that no significant differences in viral titer could be observed between autophagy-deficient atg7−/− and wild-type atg7+/+ MEFs (Fig. 3A). Subsequently, we studied the formation of DMVs in the same cells by immunofluorescence. In agreement with the TCID50 data, replication of EAV in autophagy-deficient cells was unaffected because numerous dsRNA-positive puncta could be readily detected, which were furthermore still colocalizing with LC3 (Fig. 3B). To exclude a possible role for Atg5/Atg7-independent alternative autophagy in the replication of EAV, we performed a similar experiment using MEFs lacking Rb1cc1/Fip200, a gene essential for this type of autophagy.36 Replication of EAV was normal in these cells as well (Fig. S4). All together, these results indicate that replication of EAV does not depend on the presence of an intact autophagy pathway.
Figure 3. EAV replication and association of LC3 with the DMVs does not depend on an intact autophagy machinery. (A) End point 10-fold dilutions of an EAV stock were titrated on Atg7+/+ and atg7−/− MEFs. Cytopathic effects were scored at 4 d p.i. and TCID50 units per ml were calculated. Similar titers were observed, indicating that Atg7 deletion does not affect EAV entry, replication or egress. Values presented in the graph are calculated and expressed as the log10 of TCID50 units per ml of supernatant, and the plotted values represent the average of 3 experiments. Standard deviations are indicated. (B) The Atg7+/+ and atg7−/− MEF cells were infected with EAV and fixed and processed for immunofluorescence analysis as described in Materials and Methods using antibodies against LC3 and dsRNA.
Nonlipidated LC3-I is associated with DMVs
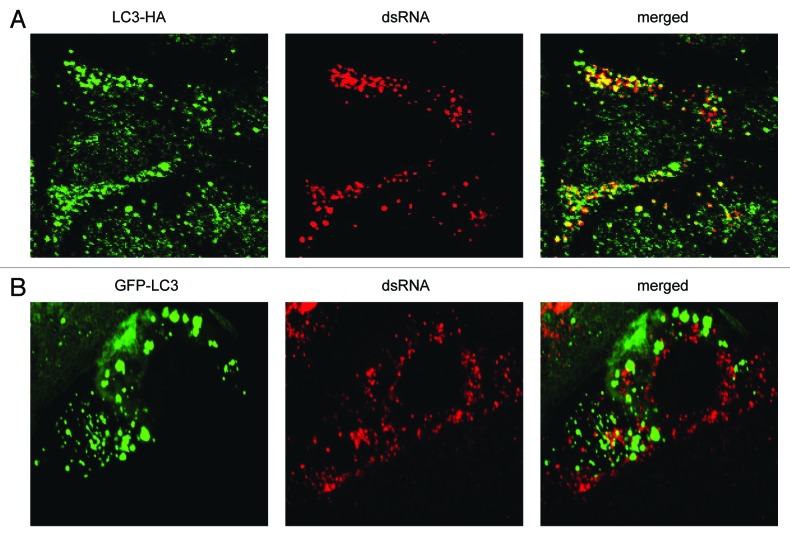
ATG7 is part of the set of ATG proteins that mediate lipidation of LC3-I into LC3-II.35 Thus, the colocalization of dsRNA with LC3 in the atg7−/− MEFs (Fig. 3B) implied that nonlipidated LC3-I associates with the EAV-induced DMVs in these cells. To confirm this assumption, we next investigated the localization of LC3 carrying a mutation at its C terminus that prevents the conjugation to phosphatidylethanolamine and thus its association with autophagosomes (Fig. S5).25,37 Importantly, this ectopically expressed C-terminally HA-tagged, nonlipidated LC3, i.e., LC3-HA, localized to the EAV-induced DMVs (Fig. 4A).25 In contrast, ectopically expressed N-terminally GFP-tagged LC3 (GFP-LC3), a conventional protein marker for autophagosome membranes37,38 did not show appreciable colocalization with the dsRNA-positive structures (Fig. 4B; Fig. S2). Taken together, these results show that lipidation is not required for LC3 recruitment onto the EAV-induced DMVs. In addition, they confirm that dsRNA puncta are not autophagosomes, in agreement with EAV replication not being dependent on a functional pathway.
Figure 4. EAV-induced DMVs are associated with LC3-I but not with N-terminally GFP-tagged LC3. Vero E6 cells were transfected with plasmids expressing either C-terminally HA-tagged nonlipidated LC3-I (LC3-HA) (A) or N-terminally GFP-tagged LC3 (GFP-LC3) (B), and subsequently infected with EAV for 16 h before being processed for immunofluorescence analysis.
LC3 is required for EAV replication
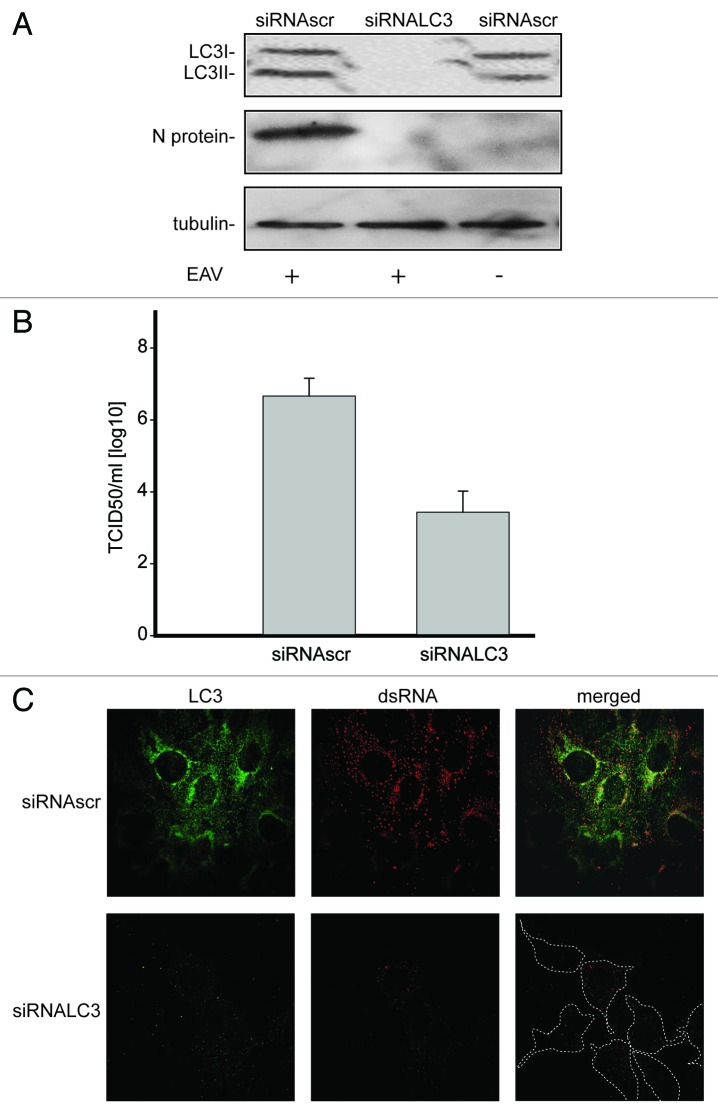
Because our data indicated that LC3-I is associated with EAV-induced DMVs, we explored whether this protein is required for viral replication. Efficient depletion of LC3A and LC3B, two of the three forms of LC3, was obtained by specific RNA interference using siRNA probes and was confirmed with antibodies recognizing these two proteins (Fig. 5A). Reduction of LC3 levels resulted in a decreased replication of EAV as judged by the much lower production of nucleocapsid (N) protein during infection (Fig. 5A). In agreement herewith a substantial drop in infectivity was also observed as measured by the TCID50 assay in LC3-depleted Vero E6 cells when compared with control cells (Figs. 5B). Furthermore, much less dsRNA was synthesized in infected cells upon depletion of LC3 as determined by immunofluorescence. This latter analysis confirmed again the efficient depletion of LC3A and LC3B after transfection with the LC3-specific siRNAs (Fig. 5C).
Figure 5. LC3 is required for EAV life cycle. Vero E6 cells were transfected with siRNAs directed against LC3A and LC3B (siRNALC3) or with control, scrambled siRNAs (siRNAscr). At 48 h post transfection, cells were inoculated with EAV. (A) Cells (mock-) infected with EAV for 16 h were lysed and protein extracts were processed for western blot analysis using antibodies recognizing LC3, N protein or tubulin. Tubulin was used as a loading control. (B) End point 10-fold dilutions of an EAV stock were titrated on the indicated transfected cells. Cytopathic effects were scored at 3 d p.i. and TCID50 units per ml were calculated. (C) Immunofluorescence examination using antibodies against the indicated proteins. Dotted lines highlight the cell contours.
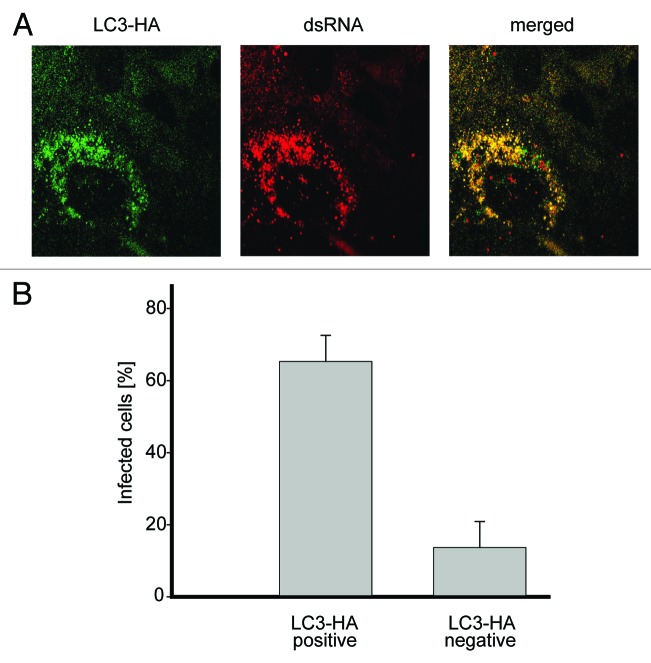
To confirm that the negative effect on EAV replication of the used siRNA probes was due to the specific depletion of LC3, a back-transfection experiment was performed. Cells transfected with the siRNAs targeting LC3A and LC3B were additionally transfected with a plasmid expressing a version of LC3-HA that carried silent mutations rendering the mRNA transcripts resistant to the action of the designed LC3A/LC3B siRNAs. EAV replication assessed by the appearance of dsRNA puncta, was detected in a large fraction of the cells expressing LC3-HA but not in the ones not carrying this construct, proving that the negative effect of the LC3-specific siRNAs on EAV replication is due to LC3A and LC3B depletion (Fig. 6A and B). We concluded that the nonlipidated form of LC3 is required for efficient replication of EAV.
Figure 6. Nonlipidated LC3 restores EAV replication in LC3-depleted cells. (A) Vero E6 cells were cotransfected with siRNA directed against LC3A and LC3B (siRNALC3) and the plasmid expressing nonlipidated LC3-HA, which is not targeted by the siRNA probes. Cells were then infected with EAV at 48 h post transfection and fixed at 16 h p.i. before being processed for immunofluorescence analysis using antibodies against the indicated proteins. (B) Statistical analysis of the experiment shown in (A). The graph illustrates the percentage of cells, positive or negative for LC3-HA that contained dsRNA puncta, i.e., productively infected by EAV.
NSP2-3–induced membrane rearrangements are not positive for LC3
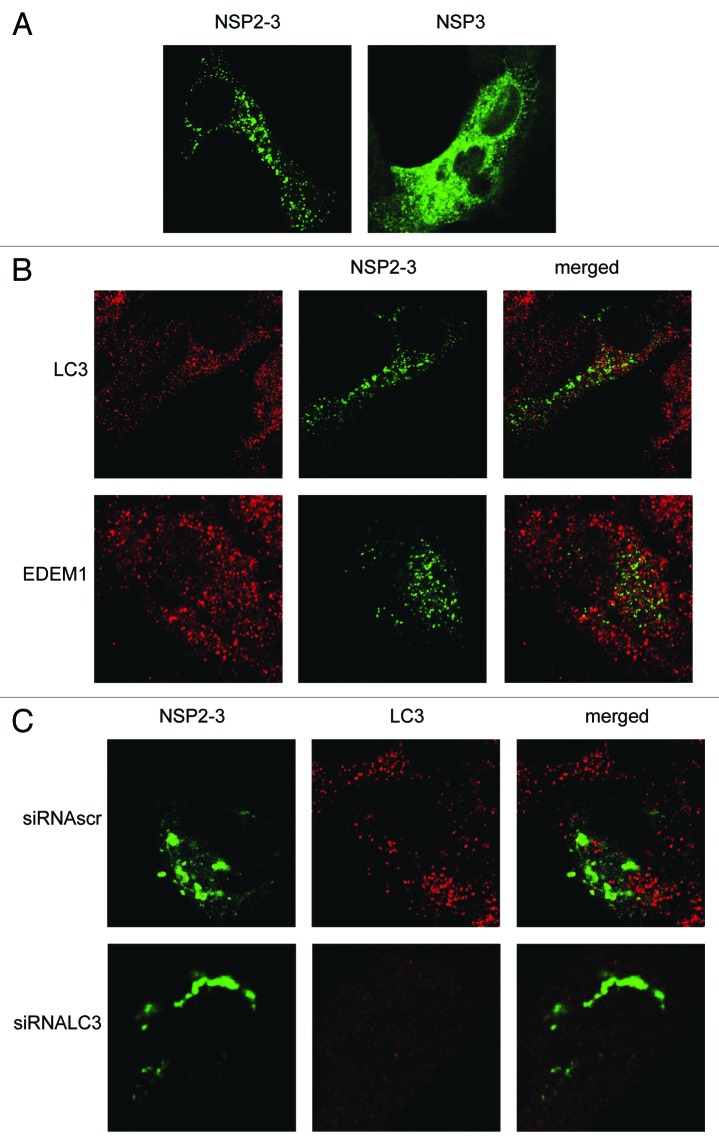
Expression of part of the EAV ORF1a, which encodes a fusion protein of NSP2 and 3 (NSP2-3), has previously been shown to induce the formation of membrane rearrangements and DMVs in the absence of EAV infection.9 Expression of NSP3 singly, in contrast, did not have the same effects. As the role of LC3 in the replication of EAV is not understood, we determined whether depletion of LC3 would inhibit the membrane modifications observed after the expression of NSP2-3. First we analyzed the localization of GFP-tagged NSP3 and NSP2-3 using the vaccinia virus T7 expression system (Fig. 7A). NSP3-GFP exhibited a reticular expression pattern, in agreement with the fact that this protein localizes to the ER when singularly expressed,9 although it was also found to some extent in puncta. In contrast, NSP2-3-GFP exhibited a very different localization pattern as it exclusively localized in puncta, in agreement with its ability to induce membrane rearrangements and DMVs.9 Subsequently, we analyzed whether LC3 and EDEM1 were recruited to NSP2-3-positive structures. As is shown in Figure 7B, no appreciable colocalization between NSP2-3-GFP and LC3 or EDEM1 was observed. Finally, we studied whether the membrane rearrangements observed after expression of NSP2-3 depend on the presence of LC3 as was observed for the DMVs present in the EAV-infected cells. Cells in which LC3 was depleted, displayed a similar NSP2-3GFP distribution pattern as cells positive for LC3 (Fig. 7C). Together, these results suggest that LC3 is not recruited to and it is not essential for the formation of membrane rearrangements induced by NSP2-3.
Figure 7. EAV NSP2-3–induced membrane rearrangements do not colocalize with LC3 and are formed independently of LC3. Vero E6 cells were infected with vTF7-3 and subsequently transfected with plasmids expressing GFP fusion proteins with either EAV NSP2-3 or NSP3. Cells were fixed at 5 h p.i. and processed for immunofluorescence analysis. (A) Comparison of the subcellular distribution of NSP2-3 and NSP3. (B) Cells expressing NSP2-3 were immuno-stained using antibodies against LC3 or EDEM1. (C) Cells were transfected with siRNAs directed against LC3A and LC3B (siRNALC3) or control siRNAs (siRNAscr) 48 h prior to infection with vTF7-3. Cells were processed for immunofluorescence analysis using the LC3-specific antibodies.
Discussion
In this work we have studied the involvement of host cellular pathways in the replication of EAV. We showed that the DMVs induced upon infection of cells by EAV contain dsRNA molecules, which are presumed to be RNA replication intermediates. Consequently, dsRNA can serve as a marker for these structures, a conclusion that is supported by data presented in a recent study that appeared during the preparation of this manuscript.8 These dsRNA-positive puncta contain the EDEMosome and autophagosome marker proteins EDEM1 and LC3, respectively, but not those of the ER (PDI) or lysosomes (LAMP1). The association of LC3 to the DMV surface was subsequently confirmed by IEM. Although infection of cells with EAV results in the induction of autophagy similarly to coronaviruses39 and LC3 is required for efficient replication of EAV, we showed that the important role of this protein in the EAV life cycle is independent of its known function in autophagy. Several lines of evidence support this conclusion: (i) LC3 associates with EAV-induced DMVs as shown by immunofluorescence and IEM; (ii) EAV replication is impaired in LC3-depleted cells; (iii) EAV life cycle is not affected in autophagy-deficient atg7−/− MEFs; (iv) N-terminally GFP-tagged LC3, a marker protein for autophagosomes, does not colocalize with the EAV-induced DMVs; (v) a nonlipidated form of LC3, which cannot be recruited onto autophagosomes, binds to EAV-induced DMVs and is able to rescue EAV replication in cells lacking endogenous LC3.
An autophagy-independent function for LC3 is not without precedent. For example, studies from several groups have indicated that LC3 is associated with the DMVs induced in cells upon infection with severe acute respiratory syndrome (SARS)-coronavirus or mouse hepatitis coronavirus (MHV).25,40-43 In addition, we have recently shown that DMV formation and virus replication in MHV-infected cells is independent of autophagy, but requires LC3.25,26 Similarly as observed for EAV, the nonlipidated form of LC3 is associated with a large proportion of the MHV-induced DMVs. DsRNA foci that do not colocalize with LC3, might represent intermediate structures or DMVs that lost their LC3 coat.25,26 Furthermore, EDEMosomes, which are vesicles that remove short-lived chaperones such as EDEM1 and OS-9 from the ER lumen and deliver them to the endolysosomal system for degradation, also display LC3-I at their limiting membranes.27 In addition, an autophagy-independent role for LC3 was recently demonstrated during the replication of the intracellular propagation of Chlamydia trachomatis.44 LC3 was shown to interact with C. trachomatis inclusions, while depletion of LC3 repressed the infectivity of this pathogen. The replication C.trachomatis, however, was enhanced rather than decreased in the absence of a functional autophagy pathway. Furthermore, the inclusions were devoid of N-terminally GFP-tagged LC3 and only labeled with endogenous LC3 similarly to EAV-, MHV- and SARS coronavirus induced DMVs.25,40 Why the GFP-tagged LC3 does not behave like endogenous LC3 is not yet clear, but may indicate an important role for the N-terminal domain in the autophagy-independent function of this protein. It has recently been shown that LC3-I is recruited to EDEMosomes and possibly MHV-induced DMVs through binding to the cytosolic tail of the transmembrane protein suppressor of lin-12-like protein 1 (SEL1L).45 One could imagine that the large GFP tag interferes with this interaction.
Although the DMVs observed in arterivirus-infected cells are reminiscent to those induced by the distantly-related coronaviruses,28,33,40,46 they display some significant differences. First, they are dissimilar in size, with the EAV-induced DMVs having a diameter approximately 3-fold smaller than their coronaviral counterparts. Second, the DMVs induced in coronavirus-infected cells are connected to additional membrane rearrangements, the so-called convoluted membranes,28,33 which have not been detected in EAV-infected cells.6-8 Despite these differences, DMVs induced by viruses of both families recruit LC3-I, while their replication does not require an intact autophagy machinery.25 Previously, the EDEMosome cargo proteins EDEM1, OS-9 and SelL1 were shown to colocalize with the MHV-induced DMVs.25,45 As EDEMosomes also display LC3-I, but not ectopically expressed GFP-LC3 or the lipidated form of LC3 (LC3-II),27 we proposed that the mechanism regulating the formation of EDEMosomes may be hijacked by MHV, and possibly other coronaviruses to ensure their efficient replication.25 The observed colocalization between EDEM1 and EAV-induced DMVs suggests that this could also be the case for arteriviruses.
The mechanism by which LC3 functions during the replication of EAV and MHV remains elusive.26,47 While our results indicate that LC3 is required for efficient replication of EAV, LC3 is not recruited to or required for the membrane rearrangements induced by expression of an EAV NSP2-3 fusion protein. Although it is not yet clear to what extent the DMVs induced by expression of EAV Nsp2-3 resemble DMVs observed in EAV-infected cells or rather correspond to biogenesis intermediates, we speculate that LC3 is not necessary for the induction of the initial membrane rearrangements per se, but may rather play a role during later stages of DMVs formation or eventually during RNA synthesis. Thus, while it seems clear that arteriviruses and coronaviruses require LC3 independently from the role that this protein has in autophagy, more investigations are needed to fully understand the contribution of LC3 and EDEMosomes in the replication of these viruses.
Materials and Methods
Cell lines and viruses
BHK-21, Vero E6, atg7+/+ and atg7−/− MEFs (the latter two cell lines are a kind gift of Masaaki Komatsu, The Tokyo Metropolitan Institute Medical Science),35 Rb1cc1/Fip200+/+ and rb1cc1/fip200−/− MEFs48 were grown in Dulbecco’s modified Eagle’s medium (Lonza, BE12-741F) supplemented with 10% (v/v) fetal bovine serum (FCS), 100 units/ml of penicillin and 0.1 mg/ml of streptomycin. BHK-21 cells were used to propagate the wild-type Bucyrus strain of EAV.49 Recombinant vaccinia virus encoding the bacteriophage T7 RNA polymerase (vTF7-3) was obtained from Bernard Moss (NIAID).50
Immunofluorescence microscopy
Cells were grown on coverslides and processed for immunofluorescence analysis as previously described.51 When indicated, cells were transiently transfected with a plasmid expressing either GFP-LC334 (a kind gift of Karla Kirkegaard, Department of Microbiology and Immunology, Stanford University) or LC3-HA25 using Lipofectamine 2000 (Invitrogen, 11668-019) according to the manufacturer’s protocol. Primary immunological reactions were performed with polyclonal antisera against LC3 (Novus Biologicals, NB600-1384), PDI (a kind gift of Masakazu Kikuchi, Department of Physiology, Kansai Medical University) or EDEM1 (Sigma, E8406), or monoclonal antibodies recognizing the HA tag (a kind gift of Guojun Bu, Washington University), dsRNA (J2; English and Scientific Consulting Bt., 10010200) or LAMP1 (Dianova, ABR-30253). The Alexa Fluor 488-conjugated goat anti-rabbit and Alexa Fluor 568-conjugated rabbit anti-mouse secondary antibodies were purchased from Invitrogen (A-11008 and A-11010). The fluorescence signals were analyzed using a confocal laser-scanning microscope (Leica, TCS SPE-II, Rijswijk, Netherlands). All experiments were repeated two or three times, and the presented images show representative fluorescence profiles.
Electron microscopy
EM and IEM experiments were performed essentially as described previously.33 Cryo-sections were labeled with specific antibodies against dsRNA and LC3 (the latter is a kind gift of Takashi Ueno, Department of Biochemistry, Jutendo University School of Medicine).
Western blot analyses
Western blot analyses were performed as described previously.25 In brief, cell extracts were prepared with lysis buffer (200 mM NaCl, 50 mM HEPES pH 6.8, 2% CHAPS and protease inhibitors) before adding sample buffer and boiling for 5 min. Proteins were then separated by SDS-PAGE and transferred onto PVDF membranes before being analyzed with antibodies against LC3 (Nanotools, 0231-100/LC3-5F10), N protein (a kind gift from Stuart Siddell, University of Bristol) or tubulin (Sigma-Aldrich, T9028). Horseradish peroxidase-congugated secondary antibodies and the ECL-TM Western Blotting Analysis System kit (Amersham, RPN2132) were used for detection.
Gene silencing by siRNA interference
Sets of three different siRNA duplexes targeting different sites within the coding sequences of LC3A and LC3B were designed by and obtained from Applied Biosystems/Ambion.25 One day after seeding, the Vero E6 cells were transfected with 40 nM of siRNA using Lipofectamine 2000. Experiments were conducted at 48 h post-transfection. Antibodies against LC3 (Nanotools, 0231-100/LC3-5F10) were used to assess by western blot the depletion of the targeted proteins.
TCID50 assay
This endpoint dilution assay quantifies the amount of virus required to produce a cytopathic effect in 50% of the wells containing inoculated cells (TCID50 unit). Cells were plated in 96-well plates after which serial dilutions of the virus were added. After 4 d of incubation, cytopathic effects were observed and recorded for each virus dilution and used to calculate the amount of TCID50 units in the stock for the particular experimental conditions or employed cell type.
Plasmid construction
The plasmid encoding a C-terminally GFP-tagged fusion of EAV NSP2-3, under the control of a bacteriophage T7 transcription-regulatory element (designated pTUG31Nsp2-3EAVGFP) was constructed by conventional cloning as follows. First, the NSP2-3 encoding fragment was amplified by PCR reaction from a plasmid containing a cDNA clone of EAV (pEAV515)52 using appropriate primers and subsequently cloned into the pGEM-T Easy vector (Promega, A1360) to generate the pGEMNsp2-3EAV plasmid. Subsequently, the NSP2-3-coding sequence was cut from pGEMNsp2-3EAV using EcoRI and cloned into the pTUG31 expression vector,53 yielding the pTUG31Nsp2-3 construct. Finally, the DNA fragment encoding GFP was excised from the pN1-EGFP vector51 by digestion with BamHI and NotI, and treated with Klenow polymerase (Invitrogen, AM2008). This fragment was cloned into SmaI-treated pTUG31Nsp2-3 thereby yielding the pTUG31Nsp2-3EAVGFP plasmid. The plasmid encoding a C-terminally GFP-tagged fusion of NSP3 was constructed in a similar way.
vTF7-3 infection and transfection
For expressions using the vTF7-3 system, subconfluent monolayers of Vero E6 cells grown in 10 cm2 tissue culture dishes were infected with vTF7-353 at a multiplicity of infection of 10. After 1 h, the medium was replaced by a transfection mixture consisting of 0.5 ml of DMEM without FCS but containing 10 units of Lipofectin (Invitrogen, 18324-020) and 1 μg of the pTUG31 expression plasmids. After a 5 min incubation at room temperature, 0.5 ml of DMEM were added, and incubation was continued at 37°C for another 3 h. Subsequently, the culture medium was replaced by fresh FCS-containing culture medium and the incubation was continued until cells were fixed and processed for immunofluorescence analysis.
Quantification of colocalized puncta
The numbers of colocalized puncta per cell were counted using the ImageJ software. The data represent the percentage of dsRNA foci colocalizing with each tested organelle protein marker per cell. Error bars represent the standard deviations from counting of 50 cells in 3 independent experiments.
Supplementary Material
Acknowledgments
The authors thank Masaaki Komatsu, Bernard Moss, Karla Kirkegaard, Guojun Bu, Takashi Ueno and Stuart Siddell for reagents. This work was supported by grants from the Netherlands Organization for Scientific Research (NWO-VIDI and NWO-ALW) and the Utrecht University (High Potential) to C.A.M. de H. and F.R.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/22743
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/22743
References
- 1.den Boon JA, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241–56. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 2.den Boon JA, Diaz A, Ahlquist P. Cytoplasmic viral replication complexes. Cell Host Microbe. 2010;8:77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackenzie J. Wrapping things up about virus RNA replication. Traffic. 2005;6:967–77. doi: 10.1111/j.1600-0854.2005.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries AAF, Horzinek MC, Rottier PJM, de Groot RJ. The genome organization of the nidovirales: Similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedersen KW, van der Meer Y, Roos N, Snijder EJ. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999;73:2016–26. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Meer Y, van Tol H, Locker JK, Snijder EJ. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J Virol. 1998;72:6689–98. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knoops K, Bárcena M, Limpens RW, Koster AJ, Mommaas AM, Snijder EJ. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol. 2012;86:2474–87. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snijder EJ, van Tol H, Roos N, Pedersen KW. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J Gen Virol. 2001;82:985–94. doi: 10.1099/0022-1317-82-5-985. [DOI] [PubMed] [Google Scholar]
- 10.Lin LT, Dawson PW, Richardson CD. Viral interactions with macroautophagy: a double-edged sword. Virology. 2010;402:1–10. doi: 10.1016/j.virol.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor MP, Kirkegaard K. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy. 2008;4:286–9. doi: 10.4161/auto.5377. [DOI] [PubMed] [Google Scholar]
- 12.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–49. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sumpter R, Jr., Levine B. Selective autophagy and viruses. Autophagy. 2011;7:260–5. doi: 10.4161/auto.7.3.14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J Virol. 2007;81:12543–53. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, et al. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008;82:9143–53. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoon SY, Ha YE, Choi JE, Ahn J, Lee H, Kweon HS, et al. Coxsackievirus B4 uses autophagy for replication after calpain activation in rat primary neurons. J Virol. 2008;82:11976–8. doi: 10.1128/JVI.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein KA, Jackson WT. Human rhinovirus 2 induces the autophagic pathway and replicates more efficiently in autophagic cells. J Virol. 2011;85:9651–4. doi: 10.1128/JVI.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein KA, Jackson WT. Picornavirus subversion of the autophagy pathway. Viruses. 2011;3:1549–61. doi: 10.3390/v3091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Li Z, Ge X, Guo X, Yang H. Autophagy promotes the replication of encephalomyocarditis virus in host cells. Autophagy. 2011;7:613–28. doi: 10.4161/auto.7.6.15267. [DOI] [PubMed] [Google Scholar]
- 20.O’Donnell V, Pacheco JM, LaRocco M, Burrage T, Jackson W, Rodriguez LL, et al. Foot-and-mouth disease virus utilizes an autophagic pathway during viral replication. Virology. 2011;410:142–50. doi: 10.1016/j.virol.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang SC, Chang CL, Wang PS, Tsai Y, Liu HS. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J Med Virol. 2009;81:1241–52. doi: 10.1002/jmv.21502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–70. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 25.Reggiori F, Monastyrska I, Verheije MH, Calì T, Ulasli M, Bianchi S, et al. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–8. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Haan CA, Molinari M, Reggiori F. Autophagy-independent LC3 function in vesicular traffic. Autophagy. 2010;6:994–6. doi: 10.4161/auto.6.7.13309. [DOI] [PubMed] [Google Scholar]
- 27.Calì T, Galli C, Olivari S, Molinari M. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem Biophys Res Commun. 2008;371:405–10. doi: 10.1016/j.bbrc.2008.04.098. [DOI] [PubMed] [Google Scholar]
- 28.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagemeijer MC, Verheije MH, Ulasli M, Shaltiël IA, de Vries LA, Reggiori F, et al. Dynamics of coronavirus replication-transcription complexes. J Virol. 2010;84:2134–49. doi: 10.1128/JVI.01716-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol. 2010;84:10438–47. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–64. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyde JL, Sosnovtsev SV, Green KY, Wobus C, Virgin HW, Mackenzie JM. Mouse norovirus replication is associated with virus-induced vesicle clusters originating from membranes derived from the secretory pathway. J Virol. 2009;83:9709–19. doi: 10.1128/JVI.00600-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulasli M, Verheije MH, de Haan CA, Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol. 2010;12:844–61. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suhy DA, Giddings TH, Jr., Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000;74:8953–65. doi: 10.1128/JVI.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 37.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cottam EM, Maier HJ, Manifava M, Vaux LC, Chandra-Schoenfelder P, Gerner W, et al. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335–47. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stertz S, Reichelt M, Spiegel M, Kuri T, Martínez-Sobrido L, García-Sastre A, et al. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–15. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Z, Thackray LB, Miller BC, Lynn TM, Becker MM, Ward E, et al. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–5. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- 42.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279:10136–41. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prentice E, McAuliffe J, Lu X, Subbarao K, Denison MR. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol. 2004;78:9977–86. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Younes HM, Al-Zeer MA, Khalil H, Gussmann J, Karlas A, Machuy N, et al. Autophagy-independent function of MAP-LC3 during intracellular propagation of Chlamydia trachomatis. Autophagy. 2011;7:814–28. doi: 10.4161/auto.7.8.15597. [DOI] [PubMed] [Google Scholar]
- 45.Bernasconi R, Galli C, Noack J, Bianchi S, de Haan CA, Reggiori F, et al. Role of the SEL1L:LC3-I complex as an ERAD tuning receptor in the mammalian ER. Mol Cell. 2012;46:809–19. doi: 10.1016/j.molcel.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 46.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol. 2002;76:3697–708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reggiori F, de Haan CA, Molinari M. Unconventional use of LC3 by coronaviruses through the alleged subversion of the ERAD tuning pathway. Viruses. 2011;3:1610–23. doi: 10.3390/v3091610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Vries AA, Chirnside ED, Bredenbeek PJ, Gravestein LA, Horzinek MC, Spaan WJ. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 1990;18:3241–7. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elroy-Stein O, Moss B. Gene expression using the vaccinia virus/T7 RNA polymerase hybrid system. Curr Protoc Mol Biol 2001; Chapter 16:Unit16.19. [DOI] [PubMed] [Google Scholar]
- 51.Verheije MH, Raaben M, Mari M, Te Lintelo EG, Reggiori F, van Kuppeveld FJ, et al. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castillo-Olivares J, Wieringa R, Bakonyi T, de Vries AA, Davis-Poynter NJ, Rottier PJ. Generation of a candidate live marker vaccine for equine arteritis virus by deletion of the major virus neutralization domain. J Virol. 2003;77:8470–80. doi: 10.1128/JVI.77.15.8470-8480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vennema H, Rijnbrand R, Heijnen L, Horzinek MC, Spaan WJ. Enhancement of the vaccinia virus/phage T7 RNA polymerase expression system using encephalomyocarditis virus 5′-untranslated region sequences. Gene. 1991;108:201–9. doi: 10.1016/0378-1119(91)90435-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.