Abstract
The notion that phosphorylation constitutes a major mechanism to induce autophagy was established 15 years ago when a conserved Atg1/ULK kinase family was identified as an essential component of the autophagy machinery. The key observation was that starved atg1Δ cells lack autophagosomes in the cytosol and fail to accumulate autophagic bodies in the vacuole. Although many studies have revealed important details of Atg1 activation and function, a cohesive model for how Atg1 regulates the autophagic machinery is lacking. Our recent findings identified conserved steps of temporal and spatial regulation of Atg1/ULK1 kinase at both the PAS and autophagosomal membranes, suggesting that Atg1 not only promotes autophagy induction, but may also facilitate late stages of autophagosome biogenesis.
Keywords: Atg1, Atg13, Atg8, LIR, PAS, ULK1, autophagosome, autophagy
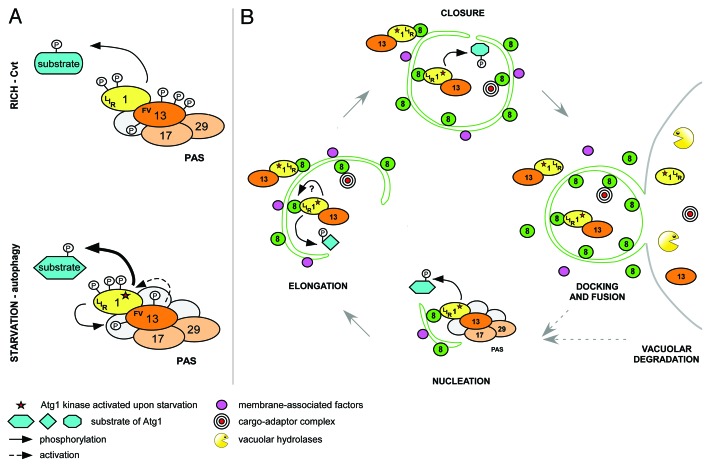
At the onset of autophagy, Atg1 and its positive regulator Atg13 are recruited to the phagophore assembly site (PAS), where they are thought to nucleate the formation of autophagosomal structures. The binding of Atg1 to Atg13 is required for kinase activity in vivo, possibly by promoting Atg1 autophosphorylation on two conserved sites in the activation loop. In contrast to metazoans, the interaction of Atg1 and Atg13 in yeast was considered starvation-regulated, since TORC1-dependent phosphorylation of Atg13 decreased Atg1 binding affinity. However, we found to our surprise that Atg1 is constitutively bound to Atg13, irrespective of the nutrient status of the cell. Thus, we speculate that Atg1 activation is mediated by additional phosphorylation and/or dephosphorylation events, by other binding partners, or by modulating its binding affinity to specific substrates (Fig. 1A). It is worth noting that Atg1 kinase activity is also required in nutrient-rich conditions for delivery of resident vacuolar enzymes by the cytoplasm to vacuole targeting (Cvt) pathway or to specifically eliminate damaged organelles or protein aggregates.
Figure 1. Model of temporal and spatial regulation of the Atg1 kinase complex. (A) Atg1 and Atg13 interact constitutively in rich and starved conditions via the FV motif. Upon starvation, phosphorylation changes on Atg1 and Atg13 activate Atg1 kinase and modulate substrate specificity by induction of conformational changes and/or recruitment of additional regulators. Atg1 and Atg13 may form a reinforcing feedback loop where Atg13 is phosphorylated in an Atg1-dependent manner to stimulate Atg1 kinase activity. (B) At the PAS, Atg1 regulates association and dissociation of Atg proteins to initiate autophagy. Activated Atg1-Atg13 complexes are then tethered to autophagosomal membranes by Atg8 via a conserved LIR motif on Atg1. This Atg1 pool promotes membrane elongation and/or closure by phosphorylation of unknown targets, or by regulating protein dynamics at the membrane. After docking and fusion of autophagosomes with the vacuole, Atg1-Atg13 complexes are degraded, modulating autophagy flux with a negative feedback mechanism.
We further compared the phenotypic and molecular consequences of ATG13 deletion with the expression of the allele that specifically abolishes the interaction with Atg1. Interestingly, although Atg13 mutant protein reduces Atg1 kinase activity in vitro to the ATG13 deletion level, loss of Atg1-Atg13 binding in vivo only partially affects the Cvt and autophagy pathways. It is thus possible that Atg13 serves additional functions, for example as an “activating platform” for autophagy induction, particularly as we observed that Atg13 is required for association of Atg17 and Atg29 with the kinase complex.
Consistent with recent studies in Arabidopsis seedlings, we found that Atg1/hsULK1 interacts with Atg8-family proteins in yeast and mammals. Apart from its involvement in autophagosome biogenesis, Atg8 participates in cargo selection by binding to a growing number of dedicated adaptors and autophagy regulators via the LC3-interacting region (LIR). Importantly, Atg1/hsULK1 binding to Atg8 is also mediated by the conserved LIR motif. Although disrupting Atg1-Atg8 interaction does not affect kinase activity or PAS recruitment of Atg1, it impairs both the Cvt pathway and autophagy, as measured by prApe1 processing and the Pho8∆60 reporter assay. In contrast, the Atg1 LIR mutant shows no clear difference in GFP-Atg8 processing upon starvation compared with the wild-type control. This surprising discrepancy may result from the different nature of these two assays. While Pho8∆60 monitors uptake and delivery of bulk cytoplasmic cargo, GFP-Atg8 cleavage measures vacuolar processing of a specific membrane-associated marker.
Interestingly, our analysis revealed that Atg1 fractionates with membranes and hsULK1 colocalizes with autophagosomal structures in a LIR-dependent manner, implying that Atg8 recruits the kinase to autophagosomal membranes. Surprisingly, among analyzed Atg1 kinase complex members, only Atg1 and Atg13 are targeted for vacuolar degradation, suggesting that some complex components may remain at the PAS or are released from membranes before vesicle completion (Fig. 1B).
We thus propose that upon starvation, Atg1 and Atg13 are recruited to the PAS for spatial kinase activation, followed by their translocation to autophagosomal membranes by binding to Atg8. It is possible that Atg1-Atg13 complexes at the PAS and autophagosomal membranes interact with different regulators and undergo distinct phosphorylation/dephosphorylation events, thereby tuning the kinase to phosphorylate different targets and regulate multiple processes. In addition to its early functions at the PAS, like vesicle nucleation, Atg1/ULK1 may also regulate late steps of autophagosome biogenesis, such as membrane expansion or closure, or fusion of autophagosomes with the vacuolar/lysosomal compartment. As the Atg1 LIR mutant decreases the volume of the cytoplasmic content delivered to the vacuole, a careful analysis of size, number and completion of autophagosomes could help to identify steps impaired by this mutation. An important future task will be to identify the specific functions and relevant substrates of Atg1 at the PAS and autophagosomes.
Importantly, our data also revealed that Atg1-Atg13 complexes are not only regulators of autophagy but also specific autophagic cargos. Degradation of activated Atg1 and Atg13 might be regarded as a “side effect” of autophagic digestion of the complexes trapped within the autophagosomal lumen. However, it may instead comprise a powerful negative feedback mechanism to regulate autophagic flux. In such a scenario, for each new autophagosome assembly, cells would need to activate a new pool of the Atg1 kinase complex at the PAS, thereby coupling Atg1 availability to its degradation. This may enable rapid response to changing nutrient levels and allow effective autophagy shut-off once nutrients are replenished (Fig. 1B). The proposed mechanism is likely to be evolutionarily conserved, as degradation of Atg1 and Atg13 has also been reported in plants. Although attractive, experimental evidence to probe the physiological importance of such a feedback loop is missing. Because Atg8 is both a positive and negative regulator of Atg1 functions, specific experimental conditions need to be designed to distinguish between these two opposing effects.
Finally, our results expand the known functions of Atg8 to include temporal and spatial regulation of Atg1 kinase, thereby coordinating the different cellular processes that govern the selective and bulk autophagy pathways. As Atg8 is involved in many aspects of autophagosome biogenesis and it interacts with various proteins, a tight regulation of Atg8 may be required to fulfill all functions. Indeed, mammalian Atg8 homologs undergo a number of post-translational modifications, including phosphorylation. It is thus attractive to speculate that Atg1 might directly phosphorylate Atg8 at autophagosomes to promote their maturation.
Acknowledgments
The Peter laboratory is supported by the European Research Council (ERC), the 7th EU framework project UNICELLSYS, and by grants from SystemsX.ch, the SNF and the ETH Zürich.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/22584