Eukaryotic cells have to overcome the complexity of chromatin organization in order to gain access to DNA to activate or repress transcription. The organization of chromatin itself acts as a physical restraint. Generally, in order to activate transcription, cells must remove repressive complexes, open the chromatin via the modification of the core and linker histones and recruit transcriptional activators and transcription machinery. A straightforward principle perhaps, but complicated by the multiplicity of enzyme activities, post-translational modifications and crosstalk mechanisms. Adding to this, these processes themselves are regulated by extracellular signals, resulting in the activation of signaling cascades that transmit the signal to the nucleus and ultimately to chromatin. The importance of chromatin-modifying enzymes and transcription factors in cancer is well-established;1 however, the complexity of signaling to chromatin makes challenging the identification of effective potential targets for the treatment of cancer.
Steroid hormones act by binding and activating specific receptors, which are hormone-dependent transcription factors, which also activate kinase-signaling pathways that target the protein components of chromatin.2 We recently demonstrated this interconnection between signaling and chromatin remodeling; since the activation of PARP1 by the cyclin-dependent kinase (CDK2) leads to PARylation of chromatin and is essential for the activation of hormone-responsive genes and cell proliferation in breast cancer cells.3 Progesterone induces a dramatic burst of nuclear PARylation, which was not observed by inhibition of either PARP1 or CDK2. Proliferation induced by progesterone is accompanied by the activation or repression of thousands of genes; indeed, microarray analysis demonstrated that 85% of progesterone target genes are dependent on PARP1 and or CDK2 with the majority (55%) dependent on the combined enzymatic activities. Prior to hormone, CDK2 is maintained in an inactive state, in complex with unliganded progesterone receptor (PR) but without cyclin A.4 Following hormone, PARP1 and cyclin A bind CDK2 facilitating the phosphorylation of Ser785 and Ser786 of PARP1. Importantly, this phosphorylation within the NAD-binding cleft of PARP1 leads to a more open catalytic domain resulting in a more active PARP1. In vitro, phosphomimetic PARP1 mutants show enhanced auto and trans-PARylation capabilities compared with wild-type and phosphor-null PARP1 mutants. In vivo, rescue of progesterone gene activation was only possible by the addition of phosphomimetic PARP1 in PARP1-knockdown cell lines.
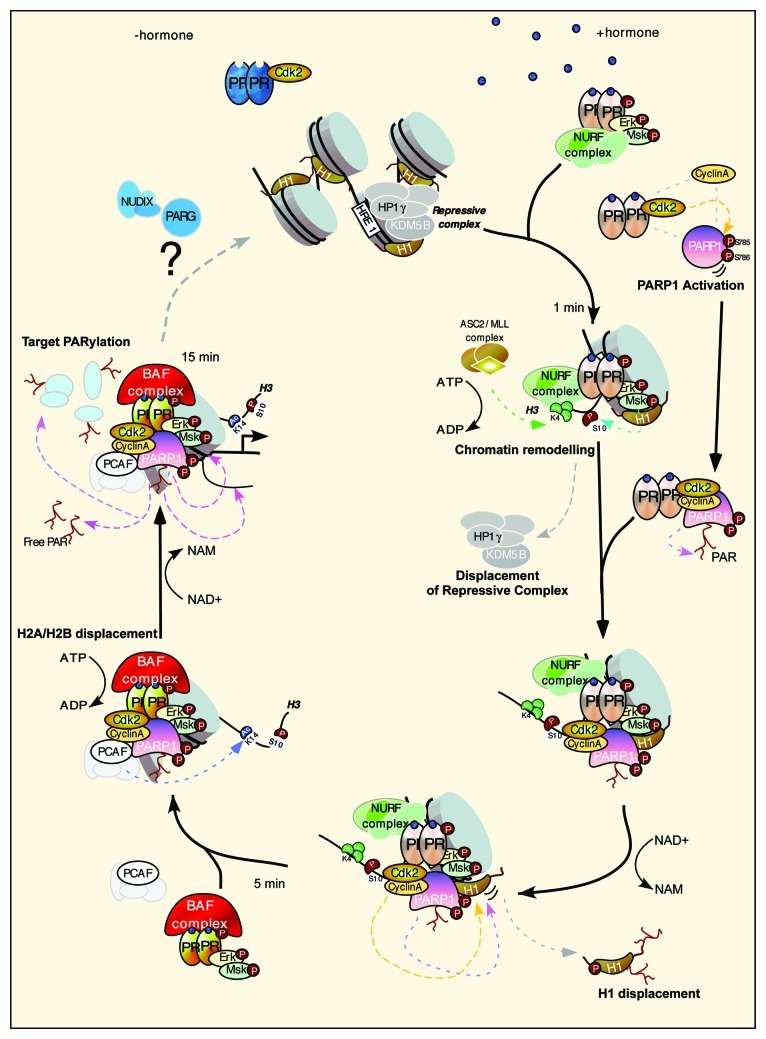
What could be the mechanism of transcriptional regulation by CDK2/PARP1? The complex cyclinA/CDK2/PARP1 is recruited to progesterone target genes via interaction with activated PR. The main target for PARP1, other than PARP1 itself, is histone H1.5 We found that the displacement of histone H1 is key for the activation of target genes.4 Global ChIP-Seq analysis demonstrated the co-recruitment of PARP1 and CDK2 to chromatin, and that the combined enzymatic activities of CDK2 and PARP1 are essential for histone H1 displacement (Fig. 1). PARylation of H1 is necessary for gene activation. One should keep in mind that not only can histones, including H1, be modified by PARP1, but also that histones have a strong affinity for the PAR polymer itself.6 Hence, the opening of chromatin via the displacement of H1 and H2A/H2B may be facilitated by both the covalent and non-covalent modification of histones. Future work will try not only to identify the residues within PARP1 and H1 that are PARylated but also aim to achieve a more global picture of all protein targets and the specific sites modified.
Figure 1. Model depicting the initial steps necessary for the activation of progesterone target genes. Prior to hormone, PR is bound to CDK2 and the promoter resides in a repressed basal state. After hormone, the activated complex of pPR, pERK and pMsk1 is recruited to chromatin concomitantly with the NURF and ASCOM complex, resulting in histone H3 phosphorylation at S10 and trimethylation at K4. These changes lead to displacement of the HP1γ/KDM5 repressive complex and to the stabilization of the NURF complex, respectively. CDK2 in complex with PR binds cyclin A and PARP1, which is activated by CDK2 phosphorylation. Activated PARP1/CDK2 complex is recruited to chromatin, facilitating the dual phosphorylation/PARylation of histone H1 and promoting its displacement. A more open chromatin facilitates the recruitment of the PCAF and the PR-BAF complex, resulting in the acetylation of H3K14, anchoring BAF to chromatin and leading to displacement of H2A/H2B in an ATP-dependent manner. PARP1 PARylates other targets in chromatin resulting in full promoter activation, which are then removed via the actions of PAR-degrading enzymes.
All parties must come to an end; indeed, global PARylation induced by hormone is a transient event, returning to basal state after 30 min. This pulse of PAR leads to some intriguing open questions; we have evidence to suggest that not only is the formation of PAR by PARP1 essential for hormone-induced gene regulation, but also its degradation via the actions of PAR-degrading enzymes, including PARG (polyADP-ribose glycohydrolase) and members of the NUDIX (nucleoside diphosphate linked to another moiety X) class of hydrolases. A future challenge is to elucidate the mechanism and function of PAR degradation and what role, if any, this plays in returning the genes and chromatin to the basal state prior to PARylation (Fig. 1). Altogether, and in view of the relatively poor response and observed toxicities of PARP1 inhibitors in cancer trials,7 combining our knowledge of CDK2 and of the interdependence of transcription activator/repressor pathways will provide insight for the treatment of breast and other cancers, applying a more combinatorial approach.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/22531
References
- 1.Dawson MA, et al. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Vicent GP, et al. Mol Endocrinol. 2010;24:2088–98. doi: 10.1210/me.2010-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright RH, et al. Genes Dev. 2012;26:1972–83. doi: 10.1101/gad.193193.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vicent GP, et al. Genes Dev. 2011;25:845–62. doi: 10.1101/gad.621811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Messner S, et al. Trends Cell Biol. 2011;21:534–42. doi: 10.1016/j.tcb.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Panzeter PL, et al. Biochemistry. 1992;31:1379–85. doi: 10.1021/bi00120a014. [DOI] [PubMed] [Google Scholar]
- 7.Kummar S, et al. BMC Med. 2012;10:25. doi: 10.1186/1741-7015-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]