Abstract
Satellite DNA sequences consist of tandemly arranged repetitive units up to thousands nucleotides long in head-to-tail orientation. The evolutionary processes by which satellites arise and evolve include unequal crossing over, gene conversion, transposition and extra chromosomal circular DNA formation. Large blocks of satellite DNA are often observed in heterochromatic regions of chromosomes and are a typical component of centromeric and telomeric regions. Satellite-rich loci may show specific banding patterns and facilitate chromosome identification and analysis of structural chromosome changes. Unlike many other genomes, nuclear genomes of banana (Musa spp.) are poor in satellite DNA and the information on this class of DNA remains limited. The banana cultivars are seed sterile clones originating mostly from natural intra-specific crosses within M. acuminata (A genome) and inter-specific crosses between M. acuminata and M. balbisiana (B genome). Previous studies revealed the closely related nature of the A and B genomes, including similarities in repetitive DNA. In this study we focused on two main banana DNA satellites, which were previously identified in silico. Their genomic organization and molecular diversity was analyzed in a set of nineteen Musa accessions, including representatives of A, B and S (M. schizocarpa) genomes and their inter-specific hybrids. The two DNA satellites showed a high level of sequence conservation within, and a high homology between Musa species. FISH with probes for the satellite DNA sequences, rRNA genes and a single-copy BAC clone 2G17 resulted in characteristic chromosome banding patterns in M. acuminata and M. balbisiana which may aid in determining genomic constitution in interspecific hybrids. In addition to improving the knowledge on Musa satellite DNA, our study increases the number of cytogenetic markers and the number of individual chromosomes, which can be identified in Musa.
Introduction
A significant part of nuclear genomes in plants, including those with small genomes such as rice, Brachypodium and Arabidopsis is occupied by various types of repetitive DNA sequences [1], [2], [3]. Repetitive DNA sequences are classified based on genomic organization of repetitive units as dispersed and tandem. Dispersed repeats, represented by various classes of transposable elements encode for proteins which facilitate their replication and integration into nuclear genome [4]. Tandem repeats usually contain non-coding sequences organized in tandem arrays [5], [6], [7], [8], [9]. The arrays may be large and consist of thousands or even millions of repetitive units arranged in head-to-tail orientation [10], [11]. The processes by which satellites arise and evolve are not well understood, and unequal crossing over, gene conversion, transposition and formation of extra chromosome circular DNA (eccDNA) were implicated [12].
The analysis of repetitive DNA and satellite DNA in particular, has been hampered by the presence of multiple copies of the same or similar sequences arranged in tandem. However, the situation changed recently with the advent of new sequencing technologies, which enable identification and reconstruction of tandem organized units even from partial genome sequencing data [13], [14], [15]. On line with the analysis at DNA level, long-range organization of satellite DNA has been studied using cytogenetic methods; fluorescence in situ hybridization (FISH) being the most frequent. Large blocks of satellite DNA are typically observed in heterochromatic regions of chromosomes and are usually located in centromeric and telomeric regions [16], [17]. Chromosome loci rich in satellite DNA usually show specific banding pattern and this makes them useful cytogenetic markers to discriminate individual chromosomes [18], [19], [20], [21]. The presence of chromosome landmarks enables identification of individual chromosomes and is a prerequisite to study structural changes accompanying evolution and speciation and to follow chromosome behavior and transmission in interspecific hybrids [22], [23].
Bananas are a staple food and important export commodity in many countries of humid tropics. Most of banana cultivars are seed sterile diploid and triploid clones originating from natural inter- and intra-specific crosses involving wild diploid species of genus Musa: M. acuminata (A genome) and M. balbisiana (B genome) [24]. Both species belong to section Eumusa (x = 11); other three sections are recognized within the genus Musa based on chromosome number and morphology: Rhodochlamys (x = 11), Australimusa (x = 10) and Callimusa (x = 10 or x = 9). Nuclear genome of Musa is relatively small (1C×∼550–750 Mbp) [25], [26] and divided to 1–2 µm long and morphologically similar chromosomes. This makes cytogenetic studies difficult and there is no reliable method to identify all chromosomes within a karyotype and discriminate parental chromosomes in hybrids. Genomic in situ hybridization (GISH) has been used occasionally to identify parental chromosomes in some hybrid cultivars [27], [28]. However, the method discriminates only (peri)centromeric chromosome regions. Apart from identifying parental chromosomes, the paucity of chromosome-specific landmarks and markers hampers studies on karyotype evolution and chromosome behavior in hybrids. To date, only a few DNA sequences and probes such as rDNA, some DNA repeats and BAC clones were found useful to study the organization of plant nuclear genomes at cytogenetic level [29], [30], [31].
Recently, a variety of repetitive DNA elements including two new satellite DNA sequences were identified in silico from 454 sequencing data of M. acuminata cv. ‘Calcutta 4’ [13]. Even though most of retrotransposons were found dispersed along all banana chromosomes, some of them (a LINE element and a CRM retrotransposon) were located in (peri)centromeric regions, while the DNA satellites were identified on specific chromosome loci [13]. These results indicated a potential of these sequences as cytogenetic markers.
In this work we characterized genomic organization of two main banana DNA satellites together with other DNA sequences (a LINE element, rRNA genes, a BAC clone) in nineteen accessions of Musa, including inter-specific hybrids. Molecular analysis of the two DNA satellites revealed their sequence conservation within and between the accessions. The mode of their genomic distribution makes them suitable as cytogenetic markers. We show that the LINE-like element is present in centromeric regions of all nineteen accessions and can be used as centromere-specific cytogenetic marker. The present work improves the knowledge of genome structure in Musa and expands the number of individual chromosomes which can be identified. Differences in fluorescent labeling patterns obtained after FISH with a set of probes can be used to support determination of genomic constitution in inter-specific hybrids.
Materials and Methods
Plant Material and Genomic DNA Extraction
In vitro rooted plants of 18 Musa accessions were obtained from the International Transit Centre (ITC, Katholieke Universitiet, Leuven, Belgium). Five rooted plants of M. balbisiana clone ‘Pisang Klutuk Wulung’ were obtained from Dr. François Côte (CIRAD, Guadeloupe). The in vitro plants were transferred to soil and all plants were maintained in a greenhouse. Table 1 lists all accessions used in the present study.
Table 1. List of Musa accessions and their genome sizes.
| Species | Subspecies | Accession name | ITC code | Genomic constitutiona | 2C nuclear DNAcontent [pg] | Mean monoploid genome size [Mbp/1Cx]b | Referencec | |
| mean | ± SD | |||||||
| M. acuminata | burmannicoides | Calcutta4 | 0249 | AA | 1.226 | 0.004 | 600 | Bartoš et al. [25] |
| burmannica | Long Tavoy | 0283 | AA | 1.238 | 0.01 | 605 | * | |
| zebrina | Maia Oa | 0728 | AA | 1.325 | 0.02 | 648 | * | |
| malaccensis | DH Pahang | 1511 | AA | 1.214 | 0.027 | 594 | * | |
| Tuu Gia | 0610 | AA | 1.261 | 0.02 | 617 | * | ||
| M. balbisiana | Cameroun | 0246 | BB | 1.121 | 0.003 | 548 | Lysák et al. [67] | |
| Honduras | 0247 | BB | 1.133 | 0.002 | 554 | * | ||
| Tani | 1120 | BB | 1.126 | 0.014 | 551 | * | ||
| Pisang Klutuk Wulung | – | BB | 1.132 | 0.025 | 554 | * | ||
| M. schizocarpa | Musa schizocarpa | 0560 | SS | 1.373 | 0.015 | 671 | * | |
| Musa schizocarpa | 1002 | SS | 1.364 | 0.003 | 667 | * | ||
| Hybrids | Obino l’Ewai | 0109 | AAB | 1.777 | 0.004 | 579 | Lysák et al. [67] | |
| Maritú | 0639 | AAB | 1.847 | 0.005 | 602 | Lysák et al. [67] | ||
| 3 Hands Planty | 1132 | AAB | 1.786 | 0.019 | 582 | * | ||
| Pelipita | 0472 | ABB | 1.751 | 0.002 | 571 | Lysák et al. [67] | ||
| Balonkawe | 0473 | ABB | 1.814 | 0.034 | 591 | * | ||
| Ato | 0820 | AS | 1.294 | 0.015 | 633 | * | ||
| Tonton Kepa | 0822 | AS | 1.318 | 0.021 | 645 | * | ||
| Umbubu | 0854 | AT | 1.397 | 0.009 | 683 | * | ||
Genome constitution as described in MGIS database (http://www.crop-diversity.org/banana/#AvailableITCAccessions).
1 pg = 0.978×109 bp [34].
The study in which DNA content was estimated.
Estimated in the present work.
Genomic DNA was prepared from nuclei isolated from healthy young leaf tissues according to Zhang et al. [32]. Isolated nuclei were incubated with 40 mM EDTA, 0.2% SDS and 0.25 µg/µl proteinase K for 5 hours at 37°C; DNA was purified by phenol/chloroform precipitation.
Estimation of Genome size
Nuclear genome size was estimated according to Bartoš et al. [25]. Suspensions of cell nuclei were prepared by chopping leaf tissues with a razor blade in a glass Petri dish containing 500 µl Otto I solution (0.1 M citric acid, 0.5% v/v Tween 20). Approximately 50 mg of young Musa leaf and 10 mg of leaf of soybean (Glycine max L. cv. Polanka, 2C = 2.5 pg DNA) [26] which served as internal standard were used for sample preparation. Crude homogenate was filtered through a 50 µm nylon mesh, nuclei were pelleted (300 g, 5 min) and resuspended in 300 µl Otto I solution. After 1 hour incubation at room temperature, 900 µl Otto II solution (0.4 M Na2HPO4 ) [33] supplemented with 50 µg/ml RNase, 50 µg/ml propidium iodide and 3 µl/ml 2-mercaptoethanol, were added. Samples were analyzed using Partec PAS flow cytometer (Partec GmbH, Münster, Germany) equipped with 488-nm argon laser. At least 5,000 nuclei were analyzed per sample. Three individuals were analyzed in each accession, and each individual was measured three times on three different days. Nuclear DNA content was calculated from individual measurements following the formula:
Mean nuclear DNA content was then calculated for each plant and converted to the number of base pairs considering 1 pg DNA equal to 0.978×109 bp [34].
Dot-plot
Satellites maTR_CL18 and maTR_CL33 (GenBank accessions: JX624137 and JX624137) were originally identified in silico after low-pass 454 sequencing genomic DNA of M. acuminata ‘Calcutta 4’ in our previous study [13] and their presence was confirmed in the recently published genome assembly of M. acuminata ‘DH Pahang’ [35]. In order to confirm tandem arrangement of both repeats prior to our experimental work, we have analyzed the sequence data using dotter [36].
Southern Hybridization
Genomic organization of DNA satellites maTR_CL18 and maTR_CL33 [13] was analyzed in all 19 Musa accessions. Aliquots of genomic DNA corresponding to 1×107 copies of monoploid (1Cx) nuclear genomes were digested using DraI whose restriction site was identified in maTR_CL18 and maTR_CL33 sequences. Digested DNA was size-fractionated by 1.2% agarose gel electrophoresis and transferred to Hybond N+ nylon membranes (Amersham, Bath, UK). Probes for satellites CL18 and CL33 were prepared after PCR amplification from genomic DNA of M. acuminata ‘Calcutta 4’ using specific primers (Table 2) and labeling by biotin. Southern hybridization was done at 68°C overnight and signals were detected using BrightStar® BioDetect™ kit under manufacturer’s instructions (Ambion, Austin, USA).
Table 2. Primers used for PCR amplification of satellite DNA and a LINE element.
| Type of repetitive DNA | DNA sequence name | Primer name | Primer sequence | PCR product length |
| Tandem repeat | ||||
| CL18 | CL18-1a | 5′-ATCATGGGCCAACACTTGAT | Ladder-like pattern | |
| 5′-TCGTGAGAGCGGGTTAGAGT | ||||
| CL18-2a | 5′- ATCATGGGCCAACACTTGAT | Ladder-like pattern | ||
| 5′-TGAGAGCGGGTTAGAGTTCC | ||||
| CL18-IN1b | 5′-CGAATGATTTGATGTCATCTCC | 491 bp | ||
| 5′-AGTGTTGGCCCATCATGTTT | ||||
| CL18-IN2b | 5′-GCAATGTTTCAACTCATTACCAA | 179 bp | ||
| 5′-GATGCTACCGGGAAAAATTG | ||||
| CL33 | CL33-1a, b | 5′-AATCGATCGAACCTCGACAT | 130 bp | |
| 5′-TCCCAATAAATTTGCCTTCG | ||||
| Non-LTR retrotransposon | ||||
| LINE element | CL1SCL8/452a | 5′-TGAAAGCAGCTTGATTTGGA | 218 bp | |
| CL1SCL8/452a | 5′-CAAGGCTTGCCAACATTTTT | |||
Primers used to prepare probes for FISH.
Primers were used to prepare probes for Southern hybridization.
Serial dilutions of genomic DNA and PCR products of satellite repeats used as standards were dot-blotted onto Hybond-N+ membranes (Amersham) with the aim to estimate copy number of satellite DNA sequences in the nuclear genomes of all evaluated Musa accessions. PCR products of satellite DNAs labeled by biotin were used as hybridization probes. Dots of genomic DNA and standards that gave the same intensity of hybridization signals were identified after visual inspection. Copy numbers of individual probes were estimated assuming that 1 pg of genomic DNA equals 0.978×109 bp [34].
Sequencing and Sequence Data Analysis of Satellite DNA
Sequences homologous to the two satellites (in the present work termed CL18-like and CL33-like satellites) were amplified from genomic DNA of all 19 Musa species using specific primers (Table 2). PCR reaction mix contained 10 ng genomic DNA, 1.5 mM MgCl2, 0.2 mM dNTPs, 1 µM primers, 1×PCR buffer and 2 U/100 of Dynazyme™ II DNA polymerase (Finnzymes, Espoo, Finland). Amplification was performed as follows: 94°C for 5 min (1 cycle), 94°C for 50 s, 57°C for 90 s, 72°C for 50 s (35 cycles) and 72°C for 10 min (1 cycle). PCR products were purified by PCR Rapid Kit (Invitek, Berlin, Germany), ligated into pCR-XL-TOPO vector and transformed into One Shot TOP10 electrocompetent E. coli (Invitrogen Life Technologies, Carlsbad, USA). For each Musa accession, 24 to 60 cloned PCR products were sequenced. Sequencing was carried out using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, USA) according to the manufacturer’s instructions and run on ABI 3730×l DNA analyzer (Applied Biosystems).
Nucleotide sequences were edited using Staden Package [37] the consensus sequences of CL33-like and the two parts of CL18-like satellites were assembled using cap3 program [38]. Multiple sequence alignments were done using MAFFT program v6.717-1 (–globalpair –maxiterate 1000) [39] and graphically displayed in SeaView v4.2.1. [40]. Sequence diversity was identified using DNA Sam program [41]. Sequence logos were generated using WebLogo tool [42]. SplitsTree4 v4.1.11 [43] was used to construct cladograms based on the Jukes-Cantor, K2P and uncorrected p-distances. Non-parametric bootstrapping with 1000 pseudoreplicates was performed to assess the nodal support. Cladograms were drawn and edited using FigTree program (http://tree.bio.ed.ac.uk/software/figtree/).
Chromosome Preparations
Metaphase spreads were prepared according to Doleželová et al. [29]. Actively growing root tips were pre-treated in 0.05% 8-hydroxyquinoline for 3 hrs and fixed in 3∶1 ethanol : acetic acid. Fixed roots were washed in a solution of 75 mM KCl and 7.5 mM EDTA (pH 4) and meristem tips were digested in a mixture of 2% (w/v) pectinase and 2% (w/v) cellulase in 75 mM KCl and 7.5 mM EDTA (pH 4) for 90 min at 30°C. Protoplast suspension was then filtered through a 150 µm nylon mesh and pelleted. The pellet was resuspended in 75 mM KCl and 7.5 mM EDTA (pH 4) and incubated for 5 min at room temperature. After pelleting, the protoplasts were washed three times with 70% ethanol, and 5 µl of suspension were dropped onto a slide. Shortly before drying out, 5 µl of 3∶1 fixative were added to the drop to induce protoplast bursting. Finally, the slide was rinsed in 100% ethanol and air-dried.
Fluorescence In Situ Hybridization (FISH)
BAC clone 2G17 [30] was labeled by Dig-11-dUTP or Bio-16-dUTP Nick Translation (Roche Applied Science, Penzberg, Germany) according to manufacturer’s instructions. FISH probes for 45S rDNA and 5S rDNA were obtained by labeling Radka1 DNA clone (45S rDNA) and Radka2 DNA clone (5S rDNA) [31] with digoxigenin-11-dUTP or biotin-16-dUTP (Roche Applied Science). Both probes were labeled by PCR using M13 forward and reverse primers (Invitrogen). Banana-specific LINE element and tandem repeats maTR_CL18 and maTR_CL33 [13] were labeled with digoxigenin-11-dUTP or biotin-16-dUTP (Roche) by PCR with specific primers (Table 2).
Hybridization mixture consisting of 50% formamide, 10% dextran sulfate in 1×SSC and 1 µg/ml labeled probe was added onto slides and denatured at 80°C for 3 min. The hybridization was carried out at 37°C overnight. The sites of probe hybridization were detected using anti-digoxigenin-FITC (Roche Applied Science) and streptavidin-Cy3 (Vector Laboratories, Burlingame, USA), and the chromosomes were counterstained with DAPI. The slides were examined with Olympus AX70 fluorescence microscope and the images of DAPI, FITC and Cy-3 fluorescence were acquired separately with a cooled high-resolution black and white CCD camera. The camera was interfaced to a PC running the MicroImage software (Olympus, Tokyo, Japan).
Results
We applied various molecular and cytogenetic approaches to study genomic organization of two major DNA satellites in Musa: maTR_CL18 and maTR_CL33. We first confirmed using dot-plot analysis that both repeats were indeed tandem organized (Figure S1). The molecular structure and variability of tandem organized regions was investigated after sequencing products obtained after PCR with specific primers. We then used Southern hybridization and fluorescence in situ hybridization to estimate the copy number of ma_TR_CL18 and maTR_CL33 and characterize distribution of the repeats on mitotic chromosomes, respectively.
Sequence Analysis of DNA Satellites
A set of primers specific for CL18 and CL33 satellites was used to amplify these repeats from all nineteen Musa accessions. The PCR products were cloned and sequenced to study diversity of repeats within and between banana accessions (Tables S1, S2, S3).
Considering the length of CL18 repetitive units (2226 bp) and heterogeneity of the sequenced regions, two parts of the CL18-like repetitive unit were amplified and assembled: CL18-part1 and CL18-part2, which corresponded to 921 bp and 825 bp regions of CL18 [13]. The sequenced region represented 1728 bp (77.6%) of CL18. In most of the Musa accessions, sequences representing both parts of CL18 repetitive units were obtained (Tables S1, S2, S3). The exceptions were M. acuminata ‘Tuu Gia’ and both representatives of M. schizocarpa, where PCR amplification followed by sequencing did not identify sequences similar to part1 of CL18. In some other clones (M. acuminata ‘DH Pahang’, ‘Long Tavoy’ and ‘Maia Oa’, M. balbisiana ‘Honduras’ and ‘Tani’, and hybrid clones ‘Balonkawe’ (ABB), ‘Ato’ (AS) and ‘Tonton Kepa’ (AS)), relatively short sequences corresponding to part1 of CL18 were obtained. On the other hand, sequences homologous to part2 of CL18 were not obtained from hybrid clone ‘3 Hands Planty’ (AAB).
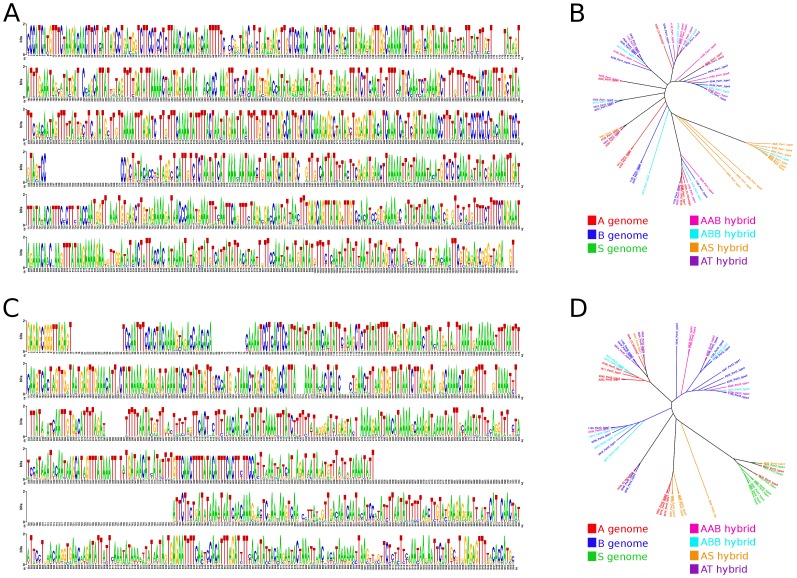
In most of the accessions, both parts of CL18-like repeat regions shared high homology to the maTR_CL18 satellite as identified in 454 data [13]. Lower sequence homology to maTR_CL18 was observed in accessions in which PCR amplification did not result in amplification of both parts of maTR_CL18 (Tables S1, S2). Sequence diversity of regions corresponding to part2 of maTR_CL18 repetitive unit was slightly lower within individual accessions as compared to part1 (Tables S1, S2). Phylogenetic analysis based on Neighbor Joining resulted in a tree in which different clades did not contain species-specific CL18-like repetitive units. Thus, we were not able to identify B genome-specific variants of CL18 suitable as B-genome specific markers (Figure 1C, D). Altogether our observations indicated that both parts homologous to CL18 satellite were highly conserved within and between the analyzed accessions (Figures 1A, B).
Figure 1. Sequence diversity of the two parts of CL18-like satellite sequences.
Consensus sequences of both parts of CL18-like satellite were reconstructed from all analyzed accessions and are displayed as sequence logo (A: CL18-part1; C: CL18-part2). Neighbor-Joining trees constructed from a Jukes-Cantor distance matrix and rooted to the midpoint show diversity of reconstructed parts of CL18-like satellite units obtained from all studied species (B: CL18-part1; D: CL18-part2).
Sequencing PCR products obtained with primers specific for the CL33 satellite confirmed its presence in all Musa accessions. In several accessions (‘Maia Oa’, ‘Tuu Gia’, ‘Pelipita’, ‘3 Hands planty’, ‘Ato’ and ‘Tonton Kepa’) 188 bp repetitive units were present in addition to 134 bp repetitive units of CL33. This significant length difference was due to a 54 bp insertion/deletion. Similarly, M. acuminata ‘Long Tavoy’ contained a 110 bp repetitive unit of CL33.
Most of the analyzed Musa accessions shared high homology to maTR_CL33 satellite [13]. Lower sequence homology to maTR_CL33 was observed in both accessions of M. schizocarpa and its interspecific hybrid (Table S3). In general, a low sequence diversity of sequences corresponding to maTR_CL33 repetitive unit was observed. The only exceptions were revealed in ‘Balonkawe’ (ABB) and one representative of M. balbisiana (Table S3). The high level of homology of sequences corresponding to maTR_CL33 satellite within and between all Musa accessions is demonstrated in Figure 2A. Similarly to the results of CL18-like repeats, cluster analysis based on Neighbor Joining showed that there were no A or B genome-specific CL33-like satellite units in the current set of Musa accessions (Figure 2B).
Figure 2. Diversity of CL33-like satellite sequence.
Consensus sequence of CL33-like satellite was reconstructed from all obtained sequence units of all analyzed accessions and is displayed as sequence logo (A). Neighbor-Joining tree constructed from a Jukes-Cantor distance matrix and rooted to the midpoint shows diversity of individual types of CL33-like repetitive units obtained from all studied accessions (B).
Nuclear DNA Content
The amount of nuclear DNA was estimated after flow cytometric analysis of propidium iodide-stained nuclei. All analyses resulted in histograms of relative DNA content with clearly defined peaks corresponding to G1 nuclei of Musa and the reference standard (Glycine max) with coefficient of variation ranging from 2.5% to 4.5%. 2C nuclear DNA content ranged from 1.121 pg to 1.397 pg for diploid accessions and from 1.751 pg to 1.847 pg for triploid accessions (Table 1). Among the diploid accessions, the lowest DNA content was found in M. balbisiana (2C = 1.121–1.133 pg), while the highest 2C DNA content of 1.397 pg was estimated in ‘Umbubu’ (AT genomic constitution).
Southern Hybridization
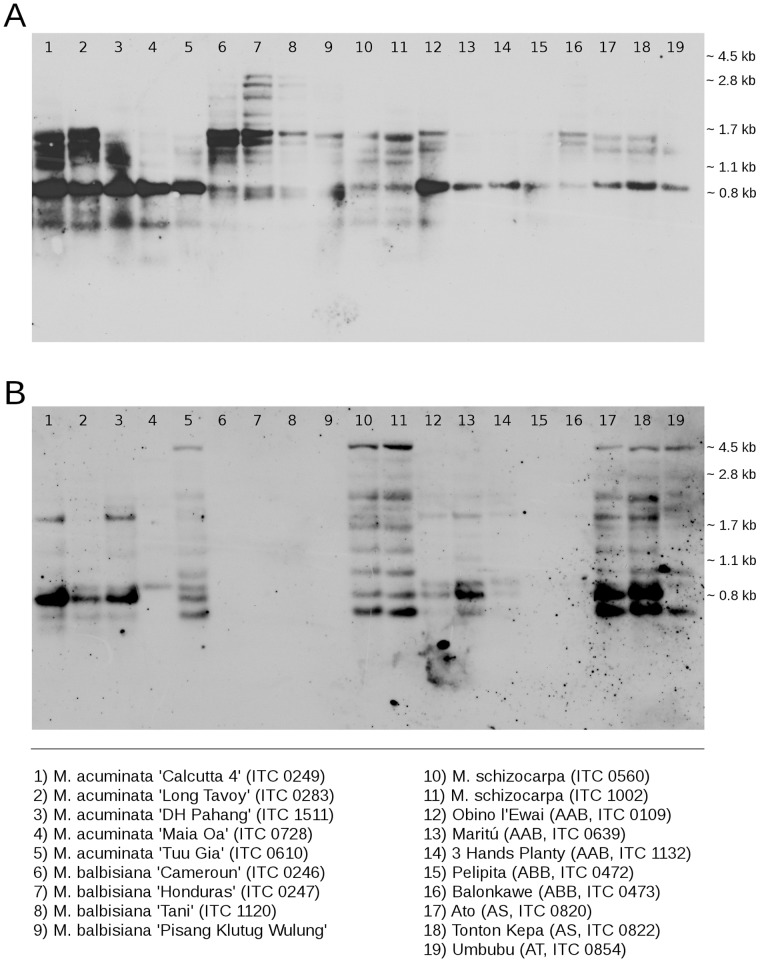
Southern hybridization with probes for maTR_CL18 and maTR_CL33 was carried out to investigate genomic organization of the satellites. Unfortunately, a typical pattern showing individual n-mers units was not observed, most probably because DraI endonuclease did not digest all repetitive units in the analyzed accessions. Both satellites gave ladder-like pattern (Figure 3A) with various hybridization signal intensities. While the signals of maTR_CL18 repeat were visible in all Musa accessions, the presence maTR_CL33 repeat was not confirmed in any of the four accessions of M. balbisiana. Accordingly, maTR_CL33 repeat resulted in weak signals in hybrid clones with ABB genome constitution (Figure 3B).
Figure 3. Southern hybridization of genomic DNA isolated from nineteen Musa accessions.
Samples of genomic DNA corresponding to 1×107 copies of monoploid (1Cx) nuclear genomes were digested using DraI restriction enzyme and hybridized with probes for CL18-like satellite (A) and CL33-like satellite (B) at hybridization stringency of 85%.
Dot-blot analysis showed a limited variation in copy number of maTR_CL18 repeat. Its abundance ranged approximately from 1×103 to 2×103 per monoploid (1Cx) genome of M. acuminata except of M. acuminata ‘DH Pahang’ with 3.5×103 to 5×103 copies. A similar estimate (1×103 to 2×103) was made also in both accessions of M. schizocarpa and in M. balbisiana ‘Honduras’. The remaining balbisiana species contained of 3.5×103 to 5×103 copies of maTR_CL18 repeat. Monoploid genome of hybrid clones ‘Ato’ and ‘Tonton Kepa’ (AS) contained 1×103 to 2×103 copies of maTR_CL18 units, while hybrid clones ‘Umbubu’ (AT), ‘Maritú’ and ‘3 Hands Planty’ contained approximately 3×103–4×103 copies of the repeat. Finally, hybrid clones ‘Pelipita’ and ‘Balonkawe’ (ABB) as well as ‘Obino l’Ewai’ (AAB) comprised ∼5×103 to 6.5×103 copies of maTR_CL18 (Table S4).
The number of copies of maTR_CL33 repeat ranged from 2.5×103 to 4×103 per monoploid genome of M. acuminata ‘Long Tavoy’, ‘Zebrina’ and ‘Tuu Gia’ as well as in both hybrid clones ‘Ato’ and ‘Tonton Kepa’. 5.5×103 to 7×103 copies of maTR_CL33 repetitive units were estimated in monoploid genomes of M. acuminata ‘Calcutta4’ and ‘DH Pahang’ and both schizocarpa accessions. The same amount of maTR_CL33 (5.5×103 to 7×103) was revealed in hybrid clones ‘Obino l’Ewai’, ‘Maritú’ and ‘3 Hands Planty’ (AAB) as well as in ‘Umbubu’ (AT). Dot-blot analysis failed to detect presence of maTR_CL33 repeat in M. balbisiana species and in hybrid clones with ABB genomic constitution.
Mapping DNA Sequences on Mitotic Chromosomes
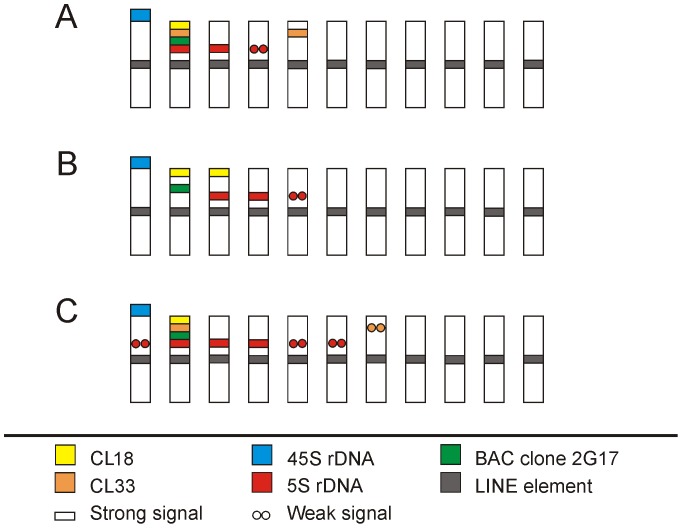
In order to study chromosome organization of the two satellites and the position of their loci in relation to other cytogenetic landmarks, multicolor FISH was done in a set of 19 Musa species and clones with probes for single-copy BAC clone 2G17, 45S rDNA and 5S rDNA. Moreover, taking into the account the relatively small size of banana chromosomes and difficulty in identifying primary constrictions, a probe for LINE element, which was identified and characterized previously in M. acuminata cv. ‘Calcutta 4’ [13], was used to label putative centromeric regions. The element was found in centromeric regions of all Musa accessions and thus was used as a centromeric marker to aid in constructing the idiograms (Figure 4, Figure S2, Figures S3A–G).
Figure 4. Idiograms of three diploid Musa accessions.
(A) M. acuminata ‘DH Pahang’ ITC 1511; (B) M. balbisiana ‘Pisang Klutuk Wulung’; (C) M. schizocarpa ITC 0560.
45S rRNA genes
FISH with a probe for 45S rDNA revealed its localization exclusively to secondary constriction; the only exception being a diploid hybrid clone ‘Ato’ (AS genome). Apart from this cultivar, in which additional 45S rDNA locus was observed on one of the nucleolar organizing chromosomes (Figure 3F), the number of 45S rDNA loci in mitotic metaphase plates corresponded to ploidy level (Table 3). 45S rDNA was detected in secondary constriction on one chromosome pair in diploid accessions, while three loci corresponding to 45S rDNA were identified in triploid accessions.
Table 3. Number of loci detected using FISH with probes for CL18 and CL33 satellites, rRNA genes and BAC clone 2G17 on mitotic metaphase plates in 19 accessions of Musa.
| Accession name | ITC* code | Genomic constitution | Chromosome number (2n) | Number of probe signals (per metaphase plate) | ||||
| 45S rDNA | 5S rDNA | 2G17 | CL18 | CL33 | ||||
| Calcutta 4 | 0249 | AA | 22 | 2 | 8 | 2 | 2 | 4 |
| Long Tavoy | 0283 | AA | 22 | 2 | 4 | 2 | 2 | 4 |
| Maia Oa | 0728 | AA | 22 | 2 | 4 | 2 | 2 | 2 |
| DH Pahang | 1511 | AA | 22 | 2 | 6 | 2 | 2 | 4 |
| Tuu Gia | 0610 | AA | 22 | 2 | 4 | 2 | 2 | 4 |
| Cameroun | 0246 | BB | 22 | 2 | 4 | 2 | 3 | − |
| Honduras | 0247 | BB | 22 | 2 | 6 | 2 | 4 | − |
| Tani | 1120 | BB | 22 | 2 | 6 | 2 | 4 | − |
| Pisang Klutuk Wulung | – | BB | 22 | 2 | 6 | 2 | 4 | − |
| Musa schizocarpa | 0560 | SS | 22 | 2 | 12 | 2 | 2 | 4 |
| Musa schizocarpa | 1002 | SS | 22 | 2 | 12 | 2 | 2 | 4 |
| Obino l’Ewai | 0109 | AAB | 33 | 3 | 9 | 4 | 4 | 4 |
| Maritú | 0639 | AAB | 33 | 3 | 8 | 4 | 4 | 4 |
| 3 Hands Planty | 1132 | AAB | 33 | 3 | 9 | 4 | 4 | 3 |
| Pelipita | 0472 | ABB | 33 | 3 | 10 | 3 | 6 | 1 |
| Balonkawe | 0473 | ABB | 33 | 3 | 10 | 4 | 6 | 1 |
| Ato | 0820 | AS | 22 | 2 | 9 | 2 | 2 | 4 |
| Tonton Kepa | 0822 | AS | 22 | 2 | 9 | 2 | 2 | 4 |
| Umbubu | 0854 | AT | 21 | 2 | 4 | 1 | 1 | 2 |
International Transit Centre (http://www.bioversityinternational.org/research/conservation/genebanks/musa_international_transit_centre.html).
5S rRNA genes
Gene loci for 5S rRNA were observed mostly in distal chromosome regions in all accessions (Figures 4, 5, Figures S3A – G, Figure S4). Diploid chromosome sets of M. acuminata ‘Long Tavoy’, M. acuminata ‘TuuGia’ and M. acuminata ‘Maia Oa’ contained four signals corresponding to 5S rRNA genes, M. acuminata ‘Pahang’ contained six signals and eight loci of 5S rRNA gene clusters were detected in mitotic metaphase plates of M. acuminata ‘Calcutta 4’. Diploid representatives of B genome showed presence of six 5S rDNA loci with the exception of M. balbisiana ‘Cameroun’, which contained only four 5S rDNA clusters. Both representatives of M. schizocarpa (SS genome) contained six chromosome pairs bearing 5S rRNA genes (Figure 4, Figure S3C, Table 3). In triploid hybrid banana clones, eight to ten 5S rDNA loci were observed (Figures S3D, E, Table 3).
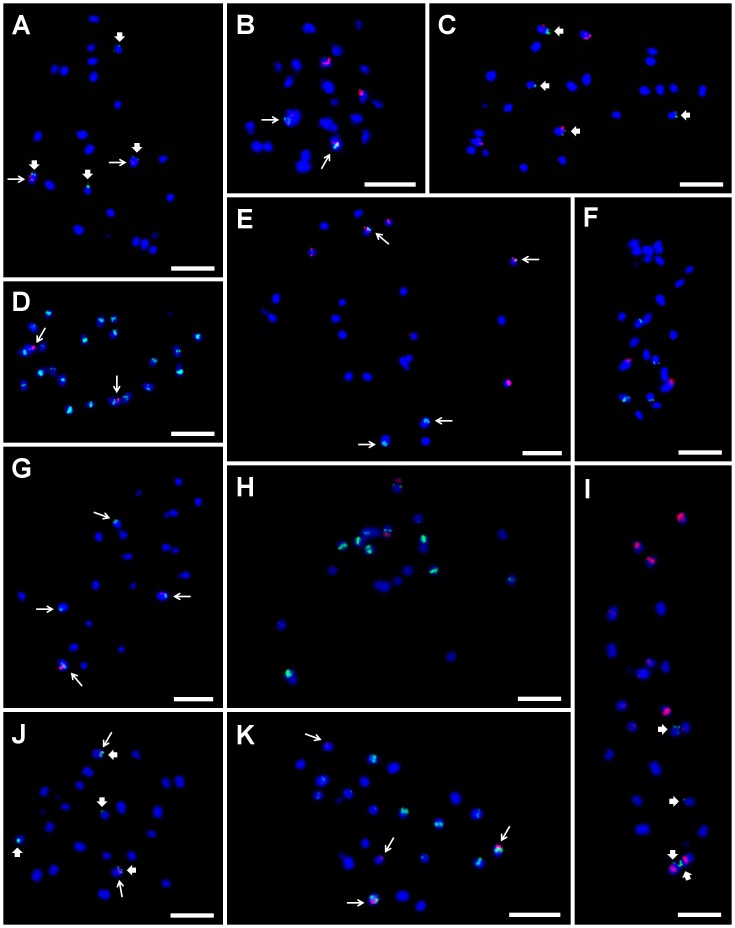
Figure 5. Examples of genomic distribution of DNA satellites as determined on mitotic metaphase chromosomes of diploid Musa accessions after FISH.
Chromosomes were counterstained with DAPI (blue). Sites of CL18 and CL33 probe hybridization are marked by long and thick arrows, respectively. (A) CL18 (red) and CL33 (green) on chromosomes of ‘Long Tavoy’. (B) 5S rDNA (red) and CL18 (green) on chromosomes of ‘Maia Oa’. (C) 5S rDNA (red) and CL33 (green) on chromosomes of ‘Long Tavoy’. (D) CL18 (red) and LINE element (green) on chromosomes of ‘Tuu Gia’. (E) 5S rDNA (red) and CL18 (green) on chromosomes of ‘Pisang Klutuk Wulung’. (F) BAC clone 2G17 (red) and 5S rDNA (green) on chromosomes of ‘Cameroun’. (G) BAC clone 2G17 (red) and CL18 (green) on chromosomes of ‘Tani’. (H) 45S rDNA (red) and 5S rDNA (green) on chromosomes of M. schizocarpa ITC 0560. (I) 5S rDNA (red) and CL33 (green) on chromosomes of M. schizocarpa ITC 1002. (J) CL18 (red) and CL33 (green) on chromosomes of M. schizocarpa ITC 1002. (K) CL18 (red) and 5S rDNA (green) on chromosomes of M. schizocarpa ITC 1002. Bar = 5 µm.
BAC clone 2G17
BAC clone 2G17, which was used in this work as additional cytogenetic marker, localized to subtelomeric regions on one chromosome pair in all diploid accessions with the exception of M. balbisiana ‘Honduras’ in which four signals were observed. Only one chromosome gave FISH signal in a diploid hybrid clone ‘Umbubu’ (AT genome), which most probably originated from a cross between M. acuminata and M. textilis (T genome). In triploid AAB and ABB clones, three or four loci containing DNA sequences homologous to BAC 2G17 were found (Figure 5, Figures S3D, E, Figure S4, Table 3).
Satellite repeats in diploid genomes
Satellite CL18 localized to subtelomeric regions on one pair of mitotic chromosomes in all accessions of M. acuminata and always co-localized with BAC clone 2G17. Satellite CL33 localized to subtelomeric regions on one chromosome pair in ‘Maia Oa’ and on two chromosome pairs of the remaining M. acuminata accessions. Further FISH experiments revealed that CL33 satellite co-localized with CL18 and BAC 2G17 on one chromosome pair (Figures 4, 5, Figure S3A, Table 3) in all M. acuminata accessions. Similar FISH patterns were observed for CL18 and CL33 in M. schizocarpa (Figures 4, 5, Figure S3C, Table 3).
Three M. balbisiana genotypes (‘Honduras’, ‘Tani’, ‘Pisang Klutuk Wulung’) contained two chromosome pairs bearing satellite CL18 in their subtelomeric regions. On one of the chromosome pairs, a probe for CL18 co-localized with BAC 2G17 and on the other co-localized with 5S rDNA (Figures 4, 5, and Figure S3B). The fourth accession of M. balbisiana - ‘Cameroun’ - had only three CL18 loci as a FISH signal was missing on the chromosome with 5S rDNA locus (Figure 5, and Figure S3B). CL33 gave no visible signals on metaphase chromosomes of M. balbisiana.
Satellite repeats in AAB and ABB hybrids
Out of a number of the existing interspecific hybrids with a combination of A and B genomes, three hybrid accessions with AAB genomic constitution (‘Obino l’Ewai’, ‘Maritú’ and ‘3 Hands Planty’) and two ABB hybrids (‘Pelipita’ and ‘Balonkawe’) were chosen for this study. All AAB interspecific hybrids and one ABB hybrid (‘Balonkawe’) gave signals of BAC 2G17 in subtelomeric regions of four different chromosomes. Among them, one chromosome was bearing an additional weak signal of BAC 2G17. In the second ABB hybrid ‘Pelipita’, BAC 2G17 localized in subtelomeric regions of three different metaphase chromosomes (Figures S3E, S4). 5S rDNA loci were found on eight metaphase chromosomes in the genome of ‘Maritú’, on nine chromosomes of ‘Obino l’Ewai’ and ‘3 Hands Planty’, while ‘Pelipita’ and ‘Balonkawe’ contained ten clusters corresponding to 5S rDNA.
Satellite CL18 localized in subtelomeric regions on four mitotic chromosomes in AAB hybrids; FISH with satellite CL33 resulted in three visible signals in subtelomeric regions of metaphase chromosomes of ‘3 Hands Planty’ and four signals were detected on chromosomes of ‘Obino l’Ewai’ and ‘Maritú’. Further analysis showed that two chromosomes carried both CL18 and CL33 satellite repeats. (Figures S3D, S4, Table 3). Hybrid ABB clones ‘Balonkawe’ and ‘Pelipita’ contained six signals of satellite CL18 which were detected on different chromosomes and one signal of satellite CL33. Multicolor FISH with the combination of probes corresponding to satellite repeats and 5S rRNA genes revealed one chromosome carrying CL18, CL33, 5S rDNA and BAC 2G17. Two other chromosomes exhibiting combination of FISH signals for 2G17 and CL18 satellite repeat and three additional chromosomes bearing signals for CL18 satellite and 5S rRNA genes (Figure S3E, Table 3 ).
Satellite repeats in AS and AT hybrids
FISH was also used to study genomic distribution of the two satellites in diploid hybrid clones ‘Ato’ and ‘Tonton Kepa’ (AS genome) and ‘Umbubu’ (AT genome). Nuclear genome of AS hybrids contained two signals of CL18 and CL33 which co-localized in subtelomeric regions on two different chromosomes, while nuclear genome of AT hybrid clone ‘Umbubu’ contained only one signal of CL18 satellite and two signals of CL33 satellite (Figure S3F, G, Table 3 ).
Chromosome Identification
Multicolor FISH experiments with combinations of probes for 45S rDNA, 5S rDNA, banana-specific LINE-like element, DNA satellites CL18 and CL33 and a single copy BAC clone 2G17 facilitated identification of various numbers of chromosomes in diploid accessions of Musa (Figures 4, S3A–G, Table 3). Within the A genome representatives, three chromosomes could be distinguished in M. acuminata ‘Maia Oa’ and four chromosomes were identified in M. acuminata clones ‘Long Tavoy’ and ‘Tuu Gia’. In M. acuminata ‘Calcutta 4’, four chromosomes exhibited a specific hybridization pattern. Although two other chromosomes carried 5S rDNA sequences in this clone, the two chromosomes could not be discriminated from each other as the probes hybridized to similar chromosome regions. Five different chromosomes could be identified in M. acuminata ‘DH Pahang’.
Within the representatives of M. balbisiana, four chromosomes could be discriminated in ‘Cameroun’, five chromosomes were distinguished in ‘Tani’ and ‘Pisang Klutuk Wulug’ and six different chromosomes were identified in ‘Honduras’. Three different chromosomes could be recognized in the two S genome representatives. Moreover, two chromosomes carrying strong signals of 5S rDNA and other two chromosomes carrying weak signals of 5S rDNA were identified in their karyotypes. Unfortunately, the probes localized to similar chromosome regions in both cases and the chromosomes could not be differentiated from each other. Due to the lack of probes specific for A, B, S and T genomes, we were not able to identify homologous chromosomes in inter-specific hybrids.
Discussion
Satellite repeats may undergo rapid changes in copy number and nucleotide sequence, which may result in the evolution of genus and species-specific repeats [18], [19], [20], [44], [45]. At the same time, their repetitive units are usually homogenized within a species due to concerted evolution [6], [46], [47], [48]. These features make them an attractive object for evolutionary and cytogenetic studies. Although about 45% of the Musa genome consists of various repetitive DNA sequences [35], satellite DNA repeats represent only ∼0.3% of the genome [13]. The presence of only two major satellite repeats in Musa and their low copy numbers may be related to relatively small genome and is on line with observations in some other small plant genomes [49], [50], [51].
Satellite DNA sequences are typical components of subtelomeric and centromeric chromosome regions, but may also form clusters in interstitial regions [14], [18], [52], [53], [54], [55], [56]. Our earlier studies [13], [31], [57] indicated that Musa does not have a typical centromeric satellite and that Musa centromeres are rather made of various types of retrotransposons, especially Ty3/Gypsy-like elements and LINE-like elements. Ty3/Gypsy-like elements were found in high copy numbers in centromeric regions in other plant species [58], [59], [60], [61], [62]. On the other hand, LINE elements are usually dispersed along chromosomes including their pericentromeric regions [63], [64], [65]. The present study confirmed preferential localization of the LINE element to centromeric regions in Musa and FISH with a probe for the LINE element appears a convenient way to label primary constrictions, which are not always easily visible on small and condensed mitotic metaphase chromosomes of Musa.
Our analyses revealed that nuclear genomes of all Musa accessions included in the present study contained the major DNA satellites CL33 and CL18. With a few exceptions, the satellites were found highly conserved (Figures 1A, B, 2A). Judging from the cluster analysis based on Neighbor Joining method, there are no A or B genome-specific satellite units in Musa (Figure 2B). These findings indicate that both satellites originated before the divergence of the section Eumusa (∼28 Mya) [66] and that their sequences remained conserved during speciation. Unfortunately, our observation means that the satellites are not suitable as FISH probes to discriminate A and B genome chromosomes and hence cannot help in characterizing genomic constitution in interspecific Musa hybrids.
One should bear in mind that our results on the length, sequence similarity to maTR_CL18 and maTR_CL33 satellites [13] as well as the nucleotide diversity may be affected by the fact that only sequences which were obtained after PCR amplification analyzed. Our results could be supported and refined after whole genome sequencing, e.g., using next generation sequencing, and reconstruction of complete repetitive elements [13].
We were not able to detect CL33 satellite by FISH on mitotic chromosomes of M. balbisiana, most probably because the copy number fell below the detection limit of the method. Supported by the results of Southern hybridization, these results indicate a low copy number of this satellite in the Musa B genome. This corresponds to lower copy number of other DNA repeats in M. balbisiana [31] and may be related to smaller genome size as compared to the A genome [26], [67].
Our findings extend significantly previous results of cytogenetic mapping of 45S and 5S rRNA genes and a single copy BAC clone 2G17 in Musa [25], [28], [29], [30], [31]. The 45S rDNA cluster localized to NORs in all accessions and the number of loci corresponded to their ploidy. The only exception was the additional locus on one satellite chromosome in diploid hybrid clone ‘Ato’ (AS genome).
5S rRNA genes are more diverse in the number of loci as well as in the genomic location. The largest variation in the number of 5S rDNA sites was observed among the accessions of M. acuminata where the number of loci ranged from 4 to 8 per mitotic metaphase plate. It is tempting to suggest different number of 5S loci in different subspecies of M. acuminata. Unfortunately, our results on a limited set of accessions do not support this idea. For example, ‘Maia Oa’ (M. acumina ssp. zebrina) and ‘Long Tavoy’ (M. acuminata ssp. burmanica) have the same number of 5S loci. On the other hand, cytogenetic observations may question the taxonomic classification of some accessions. Further work is needed to verify both options. The numbers of 5S rDNA loci in diploid accessions are on line with previous studies [25], [28], [29] except of M. balbisiana ‘Honduras’ where Bartoš et al [25] identified only four 5S rDNA loci. As the same genotype was analyzed in both studies (ITC 0247), the difference could be due to higher sensitivity of FISH in the present work. The highest number of 5S rDNA loci was found in M. schizocarpa. Twelve signals of 5S rDNA were detected in both representatives of this species, while Bartoš et al. [25] detected only six 5S rDNA sites in M. schizocarpa (ITC 0890). The discrepancy may be due to higher sensitivity of FISH. But as a different genotype was analyzed by Bartoš et al. [25] it is also possible that similarly to M. acuminata, there is a variation in the number of 5S loci in M. schizocarpa.
A BAC clone 2G17, which was originally identified as a single copy clone on mitotic chromosomes of M. acuminata ‘Calcutta 4’ [30] was localized on one chromosome pair in all diploid accessions with three exceptions. Four signals were observed on mitotic chromosomes of M. balbisiana ‘Honduras’. This could be a consequence of the locus duplication in this genotype. On the other hand, only three signals of CL18 were found on mitotic chromosomes of M. balbisiana ‘Cameroun’ which could be explained by reduction or even loss of one locus in this clone. Only one signal of CL18 was detected in metaphase plate of hybrid clone ‘Umbubu’ (AT genome) suggesting that the locus diverged between the genomes of Eumusa and Australimusa species. An alternative explanation is a reduction or loss of this region during the formation and evolution of the hybrid genome.
Due to the lack of probes specific for individual chromosomes, we were not able to identify all chromosomes within the karyotypes of eleven diploid accessions and eight inter-specific hybrids of Musa. Nevertheless, we have succeeded in expanding significantly the number of individual chromosomes which can be identified based on specific chromosome FISH patterns (Figure S3, Table 3). All chromosomes which could be identified unambiguously occurred in pairs of homologs in all accessions of M. acuminata, M. balbisiana and M. schizocarpa, indicating structural homozygosity. The only exception was found in M. balbisiana ‘Cameroun’ where only three loci of CL18 were found. However, our observation cannot exclude small chromosome exchanges and concerns only a subset of chromosomes. Structural heterozygosity influences chromosome behavior in meiosis, formation gametes, their genotype, distortions from expected Mendelian segregation and may cause sterility in hybrids [68]. Differences in distribution of cytogenetic landmarks within and between the diploid Musa species may indicate differences in chromosome structure potentially leading to aberrant meiosis and sterility in intra- and inter-specific hybrids [69], [70], [71].
The differences in FISH patterns between some chromosomes of M. acuminata and M. balbisiana provided information on the presence of some of their chromosomes in inter-specific hybrids and thus may contribute to the reconstruction of their genomic constitution. For example, triploid interspecific hybrid clones ‘Obino l’Ewai’, ‘Maritú’, and ‘3 Hands Planty’ had the expected number of CL18 loci, which is on line with the reported AAB genome constitution (Table 3). Triploid ABB clones ‘Pelipita’ and ‘Balonkawe’ were characterized by weak signals of CL33 after Southern hybridization. The presence of only one chromosome on which CL33 was detected by FISH seems to support the ABB genomic constitution with only one A genome (or its part) present.
Our results indicate that NOR-bearing satellite chromosomes are maintained in all hybrid clones we have analyzed. On the other hand, variation in the number of 5S rDNA loci was observed among the triploid hybrids (Table 3). Specific numbers of 5S rDNA loci in A and B genotypes could aid in identification of the origin of hybrids. However, this would be only possible if genomic organization of 5S loci is known in a larger set of M. acuminata and M. balbisiana accessions covering the existing diversity.
FISH analysis on interspecific hybrids revealed deviations in the number of loci of BAC clone 2G17 and satellite DNA sequences from the expected numbers based on reported genomic constitution. Instead of the expected three hybridization sites of BAC 2G17, four loci were identified in three out of four triploid hybrids. Based on our observations in diploids, there should be a maximum of five CL18 loci in a triploid hybrid. However, both ABB hybrids had six loci (Table 3). As we analyzed a limited set of diploid genotypes, we cannot exclude that other diploid genotypes have different number of CL18 loci and that the hybrids originated from them. The discrepancies between the expected and observed number of loci could be also due to backcrossing of primary hybrids to one of the parental species and a loss or gain of some chromosome types. Thus, our results may provide a support to the backcross hypothesis of De Langhe et al. [72]. Clearly, further analysis is needed to clarify the issue as tandem organized sequences can increase or decrease their copy number during the interspecific hybridization and polyploidization or can be removed completely from a hybrid genome [19], [73], [74]. Consequently they should be used with caution when determining genomic constitution in interspecific Musa hybrids.
Supporting Information
(A) Dot-plot comparison of maTR_CL18 which was identified in 454 sequence data of M. acuminata ‘Calcutta 4’ [13] and CL18-like repeat which was identified in whole genome sequence of M. acuminata ‘DH Pahang’ [35] . Both repetitive units are more than 2.2 kb long and are organized in tandem arrays. (B) Dot-plot comparison of maTR_CL33 which was identified in 454 data of M. acuminata ‘Calcutta 4’ [13] and CL33-like repeat which was identified in whole genome sequence of M. acuminata ‘DH Pahang’ [35]. Both repetitive units are 134 bp long and are organized in tandem arrays. Sequence similarities are represented by dots and diagonal lines (A, B).
(TIFF)
Examples of genomic distribution of satellite DNA as determined on mitotic metaphase chromosomes of Musa after FISH with labeled probes for CL18 (red, labeled by arrows) and banana LINE element (green). Chromosomes were counterstained with DAPI (blue). (A) ‘Maia Oa’. (B) ‘Long Tavoy’. (C) ‘Tani’. (D) M. schizocarpa ITC 0560. (E) ‘Pelipita’. (F) M. schizocarpa ITC 1002. (G) ‘Obino l’Ewai’. (H) ‘Maritú’. (I) ‘Ato’. (J) ‘Tonton Kepa’. (K) ‘Umbubu’. Bar = 5 µm.
(TIFF)
Idiograms of diploid (A – C) and hybrid (D – G) Musa accessions. (A) M. acuminata, (B) M. balbisiana, (C) M. schizocarpa, (D) Hybrids with AAB genomics constitution, (E) Hybrids with ABB genomic constitution, (F) Hybrids with AS genomic constitution, (G) Hybrids with AT genomic constitution. No attempt was made to identify homologs in the hybrids (D – G) and all chromosomes are shown. The chromosome sizes are only indicative.
(PDF)
Examples of genomic distribution of satellite DNA as determined on mitotic metaphase chromosomes of interspecific hybrids after FISH. Chromosomes were counterstained with DAPI (blue). Sites of CL18 and CL33 probe hybridization are marked by long and thick arrows, respectively. (A) CL18 (red) and CL33 (green) on chromosomes of ‘Maritú’. (B) CL18 (red) and 5S rDNA (green) on chromosomes of ‘Maritú’. (C) 5S rDNA (red) and CL33 (green) on chromosomes of ‘Maritú’. (D) CL18 (red) and BAC clone 2G17 (green) on chromosomes of ‘Pelipita’. (E) CL18 (red) and 5S rDNA on chromosomes of ‘Pelipita’. (F) CL18 (red) and CL33 (green) on chromosomes of ‘Ato’. (G) CL18 (red) and 5S rDNA (green) on chromosomes of ‘Tonton Kepa’. (H) CL18 (red) and CL33 (green) on chromosomes of ‘Umbubu’. (I) 5S rDNA (red) and CL33 (green) on chromosomes of ‘Ato’. (J) CL18 (red) and CL33 (green) on chromosomes of ‘Tonton Kepa’. (K) 5S rDNA (red) and CL18 (green) on chromosomes of ‘Umbubu’. Bar = 5 µm.
(TIFF)
Basic characteristics and nucleotide diversity of part1 of CL18-like repeats.
(DOC)
Basic characteristics and nucleotide diversity of part2 of CL18-like repeats.
(DOC)
Basic characteristics and nucleotide diversity of CL33-like repeats.
(DOC)
Copy numbers of maTR_CL18 and maTR_CL33 satellites.
(DOC)
Acknowledgments
We thank Ir. Ines van den Houwe for providing the plant material and Ms. Radka Tušková and Ms. Eva Jahnová for excellent technical assistance.
Funding Statement
This work was supported by the Academy of Sciences of the Czech Republic (award KJB500380901), by the Bioversity International (LoA CfL 2011/51), Ministry of Education, Youth and Sports of the Czech Republic (award LG12021) and the European Regional Development Fund (Operational Programme Research and Development for Innovations No. ED0007/01/01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bennetzen JL, Kellog EA (1997) Do plants have a one-way ticket to genomic obesity? Plant Cell 9: 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ingham LD, Hanna WW, Baier JW, Hannah LC (1993) Origin of the main class of repetitive DNA within selected Pennisetum species. Mol Gen Genet 238: 350–356. [DOI] [PubMed] [Google Scholar]
- 3. Shapiro JA, von Sternberg R (2005) Why repetitive DNA is essential to genome function. Biol Rev 80: 227–250. [DOI] [PubMed] [Google Scholar]
- 4. Kubis S, Schmidt T, Heslop-Harrison JS (1998) Repetitive DNA elements as a major component of plant genomes. Ann Bot 82: 45–55. [Google Scholar]
- 5. Charlesworth B, Sniegowski PE, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 37: 215–220. [DOI] [PubMed] [Google Scholar]
- 6. Elder JF Jr, Turner BJ (1995) Concerted evolution of repetitive DNA sequences in eukaryotes. Q Rev Biol 70: 297–320. [DOI] [PubMed] [Google Scholar]
- 7. Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, et al. (2002) Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14: 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellegren H (2004) Microsatellites: simple sequences with complex evolution. Nat Rev Genet 5: 435–445. [DOI] [PubMed] [Google Scholar]
- 9. Tek AL, Jiang J (2004) The centromere regions of potato chromosomes contain megabase-sized tandem arrays of telomere-similar sequence. Chromosoma 113: 77–83. [DOI] [PubMed] [Google Scholar]
- 10. Macas J, Požárková D, Navrátilová A, Nouzová M, Neumann P (2000) Two new families of tandem repeats isolated from genus Vicia using genomic self-priming PCR. Mol Gen Genet 263: 741–751. [DOI] [PubMed] [Google Scholar]
- 11. Willard HF (1991) Evolution of alpha satellite. Curr Opin Genet Dev 1: 509–514. [DOI] [PubMed] [Google Scholar]
- 13. Hřibová E, Neumann P, Matsumoto T, Roux N, Macas J, et al. (2010) Repetitive part of the banana (Musa acuminata) genome investigated by low-depth 454 sequencing. BMC Plant Biol 10: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Macas J, Neumann P, Navrátilová A (2007) Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula . BMC Genomics 8: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Macas J, Kejnovský E, Neumann P, Novák P, Koblížková A, et al. (2011) Next generation sequencing-based analysis of repetitive DNA in the model dioecious plant Silene latifolia . PLoS One 6: e27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidt T, Heslop-Harrison JS (1998) Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends Plant Sci 3: 195–199. [Google Scholar]
- 17. Zatloukalová P, Hřibová E, Kubaláková M, Suchánková P, Šimková H, et al. (2011) Integration of genetic and physical maps of chickpea (Cicer arietinum L.) genome using flow-sorted chromosomes. Chromosome Res 19: 729–739. [DOI] [PubMed] [Google Scholar]
- 18. Han YH, Zhang ZH, Liu JH, Lu JY, Huang SW, et al. (2008) Distribution of the tandem repeat sequences and karyotyping in cucumber (Cucumis sativus L.) by fluorescence in situ hybridization. Cytogenet Genome Res 122: 80–88. [DOI] [PubMed] [Google Scholar]
- 19. Macas J, Navrátilová A, Koblížková A (2006) Sequence homogenization and chromosomal localization of VicTR-B satellites differ between closely related Vicia species. Chromosoma 115: 437–447. [DOI] [PubMed] [Google Scholar]
- 21. Sharma S, Raina SN (2005) Organization and evolution of highly repeated satellite DNA in plant chromosomes. Cytogenet Genome Res. 109: 15–26. [DOI] [PubMed] [Google Scholar]
- 22. Kopecký D, Havránková M, Loureiro J, Castro S, Lukaszewski AJ, et al. (2010) Physical distribution of homoeologous recombination in individual chromosomes of Festuca pratensis in Lolium multiflorum. Cytogenet Genome Res. 120: 370–383. [DOI] [PubMed] [Google Scholar]
- 23. Kopecký D, Lukaszewski AJ, Doležel J (2008) Cytogenetics of Festulolium (Festuca×Lolium hybrids). Cytogenet Genome Res. 120: 370–383. [DOI] [PubMed] [Google Scholar]
- 24. Simmonds NW, Shepherd K (1955) The taxonomy and origins of the cultivated bananas. J Linn Soc Bot 55: 302–312. [Google Scholar]
- 25. Bartoš J, Alkhimova O, Doleželová M, De Langhe E, Doležel J (2005) Nuclear genome size and genomic distribution of ribosomal DNA in Musa and Ensete (Musaceae): taxonomic implications. Cytogenet Genome Res 109: 50–57. [DOI] [PubMed] [Google Scholar]
- 26. Doležel J, Doleželová M, Novák FJ (1994) Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and Musa balbisiana). Biol Plant 36: 351–357. [Google Scholar]
- 27. D’Hont A, Paget-Goy A, Escoute J, Carreel F (2000) The interspecific genome structure of cultivated banana, Musa spp. revealed by genomic DNA in situ hybridization. Theor Appl Genet 100: 177–183. [Google Scholar]
- 28. Osuji JO, Harrison G, Crouch J, Heslop-Harrison JS (1997) Identification of the genomic constitution of Musa L. lines (bananas, plantains and hybrids) using molecular cytogenetics. Ann Bot 80: 787–793. [Google Scholar]
- 29. Doleželová M, Valárik M, Swennen R, Horry JP, Doležel J (1998) Physical mapping of the 18S–25S and 5S ribosomal RNA genes in diploid bananas. Biol Plant 41: 497–505. [Google Scholar]
- 30. Hřibová E, Doleželová M, Doležel J (2008) Localization of BAC clones on mitotic chromosomes of Musa acuminata using fluorescence in situ hybridization. Biol Plant 52: 445–452. [Google Scholar]
- 31. Valárik M, Šimková H, Hřibová E, Šafář J, Doleželová M, et al. (2002) Isolation, characterization and chromosome localization of repetitive DNA sequences in bananas (Musa spp.). Chromosome Res 10: 89–100. [DOI] [PubMed] [Google Scholar]
- 32. Zhang HB, Zhao X, Ding X, Paterson AH, Wing RA (1995) Preparation of megabase-size DNA from plant nuclei. Plant J 7: 175–184. [Google Scholar]
- 33.Otto F (1990) DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Crissman HA, Darzynkiewicz Z, editors. Methods in Cell Biology. Academic Press, New York, Vol. 33. Pp. 105–110. [DOI] [PubMed] [Google Scholar]
- 34. Doležel J, Bartoš J, Voglmayr H, Greilhuber J (2003) Nuclear DNA content and genome size of trout and human. Cytometry A 51: 127–128. [DOI] [PubMed] [Google Scholar]
- 35. D’Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, et al. (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488: 213–217. [DOI] [PubMed] [Google Scholar]
- 36. Sonnhammer ELL, Durbin R (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: GC1–GC10. [DOI] [PubMed] [Google Scholar]
- 37. Staden R (1996) The Staden sequence analysis package. Mol Biotechnol 5: 233–241. [DOI] [PubMed] [Google Scholar]
- 38. Huan X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Galtier N, Gouy M, Gautier C (1996) SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12: 543–548. [DOI] [PubMed] [Google Scholar]
- 41. Eckert AJ, Liechty JD, Tearse BR, Pande B, Neale DB (2010) DnaSAM: Software to perform neutrality testing for large datasets with complex null models. Mol Ecol Resour 10: 542–545. [DOI] [PubMed] [Google Scholar]
- 42. Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23: 254–267. [DOI] [PubMed] [Google Scholar]
- 44. Dechyeva D, Gindullis F, Schmidt T (2003) Divergence of satellite DNA and interspersion of dispersed repeats in the genome of the wild beet Beta procumbens . Chromosome Res 11: 3–21. [DOI] [PubMed] [Google Scholar]
- 45. King K, Jobst J, Hemleben V (1995) Differential homogenization and amplification of two satellite DNAs in the genus Cucurbita (Cucurbitaceae). J Mol Evol 41: 996–1005. [DOI] [PubMed] [Google Scholar]
- 46. Dover GA, Strachan T, Coen ES (1982) Molecular drive. Science 218: 1069. [DOI] [PubMed] [Google Scholar]
- 47. Baldwin BG, Sanderson MJ, Porter MJ, Wojciechowski MF, Campbell CS, et al. (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann Mo Bot Gard 82: 247–277. [Google Scholar]
- 48. Dover G (2002) Molecular drive. Trends Genet 18: 587–589. [DOI] [PubMed] [Google Scholar]
- 49. Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- 50. International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- 51. International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- 52. Galasso I, Schmidt T, Pignone D, Heslop-Harrison JS (1995) The molecular cytogenetics of Vigna unguiculata (L.) Walp: the physical organization and characterization of 18S-5.8S-25S rRNA genes, 5S rRNA genes, telomere-like sequences, and a family of centromeric repetitive DNA sequences. Theor Appl Genet 91: 928–935. [DOI] [PubMed] [Google Scholar]
- 53. Houben A, Schubert I (2003) DNA and proteins of plant centromeres. Curr Opin Plant Biol 6: 554–560. [DOI] [PubMed] [Google Scholar]
- 54. Jiang J, Birchler JA, Parrott WA, Dawe RK (2003) A molecular view of plant centromeres. Trends Plant Sci 8: 570–574. [DOI] [PubMed] [Google Scholar]
- 55. Macas J, Neumann P, Novák P, Jiang J (2010) Global sequence characterization of rice centromeric satellite based on oligomer frequency analysis in large-scale sequencing data. Bioinformatics 26: 2101–2108. [DOI] [PubMed] [Google Scholar]
- 56. Torres GA, Gong Z, Iovene M, Hirsch CD, Buell CR, et al. (2011) Organization and evolution of subtelomeric satellite repeats in the potato genome. G3: Genes, Genomes, Genetics 1: 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hřibová E, Doleželová M, Town CD, Macas J, Doležel J (2007) Isolation and characterization of the highly repeated fraction of the banana genome. Cytogenet Genome Res 119: 268–274. [DOI] [PubMed] [Google Scholar]
- 58. Bao W, Zhang W, Yang Q, Zhang Y, Han B, et al. (2006) Diversity of centromeric repeats in two closely related wild rice species, Oryza officinalis and Oryza rhizomatis . Mol Genet Genomics 275: 421–430. [DOI] [PubMed] [Google Scholar]
- 59. Gindullis F, Desel C, Galasso I, Schmidt T (2001) The large-scale organization of the centromeric region in Beta species. Genome Res 11: 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kumekawa N, Ohmido N, Fukui K, Ohtsubo E, Ohtsubo H (2001) A new gypsy-type retrotransposon, RIRE7: preferential insertion into the tandem repeat sequence TrsD in pericentromeric heterochromatin regions of rice chromosomes. Mol Genet Genomics 265: 480–488. [DOI] [PubMed] [Google Scholar]
- 61. Neumann P, Navrátilová A, Koblížková A, Kejnovský E, Hřibová E, et al. (2011) Plant centromeric retrotransposons: a structural and cytogenetic perspective. Mob DNA 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Presting GG, Malysheva L, Fuchs J, Schubert I (1998) A TY3/GYPSY retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J 16: 721–728. [DOI] [PubMed] [Google Scholar]
- 63. Kubis SE, Heslop-Harrison JS, Desel C, Schmidt T (1998) The genomic organization of non-LTR retrotransposons (LINEs) from three Beta species and five other angiosperms. Plant Mol Biol 36: 821–31. [DOI] [PubMed] [Google Scholar]
- 64. Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33: 479–532. [DOI] [PubMed] [Google Scholar]
- 65. Schmidt T, Kubis S, Heslop-Harrison JS (1995) Analysis and chromosomal localisation of retrotransposons in sugar beet (Beta vulgaris) LINES and Ty1-copia- like elements as major components of the genome. Chromosome Res 3: 335–45. [DOI] [PubMed] [Google Scholar]
- 66. Christelová P, Valárik M, Hřibová E, De Langhe E, Doležel J (2011) A multi gene sequence- based phylogeny of the Musaceae (banana) family. BMC Evol Biol 11: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lysák MA, Doleželová M, Horry JP, Swennen R, Doležel J (1999) Flow cytometric analysis of nuclear DNA content in Musa . Theor Appl Genet 98: 1344–1350. [Google Scholar]
- 68. Cai X, Xu SS (2007) Meiosis-driven genome variation in plants. Curr Genomics 8: 151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dodds KS, Simmonds NW (1948) Sterility and parthenocarpy in diploid hybrids of Musa . Heredity 2: 101–117. [DOI] [PubMed] [Google Scholar]
- 70.Shepherd K (1999) Cytogenetics of the genus Musa. IPGRI-INIBAP. Pp.160. [Google Scholar]
- 71. Wilson GB (1946) Cytological studies in the Musae. II. Meiosis in some diploid clones. Genetics 31: 475–482. [PubMed] [Google Scholar]
- 72. De Langhe E, Hřibová E, Carpentier S, Doležel J, Swennen R (2010) Did backcrossing contribute to the origin of hybrid edible bananas? Ann Bot 106: 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tek AL, Song JQ, Macas J, Jiang J (2005) Sobo, a recently amplified satellite repeat of potato, and its implications for the origin of tandemly repeated sequences. Genetics 170: 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Urgaković D, Plohl M (2002) Variation in satellite DNA profiles-causes and effects. EMBO J 21: 5955–5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Dot-plot comparison of maTR_CL18 which was identified in 454 sequence data of M. acuminata ‘Calcutta 4’ [13] and CL18-like repeat which was identified in whole genome sequence of M. acuminata ‘DH Pahang’ [35] . Both repetitive units are more than 2.2 kb long and are organized in tandem arrays. (B) Dot-plot comparison of maTR_CL33 which was identified in 454 data of M. acuminata ‘Calcutta 4’ [13] and CL33-like repeat which was identified in whole genome sequence of M. acuminata ‘DH Pahang’ [35]. Both repetitive units are 134 bp long and are organized in tandem arrays. Sequence similarities are represented by dots and diagonal lines (A, B).
(TIFF)
Examples of genomic distribution of satellite DNA as determined on mitotic metaphase chromosomes of Musa after FISH with labeled probes for CL18 (red, labeled by arrows) and banana LINE element (green). Chromosomes were counterstained with DAPI (blue). (A) ‘Maia Oa’. (B) ‘Long Tavoy’. (C) ‘Tani’. (D) M. schizocarpa ITC 0560. (E) ‘Pelipita’. (F) M. schizocarpa ITC 1002. (G) ‘Obino l’Ewai’. (H) ‘Maritú’. (I) ‘Ato’. (J) ‘Tonton Kepa’. (K) ‘Umbubu’. Bar = 5 µm.
(TIFF)
Idiograms of diploid (A – C) and hybrid (D – G) Musa accessions. (A) M. acuminata, (B) M. balbisiana, (C) M. schizocarpa, (D) Hybrids with AAB genomics constitution, (E) Hybrids with ABB genomic constitution, (F) Hybrids with AS genomic constitution, (G) Hybrids with AT genomic constitution. No attempt was made to identify homologs in the hybrids (D – G) and all chromosomes are shown. The chromosome sizes are only indicative.
(PDF)
Examples of genomic distribution of satellite DNA as determined on mitotic metaphase chromosomes of interspecific hybrids after FISH. Chromosomes were counterstained with DAPI (blue). Sites of CL18 and CL33 probe hybridization are marked by long and thick arrows, respectively. (A) CL18 (red) and CL33 (green) on chromosomes of ‘Maritú’. (B) CL18 (red) and 5S rDNA (green) on chromosomes of ‘Maritú’. (C) 5S rDNA (red) and CL33 (green) on chromosomes of ‘Maritú’. (D) CL18 (red) and BAC clone 2G17 (green) on chromosomes of ‘Pelipita’. (E) CL18 (red) and 5S rDNA on chromosomes of ‘Pelipita’. (F) CL18 (red) and CL33 (green) on chromosomes of ‘Ato’. (G) CL18 (red) and 5S rDNA (green) on chromosomes of ‘Tonton Kepa’. (H) CL18 (red) and CL33 (green) on chromosomes of ‘Umbubu’. (I) 5S rDNA (red) and CL33 (green) on chromosomes of ‘Ato’. (J) CL18 (red) and CL33 (green) on chromosomes of ‘Tonton Kepa’. (K) 5S rDNA (red) and CL18 (green) on chromosomes of ‘Umbubu’. Bar = 5 µm.
(TIFF)
Basic characteristics and nucleotide diversity of part1 of CL18-like repeats.
(DOC)
Basic characteristics and nucleotide diversity of part2 of CL18-like repeats.
(DOC)
Basic characteristics and nucleotide diversity of CL33-like repeats.
(DOC)
Copy numbers of maTR_CL18 and maTR_CL33 satellites.
(DOC)