Abstract
Background
The minor allele of SNP rs3803662 has been shown to correlate with increased breast cancer risk and with lower expression of TOX3. The SNP is closely located to TOX3 residing within an uncharacterised gene LOC643714. The aim of the study was to examine the association of the risk allele with expression of TOX3 and LOC643714, and of mRNA levels and genotype with clinical and pathological characteristics.
Methods
The SNP was genotyped in DNA isolated from blood and normal tissue from 160 breast cancer patients and mRNA levels were measured by microarrays and quantitative real-time (qRT)-PCR in breast tumours. Association with clinical and pathological characteristics was analysed by parametric tests.
Results
An association of the risk allele of rs3803662 with lower TOX3 expression was confirmed in oestrogen receptor (ER) positive tumours. It was more often observed in lobular tumours (p = 0.04), and carriers of the risk allele who had been diagnosed with luminal A tumours had shorter overall survival (OS) than carriers of the non-risk allele (p = 0.01). Positive correlation between the mRNA levels of TOX3 and LOC643714 was observed (r = 0.44 and p < 0.001). Association analysis with tumour pathology showed that low TOX3 and LOC643714 expression correlated with high Ki67 levels (p = 0.026 and p = 0.002) and the basal subtype (p < 0.001 and p < 0.001), whereas high expression correlated with ER (p = 0.004 and p < 0.001) and progesterone receptor (PgR) (p = 0.005 and p < 0.001) expression. Furthermore, high TOX3 and LOC643714 correlated with positive lymph nodes (p < 0.001 and p = 0.01). Patients with ER positive tumours and high levels of TOX3 mRNA had shorter overall- and distant metastasis free-survival (p = 0.017 and p = 0.021), an effect mostly attributable to patients with luminal B tumours.
Conclusions
The results suggest that the effect of the risk allele of rs3803662 is strongest in luminal A tumours and that the expression levels of TOX3 and/or LOC643714 affect the progression of breast cancer. The effect may vary depending on the subtype and developmental stage of the tumour.
Keywords: TOX3, LOC643714, rs380662, Risk allele, Breast cancer, Clinical, Pathological, Survival, Oestrogen receptor
Background
Breast cancer is the most frequent cancer in women accounting for 23% of all cancers in 2008 [1]. It shows strong familial aggregation being about twice as common in first-degree relatives of individuals with the disease than in the general population [2]. Familial linkage studies have identified high penetrance, low frequency mutations in genes such as the breast cancer susceptibility genes BRCA1 and BRCA2 but these mutations only account for approximately 20% of the familial risk (reviewed in [3]). In unselected breast cancer patients, genome-wide association studies have identified low penetrance, high frequency SNPs that are associated with breast cancer risk (reviewed in [4]). SNP rs3803662, at 16q12, is one of them. It was demonstrated that the minor allele of rs3803662 conferred increased risk of breast cancer in European women [5,6], a finding that has been validated in large studies of unselected patients and patients with a familial history of breast cancer and meta-analyses [7-13]. This finding was also observed in women of East Asian descent [14-16] but not in women of African American descent [17-19]. The association was confined to oestrogen receptor positive cancer [6,20] but studies in large consortia have shown associations in ER positive and negative breast cancer, albeit stronger in ER positive disease [21,22]. The minor rs3803662 allele also increased the risk of breast cancer in BRCA1 and BRCA2 carriers [21,23,24] as well as in a population-based study of men [25].
The genes located closest to rs3803662 are TOX3 and LOC643714. The SNP is in the last exon (exon 4) of LOC643714 and near the 5’ end of TOX3. The chromosomal region spanning the 5’ end of TOX3, the intergenic region between TOX3 and LOC643714 and the entire coding part of LOC643714 are located in a 133 kb linkage disequilibrium (LD) block [13]. Other SNPs within this LD block show association with breast cancer but rs3803662 shows the strongest effect [5,13]. LOC643714 is an uncharacterised gene whose function is unknown [26]. The TOX3 gene, also known as TNRC9 and CAGF9, has a tri-nucleotide repeat motive [27] and a high mobility group (HMG)-box [28]. It is involved in the regulation of calcium dependent transcription and interacts with cAMP-response-element-binding protein CREB and CBP (CREB-binding protein) [29]. Additionally, TOX3 can interact with CITED1 and increase transcription [29,30]. CITED1 is a transcription co-regulator that enhances the activity of transcription factors such as the ER [31] and SMAD4 [32].
A correlation of the rs3803662 genotype with lower TOX3 mRNA has been observed in breast tumours where TOX3 mRNA expression decreased in an allele-dependent manner [33]. So far, one study showed an effect of TOX3 expression on breast cancer in that increased level of TOX3 mRNA predicted breast cancer metastasis to bone [34]. In the present study we report the results of association analysis between the rs3803662 genotype and expression of its closest genes, TOX3 and LOC643714, in a cohort of breast cancer patients as well as the correlation analyses between rs3803662, and expression of TOX3 and LOC643714, with known clinical and pathological characteristics.
Methods
Tumour and normal tissue samples
Freshly frozen primary breast tumour samples (n = 160) were obtained from the Department of Pathology at Landspitali University Hospital, Reykjavik, Iceland as well as normal tissue samples embedded in paraffin if a blood sample was not available from the patient. The patients were diagnosed during the years 1987–2003. Informed consent was obtained from all participants in the study. The study was approved by the Icelandic Data Protection Authority (2001/523 and 2002/463) as well as the National Bioethics Committee of Iceland (99/051 and 99/051_FS1) and a written consent was obtained from the participants in the study according to national guidelines.
DNA isolation and SNP genotyping
DNA isolation from blood samples was performed according to Miller et al. [35], and from paraffin-embedded normal tissue using a xylene treatment followed by proteinase K digestion and phenol/chloroform/isoamyl alcohol purification. Taqman assay C_25968567_10 (Applied Biosystems) was used to genotype SNP rs3803662 using 20 ng of genomic DNA isolated from the blood samples and the normal tissue. The reaction was carried out in StepOne Real-Time PCR System (Applied Biosystems). The allele frequencies of rs3803662 were in Hardy-Weinberg Equilibrium (p > 0.1).
RNA isolation and quantification
The microarray gene expression data for TOX3 were retrieved from a study including 359 breast tumours [36] via GEO (GSE22133 [37]). qRT-PCR was used to confirm the array data mRNA levels of TOX3 and LOC643714 in total RNA that was isolated from the identical fresh frozen breast tumours which were used in the first study. RNA was isolated using Trizol (Invitrogen) followed by RNeasy Midi Purification Kit (Qiagen) according to the manufacturer protocol. RNA (2 μg) was reverse transcribed with random hexamer primers using the RevertAid Minus First strand cDNA synthesis Kit (Fermentas). Quantification of gene expression was performed with Taqman Gene Expression Assays (Applied Biosystems) using 1 and 200 ng cDNA as template for TOX3 (Hs01101330_m1) and LOC643714 (Hs01000553_m1), respectively. Forty rounds of amplification were performed for TOX3 and 50 rounds for LOC643714. The gene expression was calculated relative to the expression of the reference gene TATA-binding protein (TBP, 4326322E Applied Biosystems) according to the formula: mRNA expression of target gene = 2-(mean Ct target – mean Ct reference). Expression was measured on available RNA samples; 139 and 126 RNA samples for TOX3 and LOC643714, respectively. The level of mRNA expression of LOC643714 was much lower in all samples tested and in 30 of the 126 samples LOC643714 mRNA was below the range detectable by qRT-PCR. To include these samples in the association studies the samples were given a Ct value that was 3.3 cycles higher than the highest LOC643714 Ct value measured. This calculated Ct value was used to calculate the level of LOC643714 mRNA expression in the 30 tumours relative to the expression of the reference gene TBP according to the formula given above.
Clinical information and tumour characteristics
Information regarding the date and cause of death was obtained from patient records and confirmed by the Icelandic National Register. The age at diagnosis and information about tumour size, type and nodal status as well as ER, progesterone receptor (PgR), ErbB2 and Ki67 levels, was retrieved from records at the Department of Pathology at the Landspitali-University Hospital. ER, PgR, ErbB2 and Ki67 levels were evaluated by immunohistochemical staining according to the manufacturer’s protocol (Dako): ER (M7047, clone 1D5, 1:100), PgR (M3569, clone PgR 636, 1:100), HercepTest (K5207), and Ki67 (M7240, clone MIB1, 1:100). The ER and PgR status was determined as being either positive (staining 1+ to 3+) or negative (no staining). The ErbB2 receptor status was determined as being either positive (3+ staining plus 2+ staining and positive FISH analysis) or negative (1+ plus 2+ staining and negative FISH analysis) [38]. Tumours staining for Ki67 were scored highly proliferative if the number of cells and staining intensity was above 10% and slowly proliferating below 10%. The tumours were categorized according to the WHO histological classification and the histological grade was determined by the modified Bloom-Richardson system. The molecular subtypes of the tumours were retrieved from GEO dataset GSE22133 [37].
Statistical analysis
Pearson’s χ2 analysis was used to test genotype frequencies for deviations from Hardy-Weinberg equilibrium (HWE) for SNP rs3803662. Fisher exact test was used to analyse association between the SNP genotype and clinical and pathological parameters. The association of the SNP genotype and mRNA expression was determined using ANOVA. The association of mRNA expression, as measured by qRT-PCR, to clinical and pathological parameters was determined using a t-test or ANOVA. Pearson correlation coefficient was calculated to determine the strength of the linear relationship between TOX3 and LOC643714 mRNA expression. Analyses of overall survival (OS) and distant metastasis-free survival (DMFS) were calculated by using the Kaplan-Meier method and the log-rank test. All calculations were performed using the statistical program R [39] version 2.11.1. Schematic of the analyses and significant results are depicted for the SNP and mRNA in Additional file 1: Figures S5 and S6, respectively.
Results
The rs3803662 risk allele is associated with shorter OS
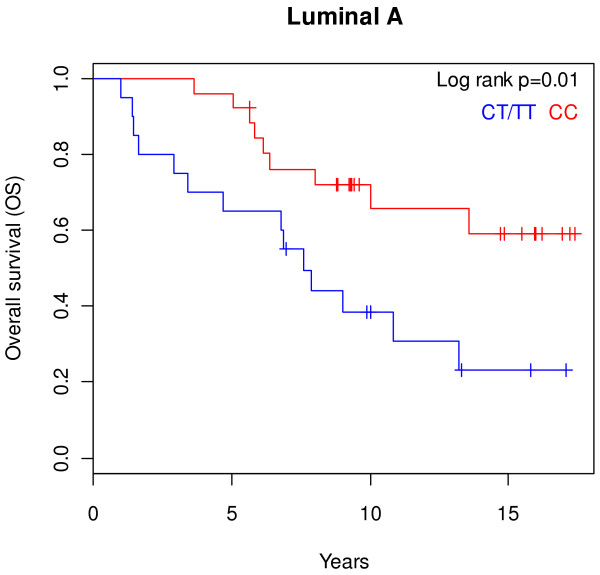
Correlation analysis between the SNP genotypes and clinical and pathological factors of breast cancer was performed. The comparison was made between patients who were homo- or heterozygous for the risk allele and patients homozygous for the non-risk allele. We observed a significant association between the risk allele and lobular tumours (p = 0.04; Additional file 2: Table S1). An association was detected between survival and the risk allele in the entire cohort. However, when survival was analysed according to molecular subtypes, patients diagnosed with luminal A tumours who carried the risk allele had shorter OS as compared to patients homozygous for the non-risk allele (p = 0.01, Figure 1). Distant metastasis-free survival (DMFS) was also shorter in this patient group but the difference did not reach significance (p = 0.24, Additional file 1: Figure S1). The rs3803662 genotype did not show correlation with OS or DMFS in other breast cancer subtypes.
Figure 1.
Patients with luminal A tumours and the risk rs3803662 allele have shorter OS. The OS was compared in patients with luminal A tumours who were homo- or heterozygous for the risk rs3803662 allele (CT/TT, n = 20) as opposed to homozygous for the non-risk rs3803662 allele (CC, n = 26), log rank p = 0.01.
rs3803662 is associated with decreased TOX3 mRNA expression in ER positive tumours
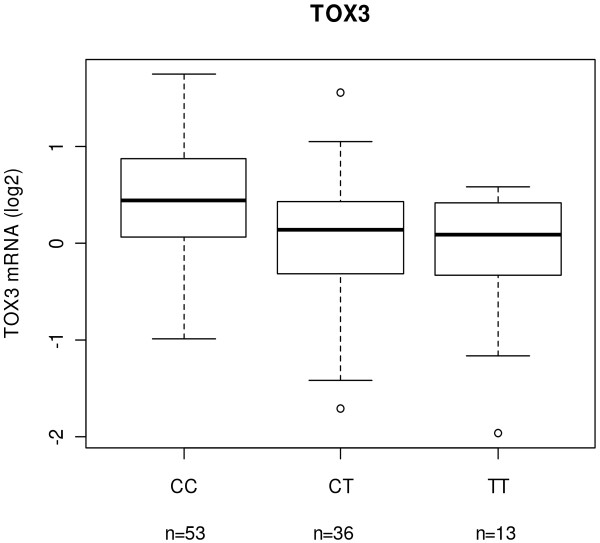
Gene expression data from microarrays were available for TOX3 in 160 breast tumours. Expression of TOX3 mRNA was compared to the rs3803662 genotype in the total set of tumours as well as in ER positive and ER negative tumours. In ER positive tumours, TOX3 expression decreased significantly with the number of risk alleles (p = 0.002, Figure 2). No difference was seen in ER negative or the entire group of tumours (p = 0.25 and p = 0.29, respectively). To confirm these results, qRT-PCR was performed to measure TOX3 mRNA levels in 139 breast tumours available from the microarray study. The qRT-PCR results correlated positively with the microarray results (r = 0.57, p < 0.001) and showed that the TOX3 mRNA level decreased with the number of risk alleles in ER positive tumours although the difference did not reach significance (p = 0.11, Additional file 1: Figure S2A). As before, no difference was observed in the ER negative or entire group of tumours (p = 0.78 and p = 0.53, respectively). Due to lack of microarray expression data of LOC643714 qRT-PCR was performed to measure the mRNA levels. There was correlation between the mRNA levels of TOX3 and LOC643714 as measured by qRT-PCR (r= 0.45, p < 0.001). Similar to TOX3, the mRNA levels of LOC643714 decreased with the number of risk alleles in ER positive tumours but the difference did not reach significance (p = 0.24, Additional file 1: Figure S2B). The majority of ER positive tumours are of the molecular subtypes luminal A and luminal B. Although not significant, a lower expression of TOX3 and LOC643714 was seen in tumours with the risk allele in luminal A tumours but not in luminal B tumours (Additional file 1: Figure S3).
Figure 2.
The minor allele of rs3803662 is associated with lower TOX3 mRNA expression. Correlation of mRNA expression level of TOX3 with genotype of rs3803662 in 102 ER positive breast cancer tumours (p = 0.002). The mRNA levels were measured on microarrays. ANOVA was used to test for significant difference between the three genotype groups.
High TOX3 and LOC643714 mRNA correlated with positive lymph nodes and ER positive tumours
Expression levels of TOX3 and LOC643714 were significantly higher in ER positive tumours than in ER negative tumours (p = 0.004 and p < 0.001, respectively; Table 1). PgR positive tumours also had significantly higher TOX3 and LOC643714 mRNA levels than PgR negative ones (p = 0.005 and p < 0.001, respectively). This was reflected in different mRNA levels of the two genes according to molecular subtypes. Luminal A and B tumours, that are mainly ER positive, expressed significantly higher mRNA than ER negative basal tumours (TOX3 and LOC653714 mRNA vs. subtypes: p < 0.001 and p < 0.001, respectively, Table 1 and Figure 3). Interestingly, two-thirds of the ERBB2 tumours were ER negative, yet they expressed significantly higher TOX3 mRNA than basal tumours (p = 0.02, Figure 3). Tumours of patients who were lymph node positive when diagnosed, had significantly higher TOX3 and LOC643714 mRNA levels than tumours of lymph node negative patients (p < 0.001 and p = 0.04, respectively, Table 1). Tumours with high expression of Ki67 had significantly lower TOX3 and LOC643714 mRNA levels than tumours with low expression of Ki67 (p = 0.026 and p < 0.001, respectively). Furthermore, low mRNA level of LOC643714 associated with a high histograde (p = 0.01) which likely echoed the strong correlation of basal tumours with high histograde (data not shown). ErbB2 negative tumours expressed more LOC643714 mRNA than ErbB2 positive tumours (p = 0.04), which was not unexpected because the majority of the ErbB2 negative tumours was ER positive.
Table 1.
TOX3 and LOC643714 mRNA expression compared with clinical and pathological characteristics
| n | TOX3 mRNA | p-value | n | LOC643714 mRNA | p-value | ||
|---|---|---|---|---|---|---|---|
| |
|
139 |
median (25% and 75%) |
|
126 |
median (25% and 75%) |
|
|
Age | |||||||
| |
< 50 |
59 (42%) |
1.92 (0.9 - 4.5) |
0.89 |
53 (42%) |
3.1 E-4 (5.6 E-6 - 1.5 E-2) |
0.07 |
| |
≥ 50 |
80 (58%) |
1.68 (1.1 - 4.0) |
|
73 (58%) |
1.7 E-3 (1.1 E-4 - 8.5 E-2) |
|
|
Tumour size (mm) | |||||||
| |
≤ 20 |
42 (31%) |
1.5 (1.0 - 4.8) |
0.45 |
39 (31%) |
6.4 E-4 (2.7 E-5 - 1.0 E-2) |
0.26 |
| |
> 20 |
95 (69%) |
2.0 (1.0 - 4.5) |
|
85 (69%) |
1.1 E-3 (9.2 E-5 - 8.5 E-2) |
|
| |
unknown |
2 |
|
|
2 |
|
|
|
Estrogen receptor | |||||||
| |
negative |
44 (33%) |
1.3 (0.4 - 3.3) |
0.004 |
41 (34%) |
9.3 E-6 (6.4 E-7 - 5.5 E-4) |
< 0.001 |
| |
positive |
91 (67%) |
2.1 (1.1 - 4.5) |
|
81 (66%) |
2.4 E-3 (4.6 E-4 - 9.8 E-2) |
|
| |
unknown |
4 |
|
|
4 |
|
|
|
Progesterone receptor | |||||||
| |
negative |
65 (49%) |
1.4 (0.6 - 4.2) |
0.005 |
59 (50%) |
1.9 E-4 (1.4 E-6 - 3.1 E-3) |
< 0.001 |
| |
positive |
67 (51%) |
2.1 (1.3 - 4.3) |
|
60 (50%) |
3.3 E-3 (4.2 E-4 - 0.5) |
|
| |
unknown |
7 |
|
|
7 |
|
|
|
ErbB2 status | |||||||
| |
negative |
114 (83%) |
1.8 (1.0 - 4.2) |
0.81 |
107 (86%) |
1.1 E-3 (6.6 E-5 - 7.7 E-2) |
0.04 |
| |
positive |
24 (17%) |
1.9 (0.8 - 5.0) |
|
18 (14%) |
2.7 E-4 (4.7 E-5 - 9.2 E-4) |
|
| |
unknown |
1 |
|
|
1 |
|
|
|
Nodal status | |||||||
| |
negative |
56 (44%) |
1.3 (0.4 - 3.4) |
< 0.001 |
53 (46%) |
5.3 E-4 (1.9 E-6 - 2.3E-2) |
0.04 |
| |
positive |
71 (56%) |
2.2 (1.2 - 5.3) |
|
62 (54%) |
1.4 E-3 (2.0 E-4 - 5.7 E-2) |
|
| |
unknown |
12 |
|
|
11 |
|
|
|
Ki67 | |||||||
| |
Low |
97 (71%) |
2.1 (1.1 - 4.4) |
0.026 |
87 (70%) |
2.4 E-3 (2.2 E-4 - 9.0 E-2) |
< 0.001 |
| |
High |
39 (29%) |
1.2 (0.7 - 2.4) |
|
37 (30%) |
9.6 E-5 (1.9 E-6 - 9.0 E-4) |
|
| |
unknown |
3 |
|
|
2 |
|
|
|
Histograde | |||||||
| |
1 |
12 (9%) |
1.6 (0.8 - 3.5) |
0.16 |
12 (9%) |
1.1 E-3 (2.7 E-4 - 3.5 E-2) |
0.01 |
| |
2 |
78 (57%) |
1.9 (1.1 - 4.4) |
|
69 (56%) |
3.5 E-3 (1.9 E-4 - 9.8 E-2) |
|
| |
3 |
47 (34%) |
1.6 (0.5 - 4.2) |
|
43 (35%) |
2.3 E-4 (3.3 E-6 - 1.5 E-3) |
|
| |
unknown |
2 |
|
|
2 |
|
|
|
Familial status 1 | |||||||
| |
BRCA2 |
26 (19%) |
2.9 (0.8 - 5.1) |
0.90 |
25 (20%) |
6.7 E-4 (1.2 E-4 - 4.2 E-3) |
0.40 |
| |
Non-BRCA2 |
112 (81%) |
1.6 (1.0 - 4.2) |
|
100 (80%) |
1.2 E-3 (6.3 E-5 - 6.4 E-2) |
|
| |
unknown |
1 |
|
|
1 |
|
|
|
Subtype | |||||||
| |
Basal |
20 (17%) |
0.5 (0.1 – 1.3) |
< 0.001 |
23 (21%) |
3.1 E-6 (2.3 E-7 - 2.0 E-4) |
< 0.001 |
| |
ERBB2 |
15 (12%) |
1.6 (1.2 - 5.0) |
|
9 (8%) |
9.7 E-4 (2.7 E-4 - 1.5 E-2) |
|
| |
Luminal A |
43 (36%) |
1.6 (1.1 - 3.1) |
|
38 (35%) |
8.9 E-3 (5.0 E-4 - 9.8 E-2) |
|
| |
Luminal B |
30 (25%) |
4.0 (1.6 - 5.4) |
|
28 (26%) |
1.2 E-3 (6.7 E-4 – 3.0 E-1) |
|
| |
Normal-like |
12 (10%) |
1.8 (1.2 - 4.4) |
|
10 (9%) |
5.8 E-4 (3.4 E-5 - 2.7 E-3) |
|
| |
Unclass/unknown |
19 |
|
|
18 |
|
|
|
Histological type | |||||||
| |
Ductal |
116 (86%) |
2.1 (1.0 - 4.5) |
0.12 |
105 (86%) |
1.1 E-3 (9.2 E-5 - 5.2 E-2) |
0.91 |
| |
Lobular |
11 (8%) |
1.3 (1.3 - 2.2) |
|
9 (7%) |
6.2 E-4 (1.2 E-4 - 7.8 E-3) |
|
| |
other |
8 (6%) |
1.0 (0.6 - 1.5) |
|
8 (7%) |
9.8 E-5 (5.2 E-6 - 2.2 E-2) |
|
| |
unknown |
4 |
|
|
4 |
|
|
|
Metastasis | |||||||
| |
negative |
63 (50%) |
1.6 (0.9 - 3.1) |
0.18 |
63 (55%) |
6.4 E-4 (6.3 E-6 - 1.9 E-2) |
0.16 |
| |
positive |
63 (50%) |
1.9 (1.1 - 4.9) |
|
52 (45%) |
1.6 E-3 (1.4 E-4 - 8.1 E-2) |
|
| unknown | 13 | 11 | |||||
The table shows the median and the 25th and 75th percentiles. The p-value is calculated with log2 transformed data using a t-test or ANOVA.
1 One tumour sample was BRCA1 positive and was not used in familial status calculations.
Figure 3.
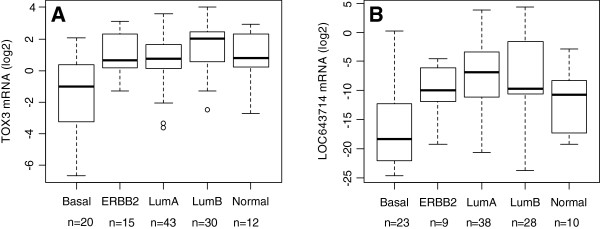
TOX3 and LOC643714 mRNA expression was lowest in tumours of the basal subtype. The mRNA expression levels in: A) TOX3 (p < 0.001) and B) LOC643714 (p < 0.001) in five breast tumour subtypes. ANOVA was used to test for differences in expression between the different subtypes. The difference in TOX3 expression between basal and ERBB2 tumours was significant (p = 0.02).
Due to the difference in expression of TOX3 and LOC643714 according to ER status, the association analysis was performed separately for the ER positive and negative subgroups. The results of the correlation analyses in the ER positive subgroup were similar to the whole group of tumours although in most cases it did not reach significance, possibly due to lower number of cases included in the analyses (Additional file 2: Table S2). The only significant correlation remaining was the one between high LOC643714 mRNA and low Ki67 (p = 0.02) and negative ErbB2 status (p = 0.01, Additional file 2: Table S2). Expression of LOC643714 correlated with that of TOX3 in ER negative tumours (r = 0.64, p < 0.001) but not in ER positive tumours (r = 0.13, p = 0.25). The result for the ER negative tumour subgroup was similar to the whole group of tumours in that high TOX3 and LOC643714 mRNA levels were associated with positive lymph nodes (p < 0.001 and p = 0.01, respectively) and a positive PgR status, although only significant for TOX3 (p = 0.002, Additional file 2: Table S3). But there was lack of correlation between low mRNA levels of TOX3 and LOC643714 and high expression of Ki67 and low mRNA level of LOC643714 and negative ErbB2 status of the tumour as well as high histograde. A significant correlation was observed between high TOX3 mRNA in tumours of patients with metastasis (p = 0.015, Additional file 2: Table S3), which was observed as a trend in the whole group of tumours (Table 1).
Elevated TOX3 mRNA expression in breast tumours is associated with poor survival
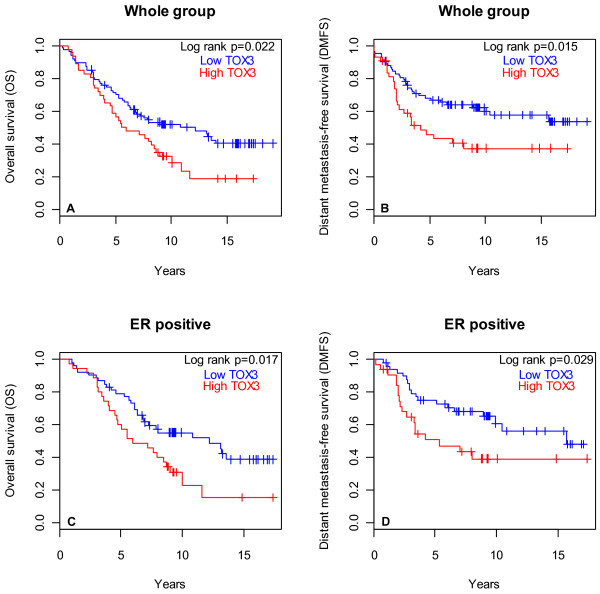
Survival was compared between patients with tumours that expressed high mRNA levels (above average) of TOX3 or LOC643714 as opposed to low levels (below average). Patients with tumours that express high TOX3 mRNA had shorter DMFS and OS (p = 0.015 and p = 0.022, respectively, Figure 4). This effect was independent of lymph node status. Survival analysis within subgroups depending on ER status revealed that high TOX3 mRNA expression in patients with ER positive tumours resulted in shorter DMFS and OS than in patients with low TOX3 mRNA (p = 0.029 and p = 0.017, respectively, Figure 4). This effect was mostly due to tumours of the luminal B subtype because their removal from the analysis resulted in loss of the effect of TOX3 mRNA on OS and DMFS (log rank p = 0.64 and log rank p = 0.86, respectively) whereas removal of luminal A tumours had small effect on survival (OS: log rank p = 0.02 and DMFS: log rank p = 0.05; Additional file 1: Figure S4). Fewer tumours are within the ER negative group and the effect of TOX3 mRNA level on survival was not statistically significant. There was no effect of high LOC643714 mRNA on DMFS or OS.
Figure 4.
High TOX3 mRNA was associated with shorter survival. Tumours were classified into high and low expressing tumours based on mRNA levels that were above or below average, respectively. Accordingly, survival was analysed in the whole group of patients: A) OS: 88 low vs. 46 high, log rank p = 0.022; B) DMFS: 83 low vs. 43 high, log rank p = 0.015 and in ER positive patients: C) OS: 53 low vs. 35 high, log rank p = 0.017; D) DMFS: 49 low vs. 33 high, log rank p = 0.029.
Discussion
In summary, the minor allele of SNP rs3803662 was associated with shorter survival in breast cancer patients with luminal A tumours, with lower mRNA expression of TOX3 in ER positive tumours as well as being more frequently observed in lobular tumours (Additional file 1: Figure S5). Also, low expression of TOX3 and the neighbouring gene LOC643714 was associated with high Ki67 and the basal tumour subtype while high mRNA expression was associated with ER status, positive lymph nodes at diagnosis and shorter survival (Additional file 1: Figure S6).
The risk allele of rs3803662 has been shown to associate with ER and PgR status [6,11,20-22,40], grade of tumours [21,22,40] and diagnosis before the age of 60 [9]. In our study cohort, that was smaller than the aforementioned studies, we did not observe a correlation between genotype at rs3803662 and age or tumour characteristics except that the risk allele was more frequent in patients with lobular tumours, which is in agreement with published data [6,21]. The Breast Cancer Association Consortium (BCAC) recently reported that ER positive patients, homozygous for the risk allele, had poorer overall and breast cancer specific survival than patients, homo- and heterozygous for the non-risk allele [40]. The BCAC study did not include information on the molecular subtypes of the tumours. In our cohort, the risk allele was significantly associated with poor overall survival only in patients with luminal A tumours. The rs3803662 genotype has been shown to associate most strongly with the ER/PgR positive and ERBB2 negative subgroups [21], which is the expression pattern of tumours of the luminal A subtype, further supporting that the effect seen in the BCAC study may be connected to luminal A tumours.
A decrease in TOX3 mRNA according to the rs3803662 genotype was observed in a large Dutch study of 1401 breast tumours [33] but analysis according to ER status was not reported. In our study, the allele-dependent decrease of TOX3 or LOC643714 mRNA level was only observed in ER positive tumours. The TOX3 mRNA levels were quantified by microarrays and confirmed by qRT-PCR. A significant association was not seen using the latter method, possibly due to the lower number of tumours that were available for qRT-PCR. Expression of LOC643714 was only measured by qRT-PCR and a positive correlation was found between TOX3 and LOC643714 mRNA levels. The closely located genes are potentially co-regulated. LOC643714 mRNA expression in breast tumours was very low and near the detection limit of the assay. This was previously observed by Udler et al. who reported negligible mRNA levels of LOC643714 in normal breast and breast tumours [13]. A larger cohort will be necessary to confirm the association between rs3803662 and LOC643714 expression.
The rs3803662 genotype may only affect expression of TOX3 and LOC643714 in certain tumour subtypes. Although we observed a reduction in gene expression in ER positive tumours, our preliminary analysis suggests that this is limited to luminal A tumours (Additional file 1: Figure S3) and another mechanism may be responsible for controlling expression of these genes in luminal B tumours. Although a higher number of tumours are required to confirm this observation, the majority of the adverse effect of high TOX3 expression in ER positive tumours was related to luminal B tumours (Additional file 1: Figure S4). Furthermore, higher TOX3 mRNA was observed in patients with luminal B tumours with metastasis (p = 0.056, data not shown) but more tumours are necessary to observe a significant association of metastasis to bone as has been reported with high TOX3 expression [34]. The risk allele of rs3803662 was highly correlated with a subgroup of basal tumours that expresses EGFR and CK5/6 but not ER, PgR and ERBB2 [40]. Expression of TOX3 and LOC643714 was significantly lower in basal tumours in our cohort but we did not observe differences in expression according to genotype or a difference in allele distribution compared to the other tumour subtypes as has been reported for rs9940048, which is located within TOX3[41].
The rs3803662 SNP may regulate expression of genes more distantly located than TOX3 and LOC643714. It lies within an evolutionary conserved region that has an open chromatin conformation suggestive of an active transcription regulatory region [13]. Our preliminary analysis of the association of the rs3803662 risk allele with the expression of genes located within 1 Mb of the SNP revealed an association with the retinoblastoma-like 2 gene (RBL2) only, where an increase in expression was seen with the number of risk alleles (p = 0.02, data not shown). This result is in agreement with Udler et al., who reported a dose-dependent association between the rs3803662 risk allele with increased expression of RBL2 mRNA expression in lymphocytes; however, an association between the rs3803662 genotype and RBL2 mRNA expression in 77 breast tumours was not seen [13]. Expression of RBL2 was recently reported to be associated with prognosis [40]. It is a member of the retinoblastoma gene family [42] and has been shown to be involved in cell cycle regulation, and it is frequently deleted in breast tumours [43].
Very low expression of TOX3 and LOC643714 was observed in the basal subtype, an observation that has been reported for TOX3[41]. The significantly lower TOX3 and LOC643714 expression in tumours expressing high Ki67 was due to the relatively high number of basal tumours expressing high Ki67. Expression correlated significantly with metastasis in ER negative tumours but not with metastasis to bone [34]. High expression of both TOX3 and LOC643714 mRNA was observed in tumours of lymph node positive patients but only TOX3 mRNA correlated with survival where high levels were seen in patients with shorter overall and distant metastasis free survival in the entire group and for patients with ER positive tumours. This discrepancy between TOX3 and LOC643714 may possibly be due to fewer LOC643714 tumours than for TOX3, different distribution of available samples into subtypes, or the fact that there was not a correlation between TOX3 and LOC643714 in ER positive tumours. It was recently reported that TOX3 expression was not associated with prognosis [40]. The study was large but the data came from 9 separate studies in publicly available databases. Thus, we suggest that the effect potentially vanished due to variability in study design and data acquisition.
The biological mechanism that potentially explains our results is the implication of TOX3 in proliferation and apoptosis. Knockdown of TOX3 expression with siRNA in the luminal A breast cancer cell line ZR-75-1 increased cellular proliferation [44]. The same study described a FOXA1 binding site at breast cancer SNP rs4784227, whose risk allele segregates with the risk allele of rs3803662; FOXA1 preferentially bound the site that included the rs4784227 risk allele resulting in a 5-fold decrease in TOX3 expression [44]. Therefore, the correlation of FOXA1 expression with luminal A tumours [45] could explain lower expression of TOX3 in our study and poorer survival of carriers of the rs3803662 risk allele and luminal A tumours. In these tumours, TOX3 would appear to be a tumour suppressor. However, a different scenario has been described in neuronal cells that potentially may explain the effect of high TOX3 mRNA in ER positive tumours which we ascribe to luminal B tumours. Increased expression of TOX3 protected neuronal cells from cell death by altering expression levels of genes that affect apoptosis [30]. Such function in ER positive breast cancer cells, where TOX3 expression is high, could promote tumour enlargement, invasion of nearby tissues, seeding of lymph nodes which would subsequently shorten the time until death or distant metastasis. The effect of TOX3 on apoptosis in neuronal cells was mediated via its interaction with CITED1 and ERE [30] and may be the case for proliferation as well. However, the TOX3 ERE-mediated transcription appeared independent of ER and oestrogen [30] unlike that of CITED1, a co-activator of ER that modulates ER responsive genes [31]. We can only speculate that LOC643714 has a role in tumour development based on results similar to TOX3 but its mRNA is not translated and its function is unknown.
Although we assume that mRNA expression of our candidate genes may differently affect breast tumour subtypes, the study is small and the analyses according to ER status or molecular subtypes further reduced the number of tumours. As such, the study is exploratory and the results should be confirmed with a larger number of tumours. Our findings provide a suitable basis for further validation studies.
Conclusions
Taken together, the data suggest that the rs3803662 genotype may affect expression of TOX3 and LOC643714 in tumours that originate in some developmental structures within the breast, such as the lobes, and in different molecular subtypes of tumours, such as luminal A and possibly in subgroups of basal tumours. Furthermore, in other types of breast cancer, possibly luminal B tumours, expression of these genes may be controlled by other mechanisms where high expression has adverse effects on prognosis. Although further analysis is needed, we conclude that genotype at rs3803662 and expression of TOX3 and LOC643714 have adverse effect on breast cancer prognosis.
Abbreviations
BRCA1: Breast cancer 1; BRCA2: Breast cancer 2; CITED1: Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1; DMFS: Distant metastasis-free survival; ERBB2: V-erb-b2-erythroblastic leukemia viral oncogene homolog 2; ER: Oestrogen receptor; LD: Linkage disequilibrium; OS: Overall survival; PgR: Progesterone receptor; RBL2: Retinoblastoma-like 2; SNP: Single nucleotide polymorphism; TOX3: TOX high mobility group box family member 3.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ETG performed the RT-PCR experiments, contributed to data analysis as well as the writing of the manuscript. RBB conceived the study, interpreted the data and contributed to the writing of the manuscript. AA participated in the microarray experiments and critically revised the manuscript. LTA interpreted the data and critically reviewed the manuscript. HG performed the genotyping, participated in the microarray experiments and analyzed the data. BAA participated in the acquisition and interpretation of the pathologic data and critical reviewing of the manuscript. OTJ participated in acquisition of the clinical data and critical reviewing of the manuscript. IR analysed and interpreted the data, and led the writing of the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Figures S1 – S6. Supplementary Figure S1. Comparison between DMFS in patients with luminal A tumours carrying the minor vs. major rs3803662 allele. Supplementary Figure S2. Correlation of the rs3803662 genotype with expression levels of TOX3 and LOC643714. Supplementary Figure S3. Correlation of the rs3803662 genotype with expression levels of TOX3 and LOC643714 in luminal tumours. Supplementary Figure S4. Patients with luminal B tumours contribute the majority of the adverse effect correlated with high TOX3 in ER positive tumours. Supplementary Figure S5. A schematic of the correlation analyses of the rs3803662 genotype with clinical and tumour characteristics. Supplementary Figure S6. A schematic of the correlation analyses of the mRNAs of TOX3 and LOC64371 with clinical and tumour characteristics.
Tables S1 – S3. Supplementary Table S1. Association between the SNP rs3803662 genotype and clinical and pathological parameters (n = 160). Supplementary Table S2. Correlation between TOX3 and LOC643714 mRNA levels in ER positive tumours with breast cancer clinical and pathological characteristics. Supplementary Table S3. TOX3 and LOC643714 mRNA expression in ER negative tumour samples compared with breast cancer clinical and pathological characteristics.
Contributor Information
Eydis Th Gudmundsdottir, Email: eydist@landspitali.is.
Rosa B Barkardottir, Email: rosa@landspitali.is.
Adalgeir Arason, Email: adalgeir@landspitali.is.
Haukur Gunnarsson, Email: hagunnarsson@actavis.is.
Laufey Th Amundadottir, Email: amundadottirl@mail.nih.gov.
Bjarni A Agnarsson, Email: bjarniaa@landspitali.is.
Oskar Th Johannsson, Email: oskarjoh@landspitali.is.
Inga Reynisdottir, Email: ingar@landspitali.is.
Acknowledgements
This work was funded by the Landspitali-University Hospital Research Fund and “Gongum saman”, a supporting group for breast cancer research in Iceland.
References
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40(1):17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet. 2008;9:321–345. doi: 10.1146/annurev.genom.9.081307.164339. [DOI] [PubMed] [Google Scholar]
- Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M. Genetic susceptibility to breast cancer. Mol Oncol. 2010;4(3):174–191. doi: 10.1016/j.molonc.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, Masson G, Jakobsdottir M, Thorlacius S, Helgason A. et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- Chen MB, Wu XY, Shen W, Wei MX, Li C, Cai B, Tao GQ, Lu PH. Association between polymorphisms of trinucleotide repeat containing 9 gene and breast cancer risk: evidence from 62,005 subjects. Breast Cancer Res Treat. 2011;126(1):177–183. doi: 10.1007/s10549-010-1114-6. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Muller-Myhsok B, Lichtner P, Engel C, Chen B, Burwinkel B, Forsti A, Sutter C, Wappenschmidt B, Hellebrand H. et al. Low-risk variants FGFR2, TNRC9 and LSP1 in German familial breast cancer patients. Int J Cancer. 2010;126(12):2858–2862. doi: 10.1002/ijc.24986. [DOI] [PubMed] [Google Scholar]
- Huijts PE, Vreeswijk MP, Kroeze-Jansema KH, Jacobi CE, Seynaeve C, Krol-Warmerdam EM, Wijers-Koster PM, Blom JC, Pooley KA, Klijn JG. et al. Clinical correlates of low-risk variants in FGFR2, TNRC9, MAP3K1, LSP1 and 8q24 in a Dutch cohort of incident breast cancer cases. Breast Cancer Res. 2007;9(6):R78. doi: 10.1186/bcr1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney N, Colleran G, Rowan A, Walther A, Barclay E, Spain S, Jones AM, Tuohy S, Curran C, Miller N. et al. Low penetrance breast cancer predisposition SNPs are site specific. Breast Cancer Res Treat. 2009;117(1):151–159. doi: 10.1007/s10549-008-0235-7. [DOI] [PubMed] [Google Scholar]
- Reeves GK, Travis RC, Green J, Bull D, Tipper S, Baker K, Beral V, Peto R, Bell J, Zelenika D. et al. Incidence of breast cancer and its subtypes in relation to individual and multiple low-penetrance genetic susceptibility loci. JAMA. 2010;304(4):426–434. doi: 10.1001/jama.2010.1042. [DOI] [PubMed] [Google Scholar]
- Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS. et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udler MS, Ahmed S, Healey CS, Meyer K, Struewing J, Maranian M, Kwon EM, Zhang J, Tyrer J, Karlins E. et al. Fine scale mapping of the breast cancer 16q12 locus. Hum Mol Genet. 2010;19(12):2507–2515. doi: 10.1093/hmg/ddq122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Woo JH, Yu JH, Lee MJ, Moon HG, Kang D, Noh DY. Common genetic variants associated with breast cancer in Korean women and differential susceptibility according to intrinsic subtype. Cancer Epidemiol Biomarkers Prev. 2011;20(5):793–798. doi: 10.1158/1055-9965.EPI-10-1282. [DOI] [PubMed] [Google Scholar]
- Long J, Shu XO, Cai Q, Gao YT, Zheng Y, Li G, Li C, Gu K, Wen W, Xiang YB. et al. Evaluation of breast cancer susceptibility loci in Chinese women. Cancer Epidemiol Biomarkers Prev. 2010;19(9):2357–2365. doi: 10.1158/1055-9965.EPI-10-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueta A, Ito H, Kawase T, Hirose K, Hosono S, Yatabe Y, Tajima K, Tanaka H, Iwata H, Iwase H. et al. A genetic risk predictor for breast cancer using a combination of low-penetrance polymorphisms in a Japanese population. Breast Cancer Res Treat. 2011;132(2):711–721. doi: 10.1007/s10549-011-1904-5. [DOI] [PubMed] [Google Scholar]
- Hutter CM, Young AM, Ochs-Balcom HM, Carty CL, Wang T, Chen CT, Rohan TE, Kooperberg C, Peters U. Replication of breast cancer GWAS susceptibility loci in the Women's Health Initiative African American SHARe Study. Cancer Epidemiol Biomarkers Prev. 2011;20(9):1950–1959. doi: 10.1158/1055-9965.EPI-11-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Narvaez EA, Rosenberg L, Cozier YC, Cupples LA, Adams-Campbell LL, Palmer JR. Polymorphisms in the TOX3/LOC643714 locus and risk of breast cancer in African-American women. Cancer Epidemiol Biomarkers Prev. 2010;19(5):1320–1327. doi: 10.1158/1055-9965.EPI-09-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Cai Q, Signorello LB, Long J, Hargreaves MK, Deming SL, Li G, Li C, Cui Y, Blot WJ. Evaluation of 11 breast cancer susceptibility loci in African-American women. Cancer Epidemiol Biomarkers Prev. 2009;18(10):2761–2764. doi: 10.1158/1055-9965.EPI-09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Chen P, Hu Z, Shen H, Wang F, Chen L, Li M, Tang J, Wang H, Shen H. Genetic variants in trinucleotide repeat-containing 9 (TNRC9) are associated with risk of estrogen receptor positive breast cancer in a Chinese population. Breast Cancer Res Treat. 2010;124(1):237–241. doi: 10.1007/s10549-010-0809-z. [DOI] [PubMed] [Google Scholar]
- Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, Apicella C, Smith LD, Hammet F, Southey MC. et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20(16):3289–3303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, Bojesen SE, Nordestgaard BG, Axelsson CK, Arias JI. et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4(4):e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Beesley J, McGuffog L, Sinilnikova OM, Healey S, Neuhausen SL, Ding YC, Rebbeck TR, Weitzel JN, Lynch HT. et al. Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res. 2010;70(23):9742–9754. doi: 10.1158/0008-5472.CAN-10-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Spurdle AB, Sinilnikova OM, Healey S, Pooley KA, Schmutzler RK, Versmold B, Engel C, Meindl A, Arnold N. et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet. 2008;82(4):937–948. doi: 10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr N, Cooke R, Jones M, Fletcher O, Dudbridge F, Chilcott-Burns S, Tomczyk K, Broderick P, Houlston R, Ashworth A. et al. Genetic variants at chromosomes 2q35, 5p12, 6q25.1, 10q26.13, and 16q12.1 influence the risk of breast cancer in men. PLoS Genet. 2011;7(9):e1002290. doi: 10.1371/journal.pgen.1002290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National center for biotechnology information. http://www.ncbi.nlm.nih.gov/gene/643714.
- Margolis RL, Abraham MR, Gatchell SB, Li SH, Kidwai AS, Breschel TS, Stine OC, Callahan C, McInnis MG, Ross CA. cDNAs with long CAG trinucleotide repeats from human brain. Hum Genet. 1997;100(1):114–122. doi: 10.1007/s004390050476. [DOI] [PubMed] [Google Scholar]
- O'Flaherty E, Kaye J. TOX defines a conserved subfamily of HMG-box proteins. BMC Genomics. 2003;4(1):13. doi: 10.1186/1471-2164-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan SH, Qiu Z, Ghosh A. TOX3 regulates calcium-dependent transcription in neurons. Proc Natl Acad Sci USA. 2009;106(8):2909–2914. doi: 10.1073/pnas.0805555106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer S, Kovacs Z, Yuan SH, Siszler G, Kogl M, Summer H, Geerts A, Golz S, Shioda T, Methner A. TOX3 is a neuronal survival factor that induces transcription depending on the presence of CITED1 or phosphorylated CREB in the transcriptionally active complex. J Cell Sci. 2010;124(Pt 2):252–260. doi: 10.1242/jcs.068759. [DOI] [PubMed] [Google Scholar]
- Yahata T, Shao W, Endoh H, Hur J, Coser KR, Sun H, Ueda Y, Kato S, Isselbacher KJ, Brown M. et al. Selective coactivation of estrogen-dependent transcription by CITED1 CBP/p300-binding protein. Genes Dev. 2001;15(19):2598–2612. doi: 10.1101/gad.906301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda T, Lechleider RJ, Dunwoodie SL, Li H, Yahata T, de Caestecker MP, Fenner MH, Roberts AB, Isselbacher KJ. Transcriptional activating activity of Smad4: roles of SMAD hetero-oligomerization and enhancement by an associating transactivator. Proc Natl Acad Sci USA. 1998;95(17):9785–9790. doi: 10.1073/pnas.95.17.9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz M, Berns EM, Sieuwerts AM, Ruigrok-Ritstier K, de Weerd V, Groenewoud A, Uitterlinden AG, Look MP, Klijn JG, Sleijfer S. et al. Correlation of breast cancer susceptibility loci with patient characteristics, metastasis-free survival, and mRNA expression of the nearest genes. Breast Cancer Res Treat. 2012;133(3):843–851. doi: 10.1007/s10549-011-1663-3. [DOI] [PubMed] [Google Scholar]
- Smid M, Wang Y, Klijn JG, Sieuwerts AM, Zhang Y, Atkins D, Martens JW, Foekens JA. Genes associated with breast cancer metastatic to bone. J Clin Oncol. 2006;24(15):2261–2267. doi: 10.1200/JCO.2005.03.8802. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson G, Staaf J, Vallon-Christersson J, Ringner M, Holm K, Hegardt C, Gunnarsson H, Fagerholm R, Strand C, Agnarsson BA. et al. Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Res. 2010;12(3):R42. doi: 10.1186/bcr2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GSE22133. http://www.ncbi.nlm.nih.gov/gds?term=GSE22133.
- Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A. et al. American society of clinical oncology/college of American pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131(1):18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- The R project for statistical computing. http://www.r-project.org.
- Fasching PA, Pharoah PD, Cox A, Nevanlinna H, Bojesen SE, Karn T, Broeks A, van Leeuwen FE, Van't Veer LJ, Udo R. et al. The role of genetic breast cancer susceptibility variants as prognostic factors. Hum Mol Genet. 2012;21(17):3926–3939. doi: 10.1093/hmg/dds159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgard SH, Johansen FE, Alnaes GI, Naume B, Borresen-Dale AL, Kristensen VN. Genes harbouring susceptibility SNPs are differentially expressed in the breast cancer subtypes. Breast Cancer Res. 2007;9(6):113. doi: 10.1186/bcr1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayol X, Grana X, Baldi A, Sang N, Hu Q, Giordano A. Cloning of a new member of the retinoblastoma gene family (pRb2) which binds to the E1A transforming domain. Oncogene. 1993;8(9):2561–2566. [PubMed] [Google Scholar]
- Naylor TL, Greshock J, Wang Y, Colligon T, Yu QC, Clemmer V, Zaks TZ, Weber BL. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005;7(6):R1186–1198. doi: 10.1186/bcr1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowper-Sal Lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, Moore JH, Lupien M. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44(11):1191–1198. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM, Dunn S, Huntsman DG, Nakshatri H. FOXA1 expression in breast cancer–correlation with luminal subtype A and survival. Clin Cancer Res. 2007;13(15 Pt 1):4415–4421. doi: 10.1158/1078-0432.CCR-07-0122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 – S6. Supplementary Figure S1. Comparison between DMFS in patients with luminal A tumours carrying the minor vs. major rs3803662 allele. Supplementary Figure S2. Correlation of the rs3803662 genotype with expression levels of TOX3 and LOC643714. Supplementary Figure S3. Correlation of the rs3803662 genotype with expression levels of TOX3 and LOC643714 in luminal tumours. Supplementary Figure S4. Patients with luminal B tumours contribute the majority of the adverse effect correlated with high TOX3 in ER positive tumours. Supplementary Figure S5. A schematic of the correlation analyses of the rs3803662 genotype with clinical and tumour characteristics. Supplementary Figure S6. A schematic of the correlation analyses of the mRNAs of TOX3 and LOC64371 with clinical and tumour characteristics.
Tables S1 – S3. Supplementary Table S1. Association between the SNP rs3803662 genotype and clinical and pathological parameters (n = 160). Supplementary Table S2. Correlation between TOX3 and LOC643714 mRNA levels in ER positive tumours with breast cancer clinical and pathological characteristics. Supplementary Table S3. TOX3 and LOC643714 mRNA expression in ER negative tumour samples compared with breast cancer clinical and pathological characteristics.