Abstract
Multicentric Castleman Disease is largely driven by increased signaling in the pathway for the plasma cell growth factor interleukin-6. We hypothesized that interleukin-6/interleukin-6 receptor/gp130 polymorphisms contribute to increased interleukin-6 and/or other components of the interleukin-6 signaling pathway in HIV-negative Castleman Disease patients. The study group was composed of 58 patients and 50 healthy donors of a similar racial/ethnic profile. Of seven polymorphisms chosen for analysis, we observed an increased frequency between patients and controls of the minor allele of interleukin-6 receptor polymorphism rs4537545, which is in linkage disequilibrium with interleukin-6 receptor polymorphism rs2228145. Further, individuals possessing at least one copy of the minor allele of either polymorphism expressed higher levels of soluble interleukin-6 receptor. These elevated interleukin-6 receptor levels may contribute to increased interleukin-6 activity through the trans-signaling pathway. These data suggest that interleukin-6 receptor polymorphism may be a contributing factor in Castleman Disease, and further research is warranted.
Introduction
Castleman Disease (CD) is a rare lymphoproliferative disorder first defined in 1954 [1]. CD may be divided into two major forms. Unicentric CD (UCD) is typically a slow-growing solitary mass occurring at a single anatomical site. Though the enlarging node may compress vital structures, UCD is rarely life-threatening, and surgical extirpation or localized radiotherapy is generally curative. Multicentric CD (MCD) affects multiple lymph node stations and often presents with lymphadenopathy, fever, weight loss, fatigue, edema, anemia and hypoalbuminemia [2]–[4]. In severe cases, patients may develop hepatosplenomegaly, massive ascites, pleural effusions, or organ failure, and MCD occasionally progresses to non-Hodgkin’s lymphoma. The clinical manifestations of MCD are largely driven by activation of the interleukin-6 (IL6) signaling cascade, which results in an acute phase reaction with elevated erythrocyte sedimentation rate, C-reactive protein (CRP), fibrinogen, thrombocytosis and hypergammaglobulinemia [2], [4], [5]. IL6 signaling enhances cell proliferation and differentiation, inhibits apoptosis, and leads to an increase in vascular endothelial growth factor and other cytokines [6], [7]. Conversely, blockade of IL6 signaling by monoclonal antibodies results in tumor regression and abrogation of symptoms [4].
Levels of IL6 have a significant impact on the prognosis of other IL6-related diseases and malignancies, including plasma cell dyscrasias (PCD).[8]–[10] Single nucleotide polymorphisms (SNPs) in the IL6 signaling pathway have been studied in acute phase reaction, auto-immune phenomena, and malignancies, and have been linked to elevated IL6 production.[11]–[14] Increased levels of IL6, along with soluble IL6R (sIL6R), have been detected in the synovial fluid of patients with rheumatoid arthritis [5], [6], [15]. IL6 promoter polymorphism may also be associated with an increased risk of PCD [14]. In MM, IL6-related polymorphism may be prognostic of survival [16], [17]. IL6 trans-signaling via sIL6R is also biologically important in that sIL6R allows cells that do not intrinsically express membrane-bound IL6R (mbIL6R) to be responsive to the effects of IL6.[5], [15], [18]–[20] The IL6R SNP rs2228145 is a functional polymorphism resulting in an asparagine to alanine substitution at the IL6R proteolytic cleavage site, just beyond the transmembrane domain of the IL6R protein. This SNP has been shown to significantly influence serum levels of sIL6R,[9], [17]–[19], [21], [22] and is thought to act by altering the efficiency of IL6R cleavage from the cell surface, which seems to be the dominant method of sIL6R generation [15], [18], [20], [23]. The IL6R SNP rs2228145 is in linkage disequilibrium with another IL6R SNP, rs4537545, and the minor allele of the rs4537545 SNP has been shown to account for nearly 20% of the variation in sIL6R levels [22].
We hypothesized that IL6/IL6R/gp130 polymorphisms contribute to increased IL6 and/or other components of the IL6 signaling pathway in CD patients. Seven well-studied SNPs in the IL6 promoter (rs1800795, rs1800796, rs1800797), IL6R (rs2228145, rs4537545) and gp130 (rs10940495, rs715180) were chosen for analysis. As CD is a prime example of an IL6-driven disease, we sought to determine whether CD patients express the minor allele of these IL6 pathway-related SNPs more frequently than healthy individuals, and assess what effect this may have on IL6/IL6R levels in CD patients versus healthy controls.
Materials and Methods
Ethics Statement
Research protocols were approved by the UAMS Institutional Review Board, and informed written consent was obtained from all participants before samples were collected, in accordance with the Declaration of Helsinki. The informed consent documents and consenting procedure were approved by the UAMS Institutional Review Board in accordance with UAMS and federal ethics guidelines.
Study Samples
Peripheral blood mononuclear cells (PBMC) and serum were collected from 58 CD patients and 50 healthy donors (HD). HD had no history of malignancy or IL6-related disease. The CD patient cohort included both MCD (42) and UCD (16) patients, matched for age, sex and race/ethnicity to exclude SNP genotype bias (Table 1). All CD patients were HIV- and HHV8-negative by serology and quantitative PCR.
Table 1. Subject characteristics.
| CD | HD | |
| Caucasian | 44 | 43 |
| African American | 7 | 2 |
| Hispanic | 4 | – |
| Median Age | 42 | 35 |
| Females | 31 | 31 |
| Males | 27 | 19 |
The majority of both CD cases and HD controls are Caucasian (n = 44, 43 respectively) with a few subjects from other ethnicities. Both cohorts were also matched for age and gender.
SNP Genotyping
DNA was extracted from PBMC using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). IL6 promoter, IL6R, and gp130 SNP genotypes were determined using TaqMan-based SNP genotyping assays (Applied Biosystems, Foster City, CA). PCR assays were performed in triplicate on the ABI Prism 7000 Sequence Detection System (SDS) instrument and analyzed using the Allelic Discrimination module of the SDS software (Applied Biosystems).
sIL6R ELISA
Serum levels of sIL6R were determined via the Human IL6sR Quantikine ELISA Kit (R&D Systems, Minneapolis, MN) according to manufacturer specifications. Samples were assayed in duplicate. Data was acquired using SoftMax Pro software (Molecular Devices, Sunnyvale, CA), and analyzed in GraphPad Prism (GraphPad Software, La Jolla, CA).
Meso-Scale Discovery Platform-based Panoptic IL6 Assay
Many capture or detection antibodies cannot recognize IL6 when in association with proteins in human serum such as alpha-2-macroglobulin, sIL6R, soluble gp130, complement proteins C3 and C4b, and albumin, thus skewing IL6 measurements. In an original report, Sehgal demonstrated that certain antibody pairs are able to recognize IL6 when in large molecular complexes that included IL6 associated proteins [24]. Given the discrepancies in methodologies for measuring IL6 and the number of variables that can affect antibody affinity for IL6 in serum, [25], [26] we developed a Panoptic IL6 assay for the measurement of IL6 in human serum. We investigated a large panel of commercially-available anti-IL6 antibodies for their ability to bind human recombinant IL6 in human serum. Top performing candidate capture and detection antibodies from our first generation screening were optimized in a sandwich ELISA format, and a second screening revealed the three best candidate capture antibodies. Immunoprecipitation experiments determined that the top three antibodies recognized and bound all known isoforms of IL6. Final antibody pairs were then developed on a Meso-Scale Discovery (MSD) platform, which resulted in a Panoptic IL6 assay for measuring IL6 in human serum. When compared to other validated IL6 platforms and antibodies, our final antibody pair of choice demonstrated a higher range of IL6 than other standard ELISA formats when measuring human IL6 in serum. (manuscript in preparation).
Flow Cytometry
PBMC were labeled with mouse anti-human antibodies to determine levels of mbIL6R (CD126; R&D Systems) and gp130 (CD130; Becton Dickinson Biosciences, San Jose, CA) expression on T cells, monocytes, B cells, and natural killer cells. PBMC subset analysis was performed with FITC-conjugated antibodies specific for CD3, CD4, CD8, CD14, CD19, and CD56 (Becton Dickinson). Data was acquired on a FACSCalibur flow cytometer (Becton Dickinson) and analyzed using FCS Express analysis software (De Novo Software, Los Angeles, CA). Median fluorescence intensity (MFI) of the positive population (mbIL6R and gp130) was recorded for each cell type.
Statistical Analysis
Significance of differences in minor allele frequencies (MinAF) between Castleman Disease (CD) patients and healthy donors (HD) was assessed by generating a z-score. The equation used was proposed by Schaid and Jacobsen [27]. This equation accounts for deviation from perfect Hardy Weinberg Equilibrium (HWE).
Where 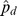 is the SNP’s major allele frequency (MajAF) in diseased subjects and
is the SNP’s major allele frequency (MajAF) in diseased subjects and 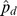 is the MajAF in controls.
is the MajAF in controls.
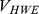 is calculated as follows:
is calculated as follows:
Where 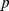 is the MajAF in all subjects,
is the MajAF in all subjects, 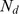 is the number of diseased subjects, and
is the number of diseased subjects, and 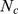 is the number of controls.
is the number of controls.
First, genotype frequencies were calculated for the entire cohort (CD + HD). Next, the MajAF and MinAF were calculated for each SNP. A Chi square was performed to determine whether the genotype frequencies were in HWE. The above formula was employed to generate the z-score for each SNP, which was then converted to a p-value utilizing GraphPad Prism (GraphPad Software).
Linkage disequilibrium (LD) was assessed utilizing the Online Encyclopedia for Genetic Epidemiology Studies’ CubeX Software (http://www.oege.org/software/cubex/). ELISA data was compared between CD patients and HD using Student’s t test.
Results and Discussion
All SNPs tested were in Hardy Weinberg Equilibrium (HWE). LD was assessed for two SNPs in the IL6 promoter region (rs1800795 and rs1800797) as well as two SNPs in the IL6R gene (rs2228145 and rs4537545), as they demonstrated similar allele frequencies. Consistent with previous reports, [14], [22] these two IL6 promoter SNPs were found to be in LD (D’ = 1.0, r2 = 0.98), as were the two IL6R SNPs (D’ = 0.977, r2 = 0.77).
Observed allele frequencies for CD patients and HD are depicted in Table 2. A significant difference in the frequency of the minor T allele of IL6R SNP rs4537545 was observed between CD patients and HD (0.49 and 0.33, respectively; p<0.018). Similarly, the minor C allele of IL6R SNP rs2228145 was overexpressed in CD patients versus controls (0.42 vs. 0.30, p<0.068). While these two SNPs exhibit high LD, we observed minor discordance in a few subjects, which resulted in the greater p-value for the rs2228145 SNP. No significant differences in allele frequencies were detected in the other SNPs tested.
Table 2. SNP allele frequencies.
| SNP | Gene | HD (n = 50) | CD (n = 58) | p-value |
| rs1800795 | IL6 | 0.35 | 0.32 | 0.6410 |
| rs1800797 | IL6 | 0.35 | 0.32 | 0.6421 |
| rs1800796 | IL6 | 0.07 | 0.06 | 0.7660 |
| rs4537545 | IL6R | 0.33 | 0.49 | 0.0175 |
| rs2228145 | IL6R | 0.30 | 0.42 | 0.0679 |
| rs10940495 | gp130 | 0.32 | 0.21 | 0.0658 |
| rs715180 | gp130 | 0.09 | 0.09 | 1.0 |
Minor allele frequencies for each of the SNPs tested are depicted for HD and CD patients. IL6 SNPs rs1800795 and rs1800797 are in LD as are the two IL6R SNPs. There is a significant difference between the minor allele frequencies of the IL6R SNP rs4537545 for HD and CD patients (p = 0.0175).
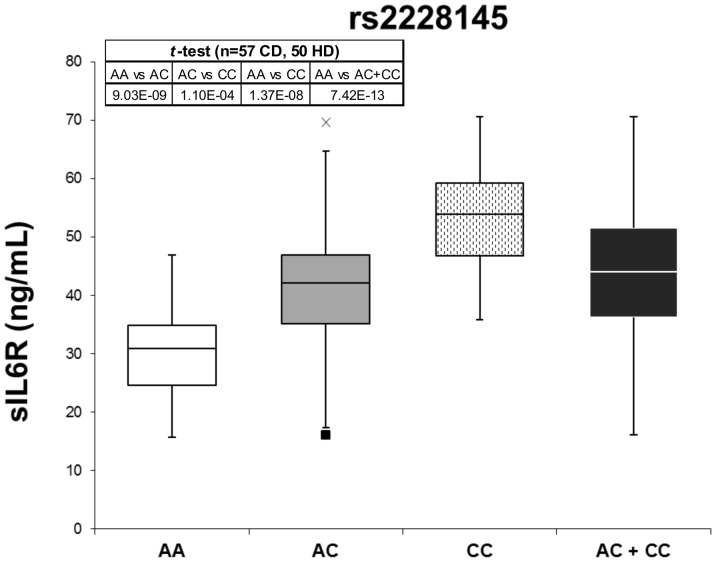
Levels of sIL6R were measured in the serum (or plasma when serum was not available) of CD patients and controls by ELISA. Individuals possessing one or more copies of the minor C allele of IL6R SNP rs2228145 produced significantly higher levels of sIL6R (averages 43.98 ng/mL vs. 29.85 ng/mL, p = 7.42×10−13, Figure 1). Furthermore, individuals possessing at least one copy of the minor T allele of the IL6R SNP rs4537545 expressed higher levels of sIL6R than those homozygous for the major C allele (averages 42.84 ng/mL and 29.36 ng/mL, respectively, data not shown), and this difference was highly significant (p = 2.98×10−12). For both IL6R SNPs, this genotype-dependent difference in sIL6R levels was also observed in both the CD patient and control cohorts independently.
Figure 1. sIL6R levels.
Levels of sIL6R in the serum of CD patients and HD were determined by ELISA. Subjects homozygous for the major allele demonstrate the lowest levels of sIL6R (white). Those who are heterozygous for the rs2228145 SNP exhibit increased levels of sIL6R in the serum (grey), while those who are homozygous for the minor allele (dots) produce the highest levels. Mean of the heterozygous and homozygous minor allele groups are also depicted (black). Min and max outliers for the heterozygous population are noted (black square and x, respectively). Differences in sIL6R levels among genotypes are highly significant.
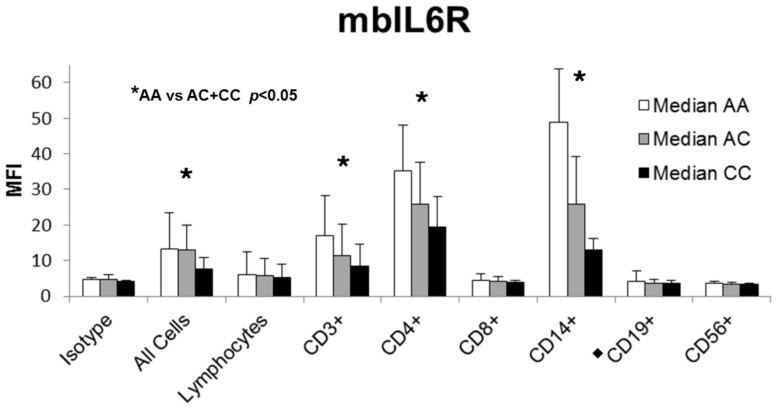
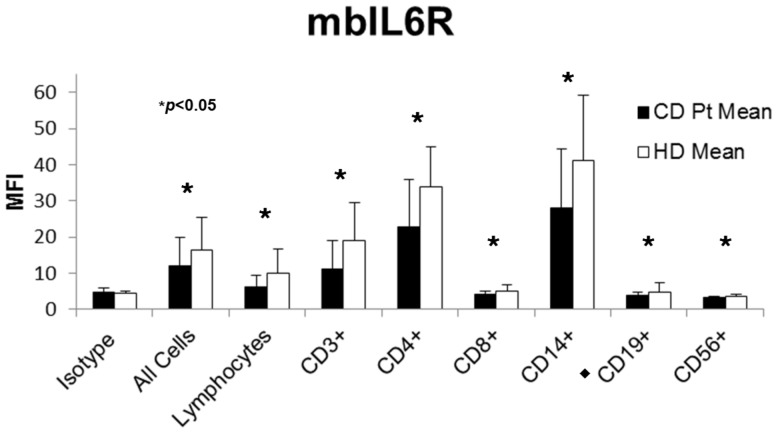
Flow cytometry was performed to assess the levels of mbIL6R as well as gp130 on CD patient and HD PBMC. Antibodies to various lymphocyte and monocyte markers were incorporated to determine if changes in mbIL6R/gp130 levels were localized to a particular cell type. Subjects who were heterozygous for the IL6R SNP rs2228145 demonstrated decreased levels of mbIL6R at the cell surface as compared to those who were homozygous for the major A allele. Subjects homozygous for the minor C allele exhibited the lowest levels of mbIL6R. As was done with ELISA data, statistics were calculated by grouping heterozygous and minor-allele homozygous individuals together, and comparing to major-allele homozygous subjects. We observed a genotype-dependent difference in mbIL6R levels on most cell types studied (Figure 2). Similar results were obtained for the IL6R SNP rs4537545 (data not shown). When mean expression levels for CD patients were compared to HD, we observed that the overall mbIL6R expression is higher on PBMC from HD than CD patients (Figure 3).
Figure 2. mbIL6R levels.
Levels of mbIL6R for CD patients (n = 58) and HD (n = 50) were measured by flow cytometry. Subjects who are homozygous for the major allele exhibit the highest levels of mbIL6R (white). Levels are decreased at the cell surface of heterozygous subjects (grey) and lowest on minor allele-homozygous subjects (black). Reductions were significant on a subset of T cells and monocytes. CD19 data is omitted from 5 patients who received rituximab (black diamond). MFI: median fluorescence intensity.
Figure 3. Mean mbIL6R levels.
Levels of mbIL6R were measured by flow cytometry. Mean mbIL6R levels for HD (n = 50, white) are significantly greater than levels for CD patients (n = 58, black) on all cell types measured. CD19 data is omitted from 5 patients who received rituximab (black diamond).
Next, we assessed whether any correlations existed between these SNPs and IL6 levels in CD patients. Given the strong link between IL6 and CRP expression and the observation that patients with elevated serum CRP (>8 mg/L) can display apparently normal serum IL6 levels (< 10 pg/mL), we hypothesized that commonly used IL6 assays were not able to accurately quantify IL6 in all patients. We therefore employed the Panoptic IL6 assay due to its distinct ability to better quantify IL6 levels (complexed and non-complexed) in human blood (manuscript in preparation). Contrary to the observations made by Reich et al. and Rafiq et al., we did not see an association between genotype and IL6 levels [21], [22]. Variations among commercially-available IL6 assays in the ability to accurately detect IL6 levels could account for our inability to reproduce the results pertaining to IL6/SNP correlations found in others’ studies.
We found that CD patients express the minor allele of two SNPs in the IL6R gene (rs4537545 and rs2228145) at higher frequencies than do HD. Our data, similar to previous reports, [17], [18], [21], [22] showed that individuals possessing one or more copies of the minor allele of these IL6R SNPs exhibited increased levels of sIL6R, and these differences were highly significant. Similar findings were reported by our group in MM patients [17]. We studied the effect of IL6R SNPs on levels of mbIL6R, and observed that individuals who are heterozygous for the IL6R SNPs expressed reduced levels of IL6R at the cell surface, and those homozygous for the minor allele expressed the lowest levels of mbIL6R. This was significant for T cells and monocytes (p<0.05, Figure 2). Further, the mean expression of mbIL6R was greater in HD than CD patients. This data shows that CD patients express IL6R SNPs rs2228145 and rs4537545 more frequently, which leads to increased shedding of IL6R from the cell surface, increased levels of sIL6R in the serum, and presumably increased levels of IL6 signaling. This study identifies for the first time a potentially predisposing factor for MCD, namely the rs2228145 polymorphism in the IL6R, in this rare and complex disorder.
Funding Statement
This work was supported by Janssen Research and Development, a Johnson and Johnson Company. The funders developed the panoptic interleukin-6 assay and performed this testing. The funders had no role in study design, decision to publish, or preparation of the manuscript.
References
- 1. Castleman B, Towne VW (1954) Case records of the Massachusetts General Hospital; weekly clinicopathological exercises; founded by Richard C. Cabot. N Engl J Med 251: 396–400. [DOI] [PubMed] [Google Scholar]
- 2. Castleman B, Iverson L, Menendez VP (1956) Localized mediastinal lymphnode hyperplasia resembling thymoma. Cancer 9: 822–830. [DOI] [PubMed] [Google Scholar]
- 3. Peterson BA, Frizzera G (1993) Multicentric Castleman’s disease. Semin Oncol 20: 636–647. [PubMed] [Google Scholar]
- 4. van Rhee F, Fayad L, Voorhees P, Furman R, Lonial S, et al. (2010) Siltuximab, a novel anti-interleukin-6 monoclonal antibody, for Castleman’s disease. Journal of Clinical Oncology 28: 3701–3708. [DOI] [PubMed] [Google Scholar]
- 5. Kishimoto T (2010) IL-6: from its discovery to clinical applications. International Immunology 22: 347–352. [DOI] [PubMed] [Google Scholar]
- 6. May LT, Santhanam U, Sehgal PB (1991) On the multimeric nature of natural human interleukin-6. Journal of Biological Chemistry 266: 9950–9955. [PubMed] [Google Scholar]
- 7. Nakahara H, Song J, Sugimoto M, Hagihara K, Kishimoto T, et al. (2003) Anti-interleukin-6 receptor antibody therapy reduces vascular endothelial growth factor production in rheumatoid arthritis. Arthritis & Rheumatism 48: 1521–1529. [DOI] [PubMed] [Google Scholar]
- 8. Bataille R, Jourdan M, Zhang XG, Klein B (1989) Serum levels of interleukin 6, a potent myeloma cell growth factor, as a reflect of disease severity in plasma cell dyscrasias. Journal of Clinical Investigation 84: 2008–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lagmay JP, London WB, Gross TG, Termuhlen A, Sullivan N, et al. (2009) Prognostic significance of interleukin-6 single nucleotide polymorphism genotypes in neuroblastoma: rs1800795 (promoter) and rs8192284 (receptor). Clinical Cancer Research 15: 5234–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salgado R, Junius S, Benoy I, Van Dam P, Vermeulen P, et al. (2003) Circulating interleukin-6 predicts survival in patients with metastatic breast cancer. International Journal of Cancer 103: 642–646. [DOI] [PubMed] [Google Scholar]
- 11. Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, et al. (1998) The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. Journal of Clinical Investigation 102: 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guerreiro CS, Ferreira P, Tavares L, Santos PM, Neves M, et al. (2009) Fatty acids, IL6, and TNFalpha polymorphisms: an example of nutrigenetics in Crohn’s disease. American Journal of Gastroenterology 104: 2241–2249. [DOI] [PubMed] [Google Scholar]
- 13. Slattery ML, Wolff RK, Herrick JS, Caan BJ, Potter JD (2007) IL6 genotypes and colon and rectal cancer. Cancer Causes & Control 18: 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cozen W, Gebregziabher M, Conti DV, Van Den Berg DJ, Coetzee GA, et al. (2006) Interleukin-6-related genotypes, body mass index, and risk of multiple myeloma and plasmacytoma. Cancer Epidemiology, Biomarkers & Prevention 15: 2285–2291. [DOI] [PubMed] [Google Scholar]
- 15. Peake NJ, Khawaja K, Myers A, Nowell MA, Jones SA, et al. (2006) Interleukin-6 signalling in juvenile idiopathic arthritis is limited by proteolytically cleaved soluble interleukin-6 receptor. Rheumatology 45: 1485–1489. [DOI] [PubMed] [Google Scholar]
- 16. Mazur G, Bogunia-Kubik K, Wrobel T, Karabon L, Polak M, et al. (2005) IL-6 and IL-10 promoter gene polymorphisms do not associate with the susceptibility for multiple myeloma. Immunology Letters 96: 241–246. [DOI] [PubMed] [Google Scholar]
- 17. Stephens OW, Zhang Q, Qu P, Zhou Y, Chavan S, et al. (2012) An intermediate-risk multiple myeloma subgroup is defined by sIL-6r: levels synergistically increase with incidence of SNP rs2228145 and 1q21 amplification. Blood 119: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Galicia JC, Tai H, Komatsu Y, Shimada Y, Akazawa K, et al. (2004) Polymorphisms in the IL-6 receptor (IL-6R) gene: strong evidence that serum levels of soluble IL-6R are genetically influenced. Genes & Immunity 5: 513–516. [DOI] [PubMed] [Google Scholar]
- 19. Mullberg J, Oberthur W, Lottspeich F, Mehl E, Dittrich E, et al. (1994) The soluble human IL-6 receptor. Mutational characterization of the proteolytic cleavage site. Journal of Immunology 152: 4958–4968. [PubMed] [Google Scholar]
- 20. Jones SA, Horiuchi S, Topley N, Yamamoto N, Fuller GM (2001) The soluble interleukin 6 receptor: mechanisms of production and implications in disease. FASEB Journal 15: 43–58. [DOI] [PubMed] [Google Scholar]
- 21. Reich D, Patterson N, Ramesh V, De Jager PL, McDonald GJ, et al. (2007) Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. American Journal of Human Genetics 80: 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rafiq S, Frayling TM, Murray A, Hurst A, Stevens K, et al. (2007) A common variant of the interleukin 6 receptor (IL-6r) gene increases IL-6r and IL-6 levels, without other inflammatory effects. Genes & Immunity 8: 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scheller J, Rose-John S (2006) Interleukin-6 and its receptor: from bench to bedside. Medical Microbiology & Immunology 195: 173–183. [DOI] [PubMed] [Google Scholar]
- 24. Sehgal PB (1996) Interleukin-6-type cytokines in vivo: regulated bioavailability. Proceedings of the Society for Experimental Biology & Medicine 213: 238–247. [DOI] [PubMed] [Google Scholar]
- 25. Ledur A, Fitting C, David B, Hamberger C, Cavaillon JM (1995) Variable estimates of cytokine levels produced by commercial ELISA kits: results using international cytokine standards. Journal of Immunological Methods 186: 171–179. [DOI] [PubMed] [Google Scholar]
- 26. Krakauer T (1998) Variability in the sensitivity of nine enzyme-linked immunosorbant assays (ELISAs) in the measurement of human interleukin 6. J Immunol Methods 219: 161–167. [DOI] [PubMed] [Google Scholar]
- 27. Schaid DJ, Jacobsen SJ (1999) Biased tests of association: comparisons of allele frequencies when departing from Hardy-Weinberg proportions. American Journal of Epidemiology 149: 706–711. [DOI] [PubMed] [Google Scholar]