Abstract
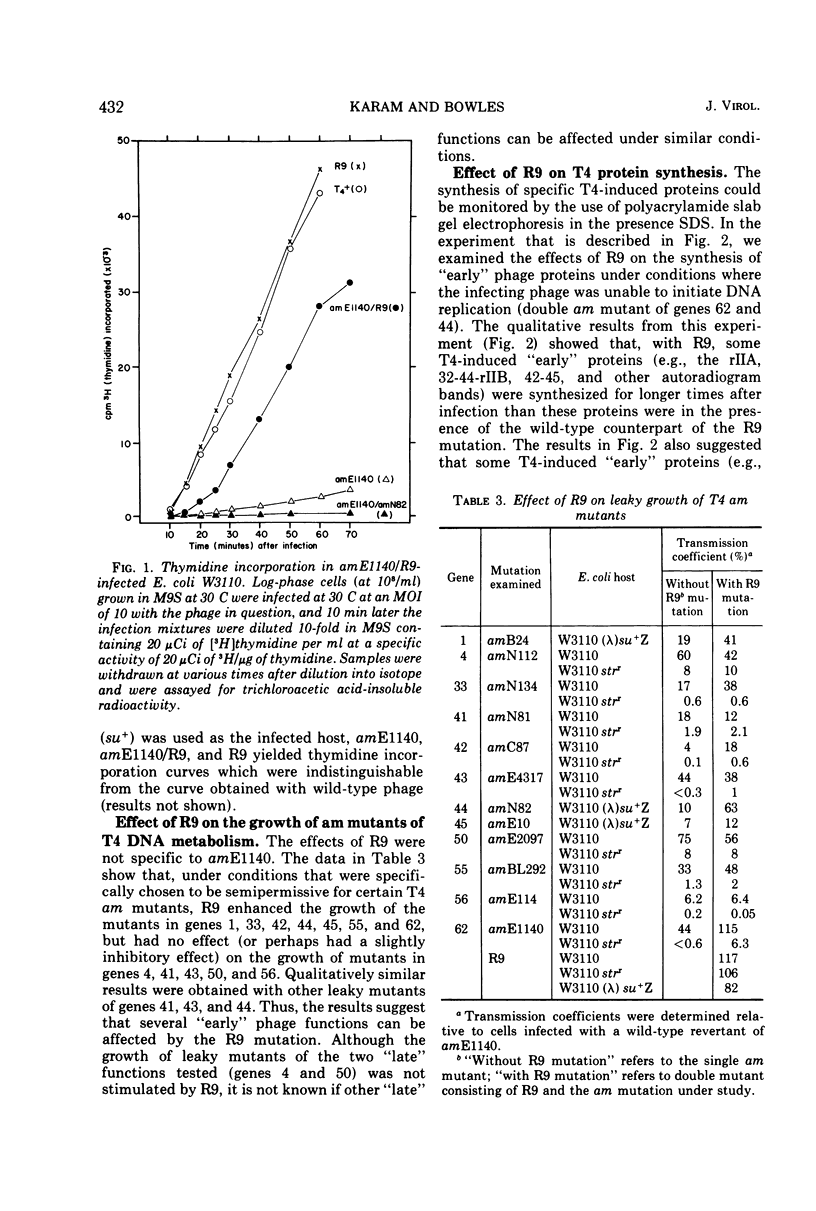
R9 was isolated as one of several mutations that enhanced the growth of a leaky amber (am) mutant of bacteriophage T4 gene 62 (product required for phage DNA synthesis) under conditions of partial suppression by ribosomal ambiguity. R9 also enhanced the growth of leaky am mutants of some, but not all, other T4 “early” gene functions. R9 mapped between mutations in genes 43 and 62. By using assays involving polyacrylamide slab gel electrophoresis in the presence of sodium dodecyl sulfate, we observed the following. (i) R9 resulted in an overproduction of many T4 “early” proteins in infected cells. The most pronounced effects of R9 were observed when phage DNA synthesis and/or the functions of maturation genes 55 and 33 were not expressed. (ii) In rifampintreated infected cells, the capacity to synthesize T4 “early” proteins decayed more slowly in the presence of the R9 mutation than in the presence of the wild-type counterpart of R9. R9 appeared to have no effect on the rates of RNA synthesis either during early or late times after infection. The results suggest that the R9 mutation leads to increased functional stability of T4 “early” messengers.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolle A., Epstein R. H., Salser W., Geiduschek E. P. Transcription during bacteriophage T4 development: synthesis and relative stability of early and late RNA. J Mol Biol. 1968 Feb 14;31(3):325–348. doi: 10.1016/0022-2836(68)90413-0. [DOI] [PubMed] [Google Scholar]
- Bremer H., Yuan D. Chain growth rate of messenger RNA in Escherichia coli infected with bacteriophage T4. J Mol Biol. 1968 Jun 28;34(3):527–540. doi: 10.1016/0022-2836(68)90178-2. [DOI] [PubMed] [Google Scholar]
- CHAMPE S. P., BENZER S. Reversal of mutant phenotypes by 5-fluorouracil: an approach to nucleotide sequences in messenger-RNA. Proc Natl Acad Sci U S A. 1962 Apr 15;48:532–546. doi: 10.1073/pnas.48.4.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. S., Wood W. B. Morphogenesis of bacteriophage T4 in extracts of mutant-infected cells. Proc Natl Acad Sci U S A. 1966 Mar;55(3):498–505. doi: 10.1073/pnas.55.3.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granboulan P., S echaud J., Kellenberger E. On the fragility of phage T4-related particles. Virology. 1971 Nov;46(2):407–425. doi: 10.1016/0042-6822(71)90042-0. [DOI] [PubMed] [Google Scholar]
- Groner Y., Pollack Y., Berissi H., Revel M. Cistron specific translation control protein in Escherichia coli. Nat New Biol. 1972 Sep 6;239(88):16–19. doi: 10.1038/newbio239016a0. [DOI] [PubMed] [Google Scholar]
- Guha A., Szybalski W., Salser W., Geiduschek E. P., Pulitzer J. F., Bolle A. Controls and polarity of transcription during bacteriophage T4 development. J Mol Biol. 1971 Jul 28;59(2):329–349. doi: 10.1016/0022-2836(71)90054-4. [DOI] [PubMed] [Google Scholar]
- Horvitz H. R. Polypeptide bound to the host RNA polymerase is specified by T4 control gene 33. Nat New Biol. 1973 Aug 1;244(135):137–140. doi: 10.1038/newbio244137a0. [DOI] [PubMed] [Google Scholar]
- Karam J. D., O'Donnell P. V. Suppression of amber mutations of bacteriophage T4 gene 43 (DNA polymerase) by translational ambiguity. J Virol. 1973 Jun;11(6):933–945. doi: 10.1128/jvi.11.6.933-945.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam J. D., Speyer J. F. Reversible inactivation of T4 ts DNA polymerase mutants in vivo. Virology. 1970 Sep;42(1):196–203. doi: 10.1016/0042-6822(70)90252-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee-Huang S., Ochoa S. Purification and properties of two messenger-discriminating species of E. coli initiation factor 3. Arch Biochem Biophys. 1973 May;156(1):84–96. doi: 10.1016/0003-9861(73)90344-5. [DOI] [PubMed] [Google Scholar]
- Mosig G. A preferred origin and direction of bacteriophage T4 DNA replication. I. A gradient of allele frequencies in crosses between normal and small T4 particles. J Mol Biol. 1970 Nov 14;53(3):503–514. doi: 10.1016/0022-2836(70)90080-x. [DOI] [PubMed] [Google Scholar]
- Nierlich D. P., Vielmetter W. Kinetic studies on the relationship of ribonucleotide precursor pools and ribonucleic acid synthesis. J Mol Biol. 1968 Feb 28;32(1):135–147. doi: 10.1016/0022-2836(68)90151-4. [DOI] [PubMed] [Google Scholar]
- Notani G. W. Regulation of bacteriophage T4 gene expression. J Mol Biol. 1973 Jan 10;73(2):231–249. doi: 10.1016/0022-2836(73)90326-4. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. Z., Gold L. M. Bacteriophage T4 gene expression. Evidence for two classes of prereplicative cistrons. J Biol Chem. 1973 Aug 10;248(15):5502–5511. [PubMed] [Google Scholar]
- Reid P., Speyer J. Rifampicin inhibition of ribonucleic acid and protein synthesis in normal and ethylenediaminetetraacetic acid-treated Escherichia coli. J Bacteriol. 1970 Oct;104(1):376–389. doi: 10.1128/jb.104.1.376-389.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski P. D., Warner H. R., Hercules K., Munro J. L., Mendelsohn S., Wiberg J. S. Mutants of bacteriophage T4 defective in the induction of T4 endonuclease II. J Biol Chem. 1971 May 25;246(10):3431–3433. [PubMed] [Google Scholar]
- Salser W., Bolle A., Epstein R. Transcription during bacteriophage T4 development: a demonstration that distinct subclasses of the "early" RNA appear at different times and that some are "turned off" at late times. J Mol Biol. 1970 Apr 28;49(2):271–295. doi: 10.1016/0022-2836(70)90246-9. [DOI] [PubMed] [Google Scholar]
- Sauerbier W., Hercules K. Control of gene function in bacteriophage T4. IV. Post-transcriptional shutoff of expression of early genes. J Virol. 1973 Sep;12(3):538–547. doi: 10.1128/jvi.12.3.538-547.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snustad D. P. Dominance interactions in Escherichia coli cells mixedly infected with bacteriophage T4D wild-type and amber mutants and their possible implications as to type of gene-product function: catalytic vs. stoichiometric. Virology. 1968 Aug;35(4):550–563. doi: 10.1016/0042-6822(68)90285-7. [DOI] [PubMed] [Google Scholar]
- Stevens A. New small polypeptides associated with DNA-dependent RNA polymerase of Escherichia coli after infection with bacteriophage T4. Proc Natl Acad Sci U S A. 1972 Mar;69(3):603–607. doi: 10.1073/pnas.69.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter G., Seifert W., Zillig W. Modified DNA-dependent RNA polymerase from E. coli infected with bacteriophage T4. Biochem Biophys Res Commun. 1968 Feb 15;30(3):240–247. doi: 10.1016/0006-291x(68)90441-5. [DOI] [PubMed] [Google Scholar]
- Wiberg J. S., Mendelsohn S., Warner V., Hercules K., Aldrich C., Munro J. L. SP62, a viable mutant of bacteriophage T4D defective in regulation of phage enzyme synthesis. J Virol. 1973 Oct;12(4):775–792. doi: 10.1128/jvi.12.4.775-792.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]