Abstract
Biotin carboxylase (BC) is a conserved component among biotin-dependent carboxylases and catalyzes the MgATP-dependent carboxylation of biotin, using bicarbonate as the CO2 donor. Studies with E. coli BC have suggested long-range communication between the two active sites of a dimer, although its mechanism is not well understood. In addition, mutations in the dimer interface can produce stable monomers that are still catalytically active. A homologous dimer for the BC domain is observed in the structure of tetrameric pyruvate carboxylase (PC) holoenzyme. We have introduced site-specific mutations in the BC domain dimer interface of S. aureus PC (SaPC), equivalent to those used for E. coli BC, and also made chimeras replacing the SaPC BC domain with the E. coli BC subunit (EcBC chimera) or the yeast ACC BC domain (ScBC chimera). We assessed the catalytic activities of these mutants and characterized their oligomerization states by gel filtration and analytical ultracentrifugation experiments. The K442E mutant and the ScBC chimera disrupted the BC dimer and were catalytically inactive, while the F403A mutant and the EcBC chimera were still tetrameric and retained catalytic activity. The R54E mutant was also tetrameric but was catalytically inactive. Crystal structures of the R54E, F403A and K442E mutants showed that they were tetrameric in the crystal, with conformational changes near the mutation site as well as in the tetramer organization. We have also produced the isolated BC domain of SaPC. In contrast to E. coli BC, the SaPC BC domain is monomeric in solution and catalytically inactive.
Introduction
Biotin-dependent carboxylases are widely distributed in nature and have important functions in many metabolic processes (1–5). They carry two distinct catalytic activities (6). A biotin carboxylase (BC) activity catalyzes the Mg2+-ATP-dependent carboxylation of biotin, using bicarbonate as the CO2 donor. Then a carboxyltransferase (CT) activity transfers the carboxyl group from carboxybiotin to the acceptor, which oftentimes is the coenzyme A (CoA) ester of an organic acid (acetyl-CoA, propionyl-CoA, etc) but can also be a small compound (pyruvate, urea). Biotin is covalently linked to a biotin carboxyl carrier protein (BCCP) component.
The BC activity is common among these enzymes, and the amino acid sequences of this component are well conserved among them. The BC subunit of the E. coli acetyl-CoA carboxylase (ACC) has been used as a model system to study this activity (7–10). It can catalyze the carboxylation of free biotin, in the absence of the CT and BCCP subunits of the ACC holoenzyme. The crystal structures of its free enzyme (11), ATP complex (12, 13), and the pentanary complex with Mg-ADP, biotin and bicarbonate (14) are all available. The BC structure contains three domains, named A, B, and C domains, and the active site is located at the interface among them (Fig. 1A). Domain B undergoes a large rearrangement and closes over the active site during catalysis (12, 14).
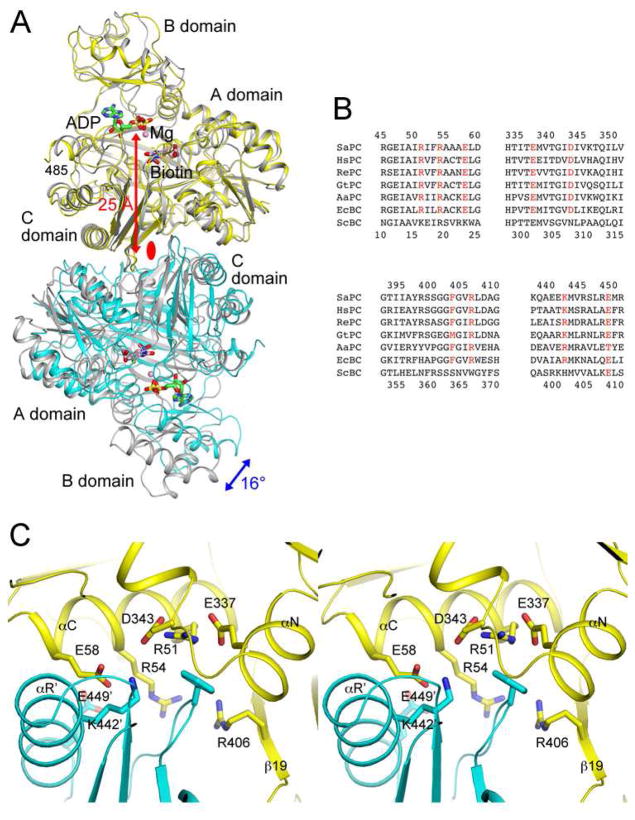
Figure 1. Conserved BC domain dimer interface for SaPC and E. coli BC.
(A). Overlay of the structures of the BC domain dimer of SaPC free enzyme (in yellow and cyan for the two monomers) (25) and the E. coli BC dimer (in gray) in complex with ADP (in green), Mg2+ (pink sphere), bicarbonate (in black), and biotin (in pink) (14). The blue arrow indicates the difference in orientation for the second monomer of the dimer between the two structures, and the red arrow indicates the distance from the dimer interface to the active site. The two-fold axis of the dimer is indicated with the red oval. (B). Alignment of amino acid sequences in the dimer interface region for SaPC, human PC (HsPC), RePC, Geobacillus thermodenitrificans PC (GtPC), Aquifex aeolicus PC (AaPC), E. coli BC (EcBC) and the BC domain of yeast ACC (ScBC). Residues having important interactions in the interface are shown in red. Residue numbers at the top are for SaPC (in human PC numbering), and those at the bottom are for E. coli BC. (C). Detailed interactions at the BC domain dimer interface of SaPC. Residues Arg54, Glu58, Phe403 and Lys442 were selected for mutagenesis. The primed residue numbers indicate the second monomer. All structure figures were produced with PyMOL (www.pymol.org).
The E. coli BC subunit is a stable dimer, formed by interaction among the A and C domains of the two monomers (Fig. 1A). The active site of each monomer is located ~25 Å away from the dimer interface, with no contribution from residues in the other monomer. However, mutagenesis studies showed that knocking out the active site of one monomer of the dimer essentially abolished catalysis at the other active site, suggesting long-range communications between the two active sites and the importance of dimerization for BC catalysis (15). An asymmetric structure of E. coli BC with only one of the two active sites occupied by an ATP analog (ADPCF2P) has been observed (13), and mathematical simulations also support this half-sites reactivity (16). In addition, the BC domain of eukaryotic ACC is monomeric in solution and is catalytically inactive (17, 18), in apparent agreement with this model.
On the other hand, mutations in the dimer interface of E. coli BC can generate stable monomers of the enzyme, and these monomeric mutants can still catalyze the BC reaction (19, 20). Moreover, the BC domain is monomeric in the α6β6 holoenzyme of propionyl-CoA carboxylase (21), shows trimeric association in the α6β6 holoenzyme of 3-methylcrotonyl-CoA carboxylase (22), and is monomeric in urea carboxylase (23). Therefore, dimerization per se may not be required for the catalytic activity of BC. The mechanism for the long-range communication between the two active sites in the E. coli BC dimer is still not fully understood.
The BC domain dimer in the structures of the tetrameric holoenzyme of pyruvate carboxylase (PC) (24–27) has a similar organization as the BC subunit dimer of E. coli ACC (Fig. 1A). Residues in the dimer interface of the two enzymes are generally conserved (Fig. 1B) and show similar interactions (Fig. 1C). To further study the importance of the BC dimer interface for catalysis, we have introduced mutations in this interface in full-length Staphylococcus aureus PC (SaPC) as well as its isolated BC domain. The mutations are equivalent to those that destabilized the dimer of E. coli BC (19), and include R54E, E58R, F403A, and K442E. In addition, we have created chimeric PCs, replacing its BC domain with the BC subunit of E. coli ACC (EcBC chimera) or the BC domain of yeast ACC (ScBC chimera). Our studies showed that the K442E mutant and the ScBC chimera have disrupted the tetramer of PC in solution and are catalytically inactive.
Materials and Methods
Mutagenesis, protein expression and purification
The dimer interface mutants of full-length SaPC were made using the QuikChange kit (Stratagene) and sequenced to verify successful mutagenesis. The mutants were over-expressed in E. coli BL21(DE3) Star cells at 20°C and purified by Ni-NTA and Sephacryl-S300 (GE Healthcare) gel filtration chromatography. The expression construct introduced an N-terminal hexa-histidine tag into the protein. The purified protein was concentrated to 20 mg/ml and flash frozen with liquid nitrogen in a buffer containing 20 mM Tris (pH 7.5), 200 mM NaCl, 2 mM dithiothreitol (DTT), and 5% (v/v) glycerol.
The EcBC and ScBC chimeras were designed based on the overlay of the structures of E. coli and S. cerevisiae BC with the BC domain of SaPC. A residue close to the C-terminus of the E. coli or S. cerevisiae BC, located in a hydrophilic segment on the surface, and structurally similar to SaPC, was chosen as the connection point for the chimera. The chimera constructs were made by overlapping PCR.
The isolated BC domain of SaPC was created by introducing a STOP codon after amino acid 455 (486 in human PC numbering) through mutagenesis. Mutations in the dimer interface were then introduced, equivalent to those for full-length SaPC. The expression and purification of these BC proteins were carried out following the same protocol as used for the full-length protein.
Analytical ultracentrifugation (AUC)
The sedimentation velocity (SV) experiments were performed using an XL-A analytical ultracentrifuge with a standard 12-mm double-sector epon charcoal-filled centerpiece (Beckman, Fullerton, CA). The sample (330 μl) and reference (370 μl) solutions were loaded into the centerpiece and mounted in an An-50 Ti rotor. The experiments were performed at 20 °C with a rotor speed of 42,000 rpm. The absorbance at 280 nm was chosen to detect the protein, which was monitored in a continuous mode with a time interval of 480 s and a step size of 0.003 cm. Three different protein concentrations (0.4 to 10 μM) were used to estimate the dynamic monomer-dimer or monomer-dimer-tetramer association. The SV results at three protein concentrations were then globally analyzed using a monomer-dimer or monomer-dimer-tetramer equilibrium model by the SEDPHAT program (28).
To characterize the size distributions of the various proteins in solution, the scans at different time intervals were fitted to a continuous c(s) distribution model using the SEDFIT program (29, 30). The size distributions for sedimentation coefficients between 0 and 20 S are solved and regularized at a confidence level of p = 0.95 by maximum entropy.
Enzyme assays
The catalytic activity of full-length wild-type and mutant SaPC was determined spectrophotometrically at 340 nm, following a published protocol that couples the production of oxaloacetate to the oxidation of NADH (31). Pyruvate titrations (up to 40 mM) were performed in the absence of acetyl-CoA. The effect of acetyl-CoA on SaPC activity was assayed at 5 mM pyruvate, which is the Km for this substrate for wild-type SaPC.
The catalytic activity of the BC domain of SaPC was determined following an established ATP hydrolysis assay (9), which couples ADP production to NADH oxidation. The reaction (0.2 ml) contained 100 mM Hepes (pH 8.5), 8 mM MgCl2, 40 mM biotin, 200 mM NaCl, 50 mM Na2CO3, 0.2 mM NADH, 0.5 mM phosphoenolpyruvate, 20 U/ml lactate dehydrogenase/pyruvate kinase, 400 nM SaPC BC domain (in terms of the monomer), and varying concentrations of ATP, which was added last to the reaction mixture.
Protein crystallization
Full-length SaPC carrying the R54E, F403A, or K442E mutation was crystallized under the same condition as wild-type SaPC (25). The protein, at 10 mg/ml, was incubated with 5 mM ATP at 4 °C for 1 h, and then mixed 1:1 with a reservoir solution that contained 200 mM ammonium tartrate and 20% (w/v) PEG3350. Crystals were grown using the sitting-drop vapor diffusion method at 21°C, cryoprotected with a solution consisting of the reservoir solution supplemented with 20% (v/v) ethylene glycol, and flash frozen in liquid nitrogen for data collection at 100K.
Extensive efforts at trying to crystallize the BC domain of SaPC (wild-type and mutants) were unsuccessful.
Data collection, structure determination and refinement
X-ray diffraction data were collected at the X29A beamline of the National Synchrotron Light Source (NSLS). The diffraction images were processed and scaled with the HKL package (32). The mutant crystals have similar unit cell parameters as those of wild-type SaPC (25), and there is one tetramer in the asymmetric unit. The structures were solved by the molecular replacement method with the program COMO (33), using the structure of wild-type SaPC as the search model (25). Structure refinement was carried out with the programs CNS (34) and Refmac (35), and manual rebuilding of the atomic models was performed with O (36) and Coot (37). The data processing and refinement statistics are summarized in Table 1. The difference between R and free R values is ~7% for the three structures, indicating possibly some over-fitting in the model.
Table 1.
Summary of Crystallographic Information
| Protein | R54E | F403A | K442E |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 96.4, 256.7, 127.0 | 96.2, 256.3, 126.7 | 96.6, 258.5, 126.9 |
| α, β, γ (°) | 90, 109.3, 90 | 90, 109.8, 90 | 90, 109.6, 90 |
| Resolution range (Å) a | 30-2.8 (2.9-2.8) | 30-2.8 (2.9-2.8) | 30-3.0 (3.1-3.0) |
| Rmerge (%) | 12.5 (44.7) | 9.2 (46.5) | 6.5 (42.9) |
| I/σI | 10.5 (3.5) | 11.7 (2.1) | 15.5 (2.5) |
| Completeness (%) | 93 (86) | 91 (94) | 98 (99) |
| Redundancy | 4.7 (4.8) | 2.8 (2.8) | 2.8 (2.7) |
| Refinement | |||
| Number of reflections | 125,986 | 122,029 | 109,041 |
| R(%) | 19.4 (26.6) | 20.9 (25.7) | 19.4 (24.5) |
| Rfree (%) | 26.2 (35.2) | 27.9 (35.3) | 26.2 (33.1) |
| Number of atoms | |||
| Protein | 34,009 | 32,422 | 32,401 |
| Ligand | 63 | 62 | 31 |
| Water | 0 | 0 | 0 |
| Average B values (Å2) | |||
| Protein | 26.2 | 49.0 | 40.2 |
| Ligand | 81.6 | 101.1 | 118.8 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.011 | 0.013 | 0.011 |
| Bond angles (°) | 1.3 | 1.4 | 1.2 |
The numbers in parentheses are for the highest-resolution shell.
Results and Discussion
Design of mutations in the BC dimer interface of SaPC
The BC domain dimer of SaPC has a similar overall organization as the E. coli BC dimer (Fig. 1A). With one monomer of the two dimers in superposition, the orientation of the other monomer differs by a 16° rotation. Residues that have interactions in the E. coli BC dimer interface are generally conserved in SaPC (Fig. 1B), and show similar interactions in its BC dimer interface as well (Fig. 1C). For example, Arg54 and Glu58 (equivalent to Arg19 and Glu23 of E. coli BC, Fig. 1B) have ion-pair interactions with Glu449′ and Lys442′ (Glu408′ and Arg401′ in E. coli BC), respectively (with the primed residue numbers indicating the other monomer) (Fig. 1C). Residues in SaPC are numbered according to their equivalents in human PC unless noted otherwise (25). Arg51 (Arg16 in E. coli BC) has ion-pair interactions with Glu337 and Asp343, which are in the binding site for Phe403′ (Phe363′ in E. coli BC).
Previous studies with E. coli BC showed that the R16E, R19E and E23R mutations greatly destabilized the BC dimer, giving rise to 5,000–8,000 fold increases in the Kd, while the F363A mutation led to only a four-fold increase in the Kd of the dimer (19, 20). The R401E mutant of E. coli BC was also produced, but the resulting protein was mostly aggregated (19).
To study the effects of the BC dimer interface on SaPC catalysis, we created four single-site mutants in this interface, R54E, E58R, F403A and K442E, which are equivalent to the R19E, E23R, F363A and R401E mutants in the E. coli dimer interface that we studied earlier (19). In addition, we created two chimeric PC proteins, replacing the BC domain of SaPC (residues 1–448 in SaPC numbering, 34–481 in human PC numbering) with the BC subunit of E. coli BC (residues 1–440) or the BC domain (residues 1–562) of yeast ACC (17). We expect that the chimera with the E. coli BC (to be referred to as the EcBC chimera) may be able to maintain the tetrameric organization of wild-type SaPC and might be catalytically active as well. In contrast, the chimera with the yeast BC domain (to be referred to as the ScBC chimera) may no longer be tetrameric, as the yeast BC domain itself is monomeric (17). Many of the residues in the dimer interface of E. coli BC are not conserved in yeast BC (Fig. 1B). More importantly, yeast BC has a different structure in this region, which is incompatible with the mode of dimerization for E. coli BC (17).
Mutations in the BC dimer interface can disrupt the SaPC tetramer
Wild-type SaPC and the various mutants were over-expressed in E. coli and purified by nickel affinity and gel filtration chromatography. The gel filtration profiles show that wild-type SaPC, the R54E and F403A single-site mutants, and the EcBC chimera migrated at the same position, indicating that they are tetrameric during purification (Fig. 2A). In contrast, the K442E mutant and the ScBC chimera migrated slower on the gel filtration column, suggesting that they may have become dimeric. Finally, the E58R mutant produced aggregated proteins, and this mutant was not studied further.
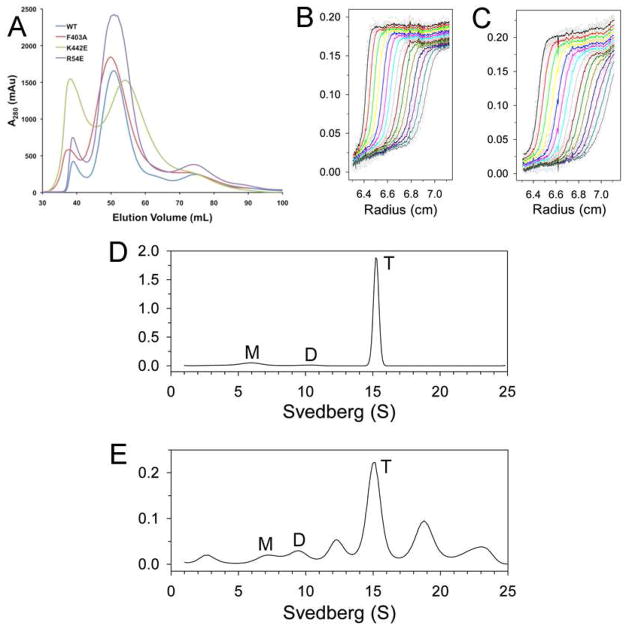
Figure 2. Oligomerization states of the dimer interface mutants in solution.
(A). Gel filtration profiles (from an S300 column) for wild-type SaPC and the dimer interface mutants. (B). Sedimentation velocity AUC data for wild-type SaPC at 1.5 μM concentration. The observed data are shown as open circles, and the theoretical fit to the data based on a rapid monomer-dimer-tetramer association model is shown as the curves. (C). Sedimentation velocity AUC data for the K442E mutant at 1.5 μM concentration. (D). Size distributions of wild-type SaPC in solution at 1 mg/ml concentration, based on the best-fit results by the continuous size distribution analysis (30). M: monomer, D: dimer, T: tetramer. (E). Size distributions of the EcBC chimera in solution at 1 mg/ml concentration.
We next carried out analytical ultracentrifugation (AUC) experiments to study in more detail the oligomerization behavior of these proteins. Sedimentation velocity data were fit to a rapid monomer-dimer-tetramer association model, from which the Kd values for monomer-dimer and dimer-tetramer association were determined. Wild-type SaPC has a Kd of 0.75 μM for the monomer-dimer equilibrium and 8 nM for the dimer-tetramer association (Fig. 2B, Table 2). The R54E and F403A mutants behaved essentially the same as the wild-type enzyme in these experiments. In contrast, the K442E mutant had a Kd of 840 μM for the dimer-tetramer association (Fig. 2C), consistent with the gel filtration data and confirming that this mutation has disrupted the tetramer of SaPC. The AUC data also showed that the EcBC chimera formed a tetramer in solution, although the Kd for the dimer-tetramer association was ~35-fold higher compared to wild-type SaPC. The ScBC chimera is dimeric in solution, consistent with our expectations and the gel filtration data.
Table 2.
Summary of analytical ultracentrifugation and kinetic data
| Protein | Kd (μM)b (monomer- dimer) | Kd (μM)b (dimer- tetramer) | kcat (s−1) | Km (mM) (for pyruvate) | kcat/Km (s−1M−1) |
|---|---|---|---|---|---|
| Full-length SaPC a | |||||
| Wild-type | 0.75±0.06 | 0.008±0.0006 | 20.2±1.4 | 3.4±0.8 | 5900±1400 |
| R54E | 0.53±0.03 | 0.003±0.0001 | N.A.c | N.A. | N.A. |
| F403A | 0.45±0.04 | 0.011±0.0006 | 1.3±0.1 | 1.4±0.4 | 930±270 |
| K442E | 0.82±0.09 | 840±190 | N.A. | N.A. | N.A. |
| EcBC chimera | 2.31±0.33 | 0.28±0.03 | 1.1±0.1 | 0.34±0.02 | 3300±200 |
| ScBC chimera | 0.56±0.06 | 68±11 | N.A. | N.A. | N.A. |
| BC domain of SaPC a | |||||
| Wild-type | 48±4 | — | N.A. | N.A. | N.A. |
| R54E | 195±7 | — | N.A. | N.A. | N.A. |
| F403A | 130±6 | — | N.A. | N.A. | N.A. |
| K442E | 86±8 | — | N.A. | N.A. | N.A. |
The protein concentrations for full-length SaPC were 0.4, 1.5 and 7.4 μM (based on monomer) in the AUC experiment, and those for the BC domain were 1, 4, and 10 μM.
For full-length SaPC, the Kd values were the best-fit results based on a rapid monomer-dimer-tetramer association model, and the local root-mean-square deviation (rmsd) was from 0.0041 to 0.0210. For BC domain alone, a rapid monomer-dimer association model was used, and the local rmsd was from 0.0037 to 0.0070.
N.A. – No activity was observed under the condition tested.
The Kd for monomer-dimer association of the two mutants that were dimeric in solution, K442E and the ScBC chimera, were essentially the same as wild-type SaPC (Table 2). The SaPC tetramer is formed by interactions at the BC, CT and PT (PC tetramerization domain) dimer interfaces (25). The AUC data on these mutants suggest that the CT and PT domains may be sufficient to mediate their dimerization.
To further characterize the oligomerization behavior of these proteins, we used AUC sedimentation velocity experiments to examine their size distributions in solution at 1 mg/ml concentration. Wild-type SaPC (Fig. 2D) and the R54E and F403A mutant were predominantly tetrameric, with roughly 10% of the species being monomeric under the experimental condition tested (Table 3). In contrast, the K442E mutant and the ScBC chimera were mostly dimeric in solution, with the K442E mutant also showing signs of aggregation (Table 3). Somewhat surprisingly, while the EcBC chimera was able to form tetramers in solution, it also showed a significant amount of aggregation (Fig. 2E, Table 3).
Table 3.
Size distributions of SaPC in solution
| Proteina | Monomer (%) | Dimer (%) | Tetramer (%) | Other (%) |
|---|---|---|---|---|
| Full-length SaPC | ||||
| Wild-type | 11 | 2 | 85 | <5 |
| R54E | 4 | 0 | 96 | <5 |
| F403A | 9 | 2 | 85 | <5 |
| K442E | 10 | 65 | — | ~25 |
| EcBC chimera | 5 | 7 | 34 | ~55 |
| ScBC chimera | 22 | 71 | — | <10 |
| BC domain of SaPC | ||||
| Wild-type | 37 | 20 | — | ~40 |
| R54E | 86 | 9 | — | ~5 |
| F403A | 67 | 12 | — | ~20 |
| K442E | 41 | 14 | — | ~45 |
The distribution was observed by AUC. The protein concentration was at 1 mg/ml (~7.5 μM for full-length SaPC and ~20 μM for the isolated BC domain, based on the monomer).
Mutations in the BC dimer interface can disrupt SaPC catalysis
We next characterized the catalytic activity of the various proteins, using a coupled enzyme assay that converted the oxaloacetate product of pyruvate carboxylation to NADH oxidation (31). Good catalytic activity was observed for the wild-type enzyme (Table 2). Among the dimer interface mutants, only the F403A mutant showed catalytic activity, with ~6-fold lower kcat/Km as compared to the wild-type enzyme. The concentration of the enzyme in these assays was ~0.1 μM (based on the monomer), and therefore the observed lack of activity for the K442E mutant and the ScBC chimera is consistent with the AUC data. These proteins are dimeric in solution at this concentration, but PC is only active in the tetrameric form. On the other hand, it is not clear why the R54E mutant is inactive, as it is a tetramer in solution. The EcBC chimera had ~2-fold lower kcat/Km compared to the wild-type enzyme, although its kcat is ~20-fold lower (Table 2). We also tested the intrinsic ATPase activity of the ScBC chimera, and found very weak (if any) activity.
Wild-type SaPC, like most other PCs, is activated by acetyl-CoA. This compound binds at the interface between two BC domains of the tetramer (24, 26), and it can also rescue the catalytic activity of a mutant carrying a change in the PT dimer interface of SaPC (25). In contrast, acetyl-CoA had no effect on the catalytic activity of the BC dimer interface mutants and the chimeras studied here.
The dimer interface mutants are still tetrameric in the crystal
To characterize the effects of mutations in the BC domain dimer interface on the structure of SaPC, we next determined the crystal structures of the R54E, F403A and K442E mutants at up to 2.8 Å resolution (Table 1). The mutants crystallized under the same condition as wild-type SaPC. The crystal structures have been deposited at the Protein Data Bank (accession codes 4HNT, 4HNU, 4HNV). The R54E and F403A mutants existed as tetramers in the crystal, as the mutations did not affect the Kd (Table 3). Surprisingly, the K442E mutant was also found to be tetrameric in the crystal, even though the Kd for its tetramer is 840 μM (Table 2). The high concentration of the protein in the crystalline environment (~50 μM in the crystallization solution, and ~4.5 mM in the crystal), and the presence of the ATP substrate, may have promoted the formation of the tetramer. It is also possible that the crystal stabilized and selectively sequestered the tetrameric form of the mutant.
The structures of the individual domains of the mutants are similar to their equivalents in the wild-type enzyme, with rms distances of 0.4–0.6 Å for Cα atoms. The overall structure of the R54E mutant tetramer is also similar to the wild-type tetramer (Fig. 3A). The second monomer of the mutant BC dimer shows only a 2° rotation relative to that of the wild-type dimer, after the first monomers of the two dimers are superimposed (Fig. 3B). In the dimer interface, the side chain of Arg406 rearranges to interact with the new Glu54 side chain, while most of the other residues show only small changes (Fig. 3C).
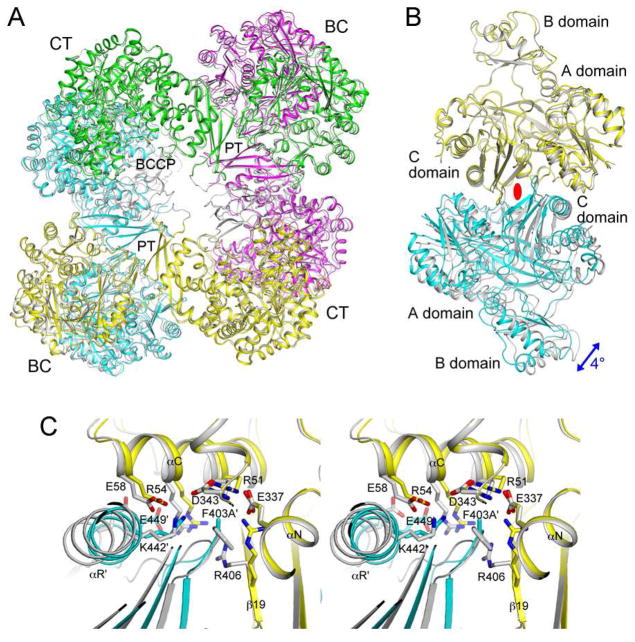
Figure 3.
The R54E mutant has only small structural differences compared to wild-type SaPC. (A). Overlay of the structures of the R54E mutant tetramer (in color for the four monomers, yellow, magenta, cyan and green) and wild-type SaPC (gray). The superposition is based on the BC domain of the first monomer (yellow). (B). Overlay of the structures of the R54E mutant BC domain dimer (yellow and cyan) and wild-type SaPC dimer (gray). The blue arrow indicates the difference in orientation for the second monomer of the dimer between the two structures. The two-fold axis of the dimer is indicated with the red oval. (C). Close-up of the BC domain dimer interface near the mutation site.
In comparison, larger changes in the organization of the tetramer (Fig. 4A) and dimer (Fig. 4B) are observed for the F403A mutant, even though the mutation did not affect the Kd of the tetramer (Table 2). A relative rotation of 4° is observed for the second monomer of the two dimers (Fig. 4B). In the absence of the Phe403 side chain, the side chain of Arg406 undergoes a large conformational change, with its guanidinium group moving by ~5.5 Å (Fig. 4C). The position of this side chain in the wild-type structure is no longer compatible with the dimer interface of the mutant as it clashes with the loop containing the mutation site (F403A). A change in the position of the αC helix is also observed (Fig. 4C), possibly coupled with the re-organization of the dimer interface.
Figure 4.
The F403A mutant shows large structural differences compared to wild-type SaPC. (A). Overlay of the structures of the F403A mutant tetramer (in color) and wild-type SaPC (gray). The superposition is based on the BC domain of the first monomer (yellow). (B). Overlay of the structures of the F403A mutant BC domain dimer (yellow and cyan) and wild-type SaPC dimer (gray). (C). Close-up of the BC domain dimer interface near the mutation site.
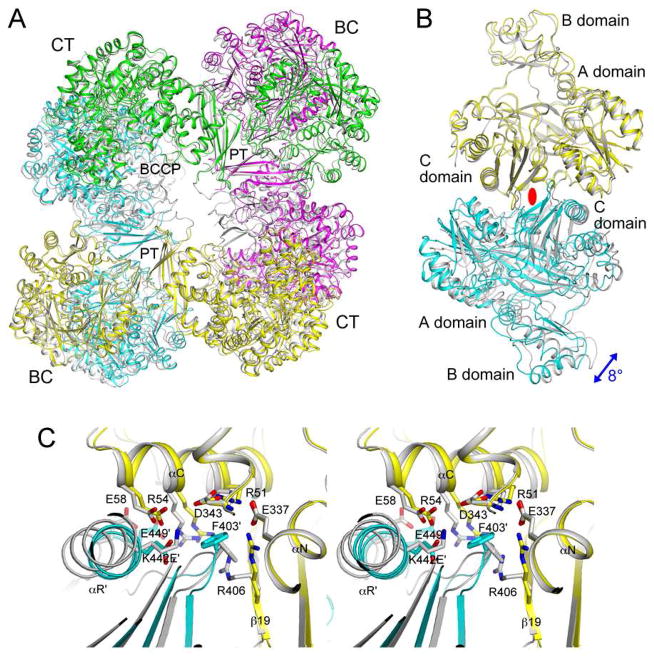
The K442E mutant also shows large changes in the organization of the tetramer (Fig. 5A) and dimer (Fig. 5B) as compared to wild-type SaPC. Rotation is 8° for the second monomer. Remarkably, the conformational changes in the dimer interface itself is rather similar to that observed for the F403A mutant (Fig. 4F), although the structures of the two mutants differ from each other away from the interface.
Figure 5.
The K442E mutant is a tetramer in the crystal but shows large structural differences compared to wild-type SaPC. (A). Overlay of the structures of the K442E mutant tetramer (in color) and wild-type SaPC (gray). The superposition is based on the BC domain of the first monomer (yellow). (B). Overlay of the structures of the K442E mutant BC domain dimer (yellow and cyan) and wild-type SaPC dimer (gray). (C). Close-up of the BC domain dimer interface near the mutation site.
The isolated BC domain of SaPC is monomeric and catalytically inactive
The tetramerization of PC involves the BC, CT and PT domains. Mutations in the BC domain dimer interface that could be detrimental may be compensated for by interactions at the CT and PT dimer interfaces. In fact, earlier studies with a human PC construct that lacked the BC domain altogether showed that it was still a tetramer (25). Therefore, to study the effects of dimer interface mutations on isolated BC, we next expressed and purified the BC domain of wild-type SaPC alone, as well as the corresponding R54E, F403A and K442E mutants. Despite having a similar dimer interface as E. coli BC, gel filtration experiments showed that the isolated BC domain of wild-type SaPC is monomeric in solution. This is confirmed by sedimentation velocity AUC experiments, which gave a Kd of 48 μM for the dimer (Table 2). The R54E, F403A and the K442E mutants have roughly the same Kd as the wild-type BC domain (Table 2). On the other hand, most of the isolated BC domain proteins have a significant amount of aggregation (Table 3), suggesting that this domain on its own may not be very stable. Possibly in agreement with this, extensive efforts at crystallizing these proteins were unsuccessful.
To test whether the isolated BC domains have catalytic activity, we used a coupled-enzyme assay that monitored the hydrolysis of ATP (9). The proteins were at 0.4 μM concentration, and 40 mM biotin was also included in the assay. No catalytic activity was observed with any of the proteins, including the wild-type isolated BC domain, under the condition tested. This indicates that the monomeric BC domain of SaPC is catalytically inactive, in contrast to observations with the monomeric mutants of E. coli BC subunit (19).
These results on the isolated BC domain of SaPC are also in contrast with those earlier on the BC domain of other pyruvate carboxylases. The crystal structure of the BC subunit of Aquifex aeolicus PC showed a dimeric organization that is rather similar to that of E. coli BC (38). On the other hand, the BC domain of Geobacillus thermodenitrificans PC showed a very different dimeric association in the crystal (39), even though residues in the dimer interface are conserved in this protein (Fig. 1B). This BC domain existed in a monomer-dimer equilibrium in solution, and was able to catalyze the hydrolysis of ATP in the presence of biotin, likely in its monomeric form (40). A chimera replacing the BC domain of G. thermodenitrificans PC with the BC subunit of A. aeolicus PC was also catalytically active (41).
Our studies show that proper dimerization of the BC domain in the tetramer of PC is important for its catalytic activity. Mutations in this interface that disturb the dimer have a negative impact on activity. On the other hand, the R54E mutant is still a tetramer but is catalytically inactive. This appears to be consistent with a long-range communication between the dimer interface and the active site, although the mechanism of this communication is not known. A recent study on E. coli ACC showed that while the BC dimer interface mutants are active as a monomer in vitro, they are insufficient to rescue cells that are defective in BC (42). This suggests that dimerization of the BC subunit is important for ACC activity in vivo, most likely dimerization is important for the assembly of this ACC holoenzyme.
Acknowledgments
This research was supported in part by a grant from the NIH (DK067238) to LT, and by grants from National Science Council, Taiwan (98-2320-B-010-026-MY3) and National Health Research Institute, Taiwan (NHRI-EX101-9947SI) to CYC. LPCY was also supported by an NIH training program in Cellular and Molecular Foundations of Biomedical Science (GM008798). PHC was also supported by an NIH Medical Scientist Training Program (GM007367).
We thank Christine Huang for help with the kinetic assays, and Neil Whalen for setting up the X29A beamline at the NSLS.
References
- 1.Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Ann Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 2.Cronan JE, Jr, Waldrop GL. Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res. 2002;41:407–435. doi: 10.1016/s0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- 3.Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci. 2005;62:1784–1803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jitrapakdee S, St Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Structure, mechanism and regulation of pyruvate carboxylase. Biochem J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong L. Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knowles JR. The mechanism of biotin-dependent enzymes. Ann Rev Biochem. 1989;58:195–221. doi: 10.1146/annurev.bi.58.070189.001211. [DOI] [PubMed] [Google Scholar]
- 7.Guchhait RB, Polakis SE, Dimroth P, Stoll E, Moss J, Lane MD. Acetyl coenzyme A carboxylase system from Escherichia coli. Purification and properties of the biotin carboxylase, carboxyltransferase, and carboxyl carrier protein components. J Biol Chem. 1974;249:6633–6645. [PubMed] [Google Scholar]
- 8.Tipton PA, Cleland WW. Catalytic mechanism of biotin carboxylase: steady-state kinetic investigations. Biochem. 1988;27:4317–4325. doi: 10.1021/bi00412a019. [DOI] [PubMed] [Google Scholar]
- 9.Blanchard CZ, Lee YM, Frantom PA, Waldrop GL. Mutations at four active site residues of biotin carboxylase abolish substrate-induced synergism by biotin. Biochem. 1999;38:3393–3400. doi: 10.1021/bi982660a. [DOI] [PubMed] [Google Scholar]
- 10.Sloane V, Blanchard CZ, Guillot F, Waldrop GL. Site-directed mutagenesis of ATP binding residues of biotin carboxylase. J Biol Chem. 2001;276:24991–24996. doi: 10.1074/jbc.M101472200. [DOI] [PubMed] [Google Scholar]
- 11.Waldrop GL, Rayment I, Holden HM. Three-dimensional structure of the biotin carboxylase subunit of acetyl-CoA carboxylase. Biochem. 1994;33:10249–10256. doi: 10.1021/bi00200a004. [DOI] [PubMed] [Google Scholar]
- 12.Thoden JB, Blanchard CZ, Holden HM, Waldrop GL. Movement of the biotin carboxylase B-domain as a result of ATP binding. J Biol Chem. 2000;275:16183–16190. doi: 10.1074/jbc.275.21.16183. [DOI] [PubMed] [Google Scholar]
- 13.Mochalkin I, Miller JR, Evdokimov A, Lightle S, Yan C, Stover CK, Waldrop GL. Structural evidence for substrate-induced synergism and half-sites reactivity in biotin carboxylase. Prot Sci. 2008;17:1706–1718. doi: 10.1110/ps.035584.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou CY, Yu LPC, Tong L. Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism. J Biol Chem. 2009;284:11690–11697. doi: 10.1074/jbc.M805783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janiyani K, Bordelon T, Waldrop GL, Cronan JE., Jr Function of Escherichia coli biotin carboxylase requires catalytic activity of both subunits of the homodimer. J Biol Chem. 2001;276:29864–29870. doi: 10.1074/jbc.M104102200. [DOI] [PubMed] [Google Scholar]
- 16.de Queiroz MS, Waldrop GL. Modeling and numerical simulation of biotin carboxylase kinetics: implications for half-sites reactivity. J Theoretical Biol. 2007;246:167–175. doi: 10.1016/j.jtbi.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y, Volrath SL, Weatherly SC, Elich TD, Tong L. A mechanism for the potent inhibition of eukaryotic acetyl-coenzyme A carboxylase by soraphen A, a macrocyclic polyketide natural product. Mol Cell. 2004;16:881–891. doi: 10.1016/j.molcel.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 18.Weatherly SC, Volrath SL, Elich TD. Expression and characterization of recombinant fungal acetyl-CoA carboxylase and isolation of a soraphen-binding domain. Biochem J. 2004;380:105–110. doi: 10.1042/BJ20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y, Chou CY, Chang GG, Tong L. Is dimerization required for the catalytic activity of bacterial biotin carboxylase? Mol Cell. 2006;22:807–818. doi: 10.1016/j.molcel.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 20.Chou CY, Tong L. Structural and biochemical studies on the regulation of biotin carboxylase by substrate inhibition and dimerization. J Biol Chem. 2011;286:24417–24425. doi: 10.1074/jbc.M111.220517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang CS, Sadre-Bazzaz K, Shen Y, Deng B, Zhou ZH, Tong L. Crystal structure of the a6b6 holoenzyme of propionyl-coenzyme A carboxylase. Nature. 2010;466:1001–1005. doi: 10.1038/nature09302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang CS, Ge P, Zhou ZH, Tong L. An unanticipated architecture of the 750-kDa a6b6 holoezyme of 3-methylcrotonyl-CoA carboxylase. Nature. 2012;481:219–223. doi: 10.1038/nature10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan C, Chou CY, Tong L, Xiang S. Crystal structure of urea carboxylase provides insights into the carboxyltransfer reaction. J Biol Chem. 2012;287:9389–9398. doi: 10.1074/jbc.M111.319475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.StMaurice M, Reinhardt L, Surinya KH, Attwood PV, Wallace JC, Cleland WW, Rayment I. Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme. Science. 2007;317:1076–1079. doi: 10.1126/science.1144504. [DOI] [PubMed] [Google Scholar]
- 25.Xiang S, Tong L. Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction. Nat Struct Mol Biol. 2008;15:295–302. doi: 10.1038/nsmb.1393. [DOI] [PubMed] [Google Scholar]
- 26.Yu LPC, Xiang S, Lasso G, Gil D, Valle M, Tong L. A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A. Structure. 2009;17:823–832. doi: 10.1016/j.str.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lietzan AD, Menefee AL, Zeczycki TN, Kumar S, Attwood PV, Wallace JC, Cleland WW, StMaurice M. Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli. Biochem. 2011;50:9708–9723. doi: 10.1021/bi201277j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 29.Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J. 2006;90:4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modak HV, Kelly DJ. Acetyl-CoA-dependent pyruvate carboxylase from the photosynthetic bacterium Rhodobacter capsulatus: rapid and efficient purification using dye-ligand affinity chromatography. Microbiol. 1995;141:2619–2628. doi: 10.1099/13500872-141-10-2619. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Jogl G, Tao X, Xu Y, Tong L. COMO: A program for combined molecular replacement. Acta Cryst. 2001;D57:1127–1134. doi: 10.1107/s0907444901006783. [DOI] [PubMed] [Google Scholar]
- 34.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan KD. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Kondo S, Nakajima Y, Sugio S, Yong-Biao J, Sueda S, Kondo H. Structure of the biotin carboxylase subunit of pyruvate carboxylase from Aquifex aeolicus at 2.2 Å resolution. Acta Cryst. 2004;D60:486–492. doi: 10.1107/S0907444904000423. [DOI] [PubMed] [Google Scholar]
- 39.Kondo S, Nakajima Y, Sugio S, Sueda S, Islam MN, Kondo H. Structure of the biotin carboxylase domain of pyruvate carboxylase from Bacillus thermodenitrificans. Acta Cryst. 2007;D63:885–890. doi: 10.1107/S0907444907029423. [DOI] [PubMed] [Google Scholar]
- 40.Sueda S, Islam MN, Kondo H. Protein engineering of pyruvate carboxylase. Investigation on the function of acetyl-CoA and the quaternary structure. Eur J Biochem. 2004;271:1391–1400. doi: 10.1111/j.1432-1033.2004.04051.x. [DOI] [PubMed] [Google Scholar]
- 41.Islam MN, Sueda S, Kondo H. Construction of new forms of pyruvate carboxylase to assess the allosteric regulation by acetyl-CoA. Protein Eng. 2005;18:71–78. doi: 10.1093/protein/gzi011. [DOI] [PubMed] [Google Scholar]
- 42.Smith AC, Cronan JE. Dimerization of the bacterial biotin carboxylase subunit is required for acetyl coenzyme A carboxylase activity in vivo. J Bacteriol. 2012;194:72–78. doi: 10.1128/JB.06309-11. [DOI] [PMC free article] [PubMed] [Google Scholar]