Summary
Targeting tumor vasculature represents a rational strategy for controlling cancer. (Z)-(+/−)-2-(1-benzylindol-3-ylmethylene)-1-azabicyclo[2.2.2]octan-3-ol (denoted VJ115) is a novel chemical entity that inhibits the enzyme ENOX1, a NADH oxidase. Genetic and small molecule inhibition of ENOX1 inhibits endothelial cell tubule formation and tumor-mediated neo-angiogenesis. Inhibition of ENOX1 radiosensitizes tumor vasculature, a consequence of enhanced apoptosis. However, the molecular mechanisms underlying these observations are not well understood. Herein, we mechanistically link ENOX1-mediated regulation of cellular NADH concentrations with proteomics profiling of endothelial cell protein expression following exposure to VJ115. Pathway Studios network analysis of potential effector molecules identified by the proteomics profiling indicated that a VJ115 exposure capable of altering intracellular NADH concentrations impacted proteins involved in cytoskeletal reorganization. The analysis was validated using RT-PCR and immunoblotting of selected proteins. RNAi knockdown of ENOX1 was shown to suppress expression of stathmin and lamin A/C, proteins identified by the proteomics analysis to be suppressed upon VJ115 exposure. These data support the hypothesis that VJ115 inhibition of ENOX1 can impact expression of proteins involved in cytoskeletal reorganization and support a hypothesis in which ENOX1 activity links elevated cellular NADH concentrations with cytoskeletal reorganization and angiogenesis.
Keywords: ENOX1, Angiogenesis, Proteomics, 2D-DIGE, NADH
Introduction
It is well established that neovascularization is required for the continued growth of a solid tumor [1] and can occur via angiogenesis, vasculogenesis, intussusceptions, vasculogenic mimicry and/or mosaic vessel formation [2]. Because of its central role in promoting tumor growth, the vascularization pathway represents a rationale therapeutic target. Unfortunately, therapeutic targeting of VEGF/VEGF-receptors has not resulted in the expected clinical benefit [3]. Thus, there is a need to identify new antiangiogenic targets [3].
ENOX1 is a 70 kDa copper-binding protein that exhibits alternating NADH oxidase and disulfide-thiol interchange activity [4]. Our recent studies support the hypothesis that ENOX1 activity is proangiogenic. RNAi-mediated inhibition of ENOX1 suppresses endothelial cell migration and the ability of endothelial cells to form tubule-like structures [5]. Using a zebrafish model of vasculogenesis, we found that Enox1 and Fli-1 are co-expressed in the intersegmental vasculature and in heart 24 hrs post-fertilization [6]. Injection of morpholino antisense oligonucleotides directed against Enox1 into zebrafish embryos suppressed Enox1 expression, with subsequent inhibition of Fli-1, Flk and vasculogenesis [6].
(Z)-(+/−)-2-(1-benzylindol-3-ylmethylene)-1-azabicyclo[2.2.2]octan-3-ol (VJ115) is a small molecule ENOX1 inhibitor [5]. In endothelial cell culture models, VJ115 phenocopies the results produced by shRNA-mediated suppression of ENOX1 [5]. Similarly, VJ115 phenocopies ENOX1 morphants in the zebrafish model [6]. Using a mouse window chamber model, we found that VJ115 inhibited tumor-driven neoangiogenesis [5]. Suppression of ENOX1 activity via ENOX1 RNAi or VJ115 increased radiation-induced caspase3-mediated endothelial cell apoptosis [5]. Administration of VJ115 with fractionated x-irradiation significantly reduced the number and density of tumor microvessels, as well as significantly delayed syngeneic and xenograft tumor growth compared to results obtained with radiation alone [5]. These data support the hypothesis that VJ115-mediated inhibition of ENOX1 activity represents a rationale and novel approach for inhibiting tumor angiogenesis.
The mechanism by which ENOX1 inhibition impairs endothelial function is not well understood. Therefore, this current study was undertaken to identify ENOX1 targets. Use of biochemical, cell biological and proteomic approaches demonstrate that ENOX1 activity regulates the expression of the cytoskeleton proteins lamin A/C and stathmin, which are involved in cell polarization [7, 8]. These results suggest a mechanistic link between NADH cellular redox and cellular polarization, regulated by ENOX1 activity.
Materials and Methods
Cell Culture and Total Cell Lysate Preparation
HUVECs were cultured in endothelial growth medium (EGM2, Lonza) supplied with SingleQuots supplements as per manufacturers’ instructions. Passage 3 HUVECs were grown to 50% confluency before fresh medium containing 50 µM final concentration of VJ115 was added and cells maintained at 37°C for up to 6 hrs. Control cells were treated with equal volume of DMSO. Quadruplet control and VJ115 treated cell cultures were washed 3 times with cold Phosphate Buffered Saline (PBS) and cell lysates were obtained using protein lysis buffer (50 mM Tris HCl pH 8,150 mM NaCl, 1% NP40, 10 mM EDTA, 0.1% Sodium Dodecyl Sulfate, 0.5% deoxycholic acid) containing protease and phosphatase inhibitor cocktails. Protein concentration was measured using Biorad protein assay reagent. The cell lysates were used for 2D- DIGE analysis.
Antibodies and Reagents
Primary polyclonal antibodies used in this study were rabbit anti-stathmin (Cell signaling, catalog #3352), mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz Biotechnology, Catalog #SC-32233), mouse antiphospho c-Jun (Santa Cruz Biotechnology, Santa Cruz, CA, Catalog #SC-822), rabbit anti-phospho p70s6K (Millipore, Billerica, MA, Catalog # 04-392), rabbit anti-phosphoribosomal 6 kinase (Cell Signaling, Danvers, MA, Catalog # 2211), rabbit anti-total ribosomal S6 protein (Cell Signaling, Danvers, MA, Catalog # 2217), rabbit monoclonal anti-HUP1 (Epitomics, Burlingame, CA, Catalog # 6643-1) and rabbit anti-laminA/C (Cell Signaling, Danvers, MA, Catalog # 2032). Secondary antibodies used for immunofluorescence was anti-rabbit IgG Alexa Fluor 647 (Invitrogen, Carlsbad, CA). For Western blots, horseradish peroxidase (HRP)-coupled goat antibodies to mouse, rabbit or goat IgG (Santa Cruz Biotechnology, Santa Cruz, CA) were used. VJ115 (Z)-(+/−)-2-(1-benzylindol-3-ylmethylene)-1-azabicyclo[2.2.2]octan-3-ol) was synthesized as described previously (13).
In vivo multiphoton microscopy of NADH
Microscopy was performed as described in ref [9]. A custom built, commercial multi-photon fluorescence microscope (Prairie Technologies) was used to acquire autofluorescence images of NADH with a 40X water-immersion objective (1.15 NA). A titanium:sapphire laser (Coherent Inc.) provided excitation light at 750 nm with an average power of 7.5–7.8 mW. A pixel dwell time of 4.8 µs was used to acquire a 256×256 pixel image. Each image was captured and averaged 8 times to reduce noise. A GaAsP PMT (H7422P-40, Hamamatsu) detected emitted photons through a 400–480 nm bandpass filter. For analysis, NADH fluorescence images were thresholded to remove background and nuclear fluorescence and the average NADH fluorescence intensity per cell was computed (ImageJ) [10].
Custom Enox1 Antibody
In collaboration with the Vanderbilt Antibody Core and the company Covance Inc., (Denver, PA), a custom ENOX1 antibody was raised by injecting rabbits with antigen conjugated with ENOX1 specific peptide [H]-CKEEQSHTQALLKVLQEQLKGTK -[NH2].
RNA Extraction and RT- PCR
A commercial RNA extraction kit (RNeasy Mini Kit, Qiagen, Chatsworth, CA) was used for extraction of total RNA from HUVECs according to the instructions of the manufacturer. SuperScript One-Step RT_PCR kit (Qiagen, Chatsworth, CA) was used for cDNA synthesis and followed by PCR using 200 ng of starting RNA. The following gene specific primers were used for RT-PCR: Human GAPDH sense, 5’-CATCACCATCTTCCAGGAGCGA-3’; and antisense, 5’-GTCTTCTGGGTGGCAGTGATGG-3’ (amplification product size was 332 bp); stathmin primers; sense, 5’- TCTTGAAGCCACCATCTCCT-3’ and antisense 5’- CAGCCTCCAGTTTCTTCTGG-3’ (amplification product size was 102 bp); Hypoxia Upregulated protein 1; sense, 5’- GGTAGAGGACAGCGCAGAAG–3’ and antisense 5’-ATTCTCCTTGGCATCTGGTG-3’ (amplification product size was 100 bp); lamin A/C; sense, 5’-TGGAGATGATCCCTTGCTGA-3’ and antisense 5’-GCATGGCCACTTCTTCCCA-3’.
Cy-dye Labeling and 2D Gel Electrophoresis and Imaging
The experiment was performed in quadruplicate, producing treated and control samples from 2h and 6h time points each with n=4. The NHS-ester dyes Cy2/3/5 were used for minimal labeling using the mixed internal standard methodology of Alban et al. [10]. For each sample, 0.15-mg of protein was precipitated separately with methanol and chloroform, as previously described [11], and resuspended in 30-µl of labeling buffer (7-M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris, 5 mM magnesium acetate). Two-thirds (100µg) of each of the sixteen experimental samples was individually labeled with 200-pmoles of either Cy3 or Cy5 such that two members of each group were labeled with Cy3 and the other two with Cy5 to compensate for any dye-specific labeling artifacts. In similar fashion, the remaining third of each of the sixteen experimental samples was combined and labeled en masse with 800-pmoles of Cy2 to generate the mixed internal standard. Labeling was performed for 30 minutes on ice in the dark, after which the reaction was quenched by the addition of 10-mM lysine for 10 minutes, followed by the addition of an equal volume of 2X rehydration buffer (7M urea, 2M thiourea, 4% CHAPS, 4mg/ml DTT).
Pairs of Cy3/Cy5 labeled samples were mixed with an equal aliquot of Cy2-labeled mixed internal standard, providing 300-µg total protein resolved on each gel. Tripartate-labeled samples were brought up to 450-µl with 1X rehydration buffer (7M urea, 2M thiourea, 4% CHAPS, 2mg/ml DTT, 0.5% IPG buffer pH 4–7) and loaded via passive rehydration into 24 cm pH 4–7 immobilized pH gradient strips (GE Healthcare) for isolectric focusing using a manifold-equipped IPGphor unit (GE Healthcare). Second-dimensional 12% SDS-PAGE was performed using hand-cast gels for which one plate was pre-silanized using an Ettan DALT 12 unit (GE Healthcare), according to manufacturers’ protocols. Cy2/3/5-specific 16-bit data files were acquired at 100µm resolution separately by dye-specific excitation and emission wavelengths using a Typhoon 9400 Variable Mode Imager (GE Healthcare), and gels were stained for total protein content with SyproRuby (Molecular Probes/Invitrogen) per the manufacturer’s instructions.
DIGE Image Analysis
The DeCyder v6.5 suite of software tools (GE Healthcare) was used for DIGE analysis. The normalized volume ratio of each individual protein spot-feature from a Cy3- or Cy5-labeled sample was directly quantified relative to the Cy2-signal from the pooled-sample internal standard corresponding to the same spot-feature. This was performed for all resolved features in a single gel where no gel-to-gel variation exists between the three co-resolved signals. The individual signals from the Cy2-standard were then used to normalize and compare Cy3:Cy2 and Cy5:Cy2 abundance ratios independently for each resolved feature that was matched across the eight-gel set, enabling statistical confidence to be associated with each change in abundance or charge-altering post-translational modification using Analysis of Variants (ANOVA) and Student's t-test univaiate analyses. Principle components analysis (PCA) was performed using the DeCyder Extended Data Analysis (EDA) module.
In-gel Digestion, Mass Spectrometry and Database Interrogation
The DIGE analytical gels were post-stained with SyproRuby (Invitrogen, Carlsbad CA) and used to robotically excise proteins of interest for digestion into peptides in-gel with modified porcine trypsin protease (Promega, Madison, Wisconsin) using an integrated Spot Handling Workstation (GE Healthcare, Piscataway, NJ). Digestion was performed in 20µL 20mM NH4HCO3 containing 0.01µg/µL trypsin Gold (Promega, Madison, Wisconsin) for 3h at 37°C, after which peptides were extracted by two rounds of incubation with 60% acetonitrile, 0.1% trifluoroacetic acid, dried and reconstituted in 15µL 0.1% formic acid and placed into autosampler vials. 5µL of each peptide hydrosylate was separately analyzed by C18 reverse-phase LC-MS/MS using a Thermo LTQ ion trap mass spectrometer equipped with a Thermo MicroAS autosampler and Eksigent HPLC nanoLC pump system, nanospray source, and Xcalibur 2.0 instrument control using standard data-dependent methods. Peak lists were generated and charge-states determined using ScanSifter (v2.1.25). Tandem MS data were analyzed with the Sequest algorithm using strict trypsin cleavage rules and peptide and fragment ion mass tolerances of 2.5 and 0.5, respectively. Oxidation of methionine (+15.99) and carbamidomethylation of cysteine (+57.02) were also allowed as variable modifications. The uniprotKB human_1.fasta database (UniProtKB/TrEMBL Release 40.5 and UniProtKB/Swiss-Prot Release 57.5, July 2009; 173736 entries) containing common contaminants and a concated reverse-database to enable false-discovery rates. IDPicker (version 2.6.165) was used to filter raw Sequest identifications by adjusting cross-correlation and deltaCn scores to produce a maximum 5% false discovery rate at the peptide level after requiring 2 or more unique peptides per protein identification and at least one peptide for each group within a cluster of related proteins. Proteins were also organized into groups and clusters based on peptide sequences that were shared between multiple database entries [12].
Gene Ontology, Construction and Visualization of Interactome Networks
Pathway Studio 7.1 software was employed to study functional interactions and possible pathways of downregulated [vimentin, F-actin capping protein subunit alpha-1, lamin/AC, dihydropyrimidinase-like 2 (DPYSL2) and stathmin] and upregulated [programmed cell death 6 –interacting protein, filamin-A, Hsp70 (grp78), Hsp90, dipeptidyl-peptidase 3] proteins.
Results
VJ115 increases cellular NADH and inhibits tubule formation by HUVECs
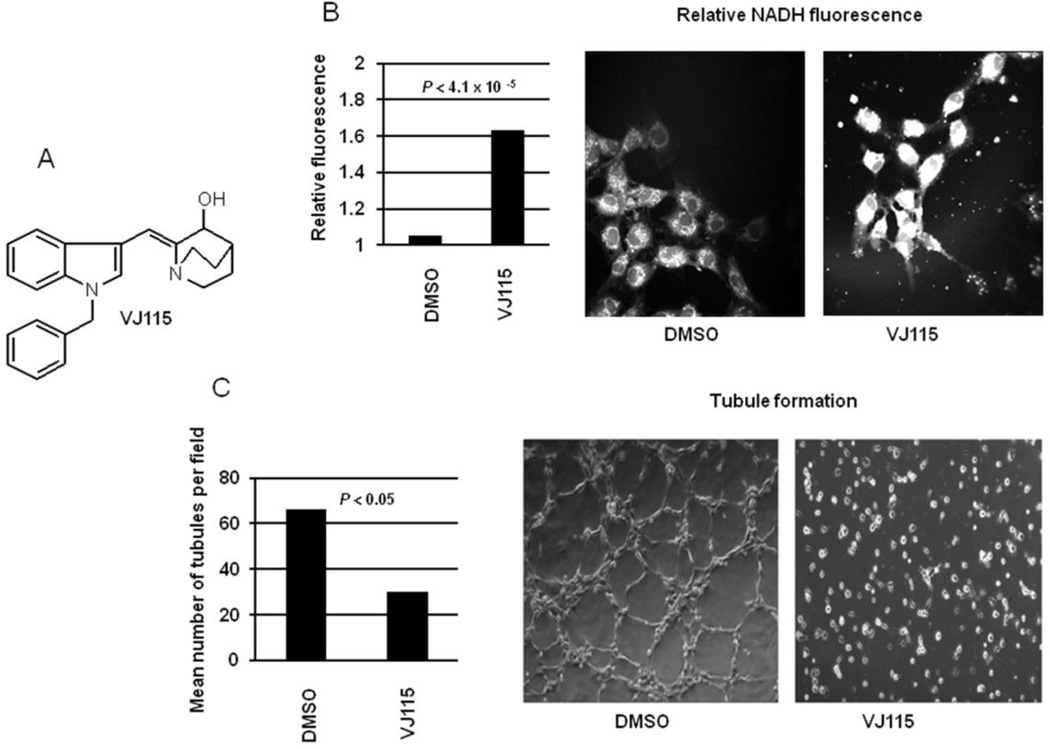
The chemical structure of VJ115 is shown in Figure 1A. As ENOX1 is a NADH oxidase [4], one would predict that VJ115-mediated inhibition of ENOX1 would increase cellular NADH levels. This hypothesis was tested using in vivo multiphoton microscopy, which allows single cell fluorescence imaging of NADH [9]. Although NADH and NADPH fluorescence cannot be distinguished, previous studies have shown that the concentration of NADH is about 5 times greater than NADPH [13], and the NADH quantum yield is 1.25 to 2.5 times greater than NAPDH [14]. Therefore the majority of the fluorescence signal can be attributed to NADH. The fluorescence intensity images obtained from cells demonstrated that exposure to VJ115 for 6 hrs increased NADH fluorescence by 1.63 fold (P = 4.1 × 10−5, n = 9 independent fields, Figure 1B), congruent with the knowledge that VJ115 inhibits ENOX1 activity with an EC50 of 50 µM [5].
Figure 1.
VJ115 increases cellular NADH and inhibits tubule formation by HUVECs. A) The chemical structure of VJ115. B) Quantification of NADH fluorescence in single cells using In vivo multiphoton microscopy (Image Dimensions: 270 um × 270 um). Also shown are representative cellular images of that fluorescence. HUVECs were added to 60 mm dishes and cells exposed to 0 or 50 µM of VJ115. NADH fluorescence (n= 9 fields) was quantified after 6 hrs of exposure to VJ115. C) Cells were inoculated into 24-well plates coated with Matrigel and were exposed to 0 or 50 µM of VJ115 for 6 hrs. The number of tubules per field (n = 4) was quantified. Also shown are representative images of tubule formation in the absence and presence of VJ115.
Consistent with previous results obtained using murine 3B11 tumor vascular endothelial cells, HUVECs, and human microvascular endothelial cells [5], exposing HUVECs to 50 M VJ115 for 6 hrs inhibited formation of capillary-like structures (P < 0.05, n = 4 independent fields) under the experimental conditions used here (Figure 1C). Of note, this 6 hr exposure to 50 M VJ115 yielded plating efficiencies of 70% or greater [5], demonstrating that inhibition of tubule formation was not a consequence of cell death.
It was of interest to determine if VJ115 would affect NADPH oxidase enzymatic activity, as measured by generation of superoxide. Exposure of HUVECs to 50 µM VJ115 for 6 hrs did not affect NAPDH oxidase activity (P > 0.05, data not shown).
Proteomics profiling of global protein expression following exposure to VJ115
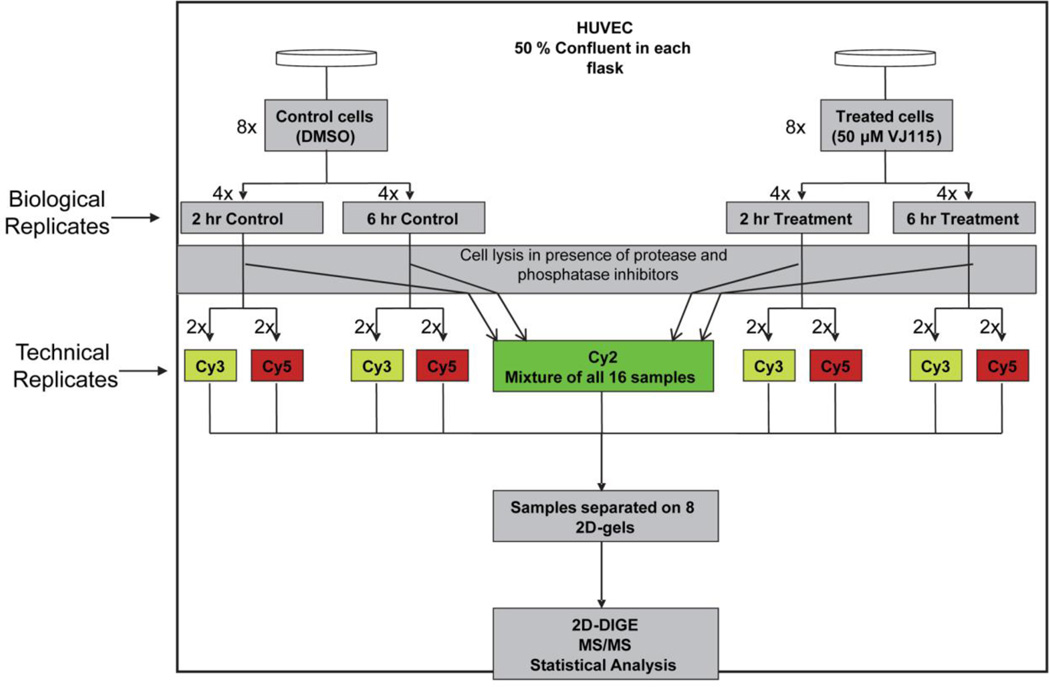
HUVECs were exposed to 50 µM of VJ115 or solvent control for 2 or 6 hours and then processed for 2 dimensional difference gel electrophoresis (DIGE). A schematic depiction of the experimental design is shown in Figure 2. Four biological replicates per treatment (DMSO or VJ115) were obtained for each time point (2h vs 6h) and the resulting 16 samples were randomized with respect to labeling with Cy3 or Cy5 and position within the eight-gel matrix (detailed in Materials and Methods section and Figure 2). Each of the eight DIGE gels co-resolved two of the 16 samples along with an equal aliquot of the mixed-sample internal standard (labeled with Cy2), which allowed for the normalization of all 16 Cy3/Cy2 or Cy5/Cy2 ratios across the dataset for any given protein feature. This also enabled univariate analysis (ANOVA) for individual protein features as well as multivariate analysis (PCA) on the global dataset (Materials and Methods). A representative image illustrating an overlay of Cy3, Cy5, and Cy2 fluorescence is shown in Figure 3A.
Figure 2.
Schematic illustration of experimental setup for studying global proteome dynamics in HUVECs exposed to VJ115. Passage 3 HUVECs at 50% confluency were exposed to VJ115 (50 µM). Samples were taken for analysis at 2 time points. At each time point 4 biological replicates were obtained. For 2D-DIGE analysis, 2 of the 4 biological replicates were labeled with Cy3 and the other 2 were labeled with Cy5. All 16 samples were separated by 2D gel electrophoresis using 8 DIGE gels, with each gel containing two of the 16 Cy3/Cy5-labeled samples along with an equal aliquot of the Cy2-labeled internal standard, which is a mixture of equal amounts of total protein from every sample, and is used to normalize ratios between gels and minimize technical variation
Figure 3.
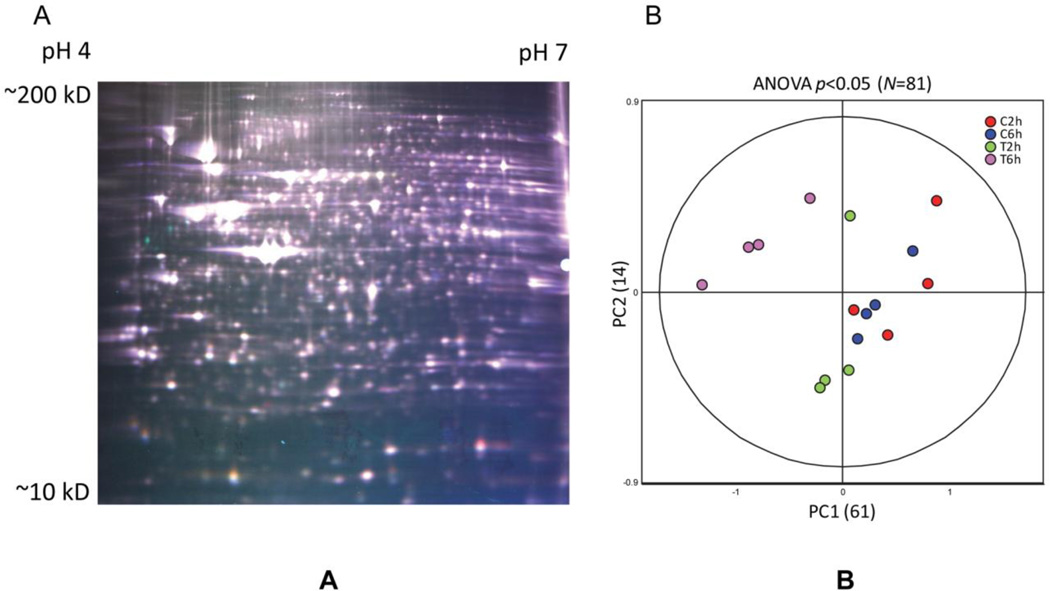
Quality of 2D-DIGE Data. A) False-colored representative DIGE gel from the resulting 8-gel experiment containing three differentially-labeled samples. Cy3-labeled sample-n (false-colored green), Cy5-labeled sample-n (red), and Cy2-labeled pooled-sample internal standard (blue) are overlayed for illustrative purposes, actual data are acquired on each dye separately and quantified relative to the internal standard as described in Materials and Methods. B) Principle analysis (PCA) properly groups the 16 individual DIGE expression maps differentiated by two principle components, PC1 and PC2, and demonstrates high reproducibility between the replicate samples within each group. PC1 distinguished 61% of the variance, with 14% additional variation distinguished by PC2. PCA was performed using expression values from 81 proteins exhibiting ANOVA P-values <0.01
Quality of 2D-DIGE Data
We assessed the major sources of variation between the 16 samples, using PCA analysis of all 1350 ratios that were matched across the eight DIGE gels. PCA analysis on the unfiltered dataset did not meaningfully organize the 16 samples, but the major sources of variation within a subset of protein features that were differentially-expressed (ANOVA P <0.05, N=81) indicated clustering between the treated and control samples, as well as distinctions between the 2h and 6h treatment time points (Figure 3B). This indicated that a low but significant contribution to the variance in the system could be attributed to biologically-relevant changes in expression, and that unanticipated sources of technical or non-biologically-relevant variation were not apparent.
Statistical Analysis and Identification of Proteins
Univariate analysis was used to determine the statistical significance for each protein feature resolved by 2D-DIGE. Following a 6 hr VJ115 exposure only 22 protein features exhibited abundance changes >20% (P < 0.05). These protein features were selected for identification by mass spectrometry after excision from the DIGE analytical gels (Materials and Methods). The locations of these excised features are shown in Figure 4. The corresponding MS/MS identification of the corresponding polypeptides is reported in Table 1 and the relative change in protein expression over time is shown in Supplemental Figure 1. In some cases proteins were identified from more than one feature, indicating the presence of VJ115-mediated post translational modification that altered the charge and/or processing of the intact protein and thus its migration in the 2D gels. Overall, 20 unique proteins were represented in this analysis that were altered in expression by VJ115 exposure. It is of note that Enox1 was not one of the proteins whose expression was affected by VJ115.
Figure 4.
The location of the 22 spots on a representative 2D-DIGE gel stained with SyproRuby is shown. Their corresponding MS/MS identification is reported in Table 1
Table 1.
Differentially Regulated Proteins
| Protein Name | Accession Number |
Spot Number | MW (kD) |
||
|---|---|---|---|---|---|
| pI | Av ratio | t-test | |||
| 6 hr vs Con | |||||
| ER proteins | |||||
| Hypoxia up-regulated protein 1(grp170) | HYOU1_HUMAN | 1 | 111 | ||
| 5.2 | 1.24 | 0.0014 | |||
| GRP78 (BiP or HSPA5) | GRP78_HUMAN | 8, 9 & 10 | 72 | ||
| 5.1 | 1.69 | 0.0037 | |||
| PDI isoenzyme A1 | PDIA1_HUMAN | 12 | 57 | ||
| 4.8 | 1.53 | 8.40E-05 | |||
| Endoplasmin (grp 94) | ENPL_HUMAN | 12 | 92 | ||
| 4.7 | 1.53 | 8.40E-05 | |||
| Procollagen lysyl-hydroxylase | PLOD2_HUMAN | 3 &4 | 85 | ||
| 6.2 | 1.66 | 0.00078 | |||
| Cytoskeleton-associated proteins | |||||
| Filamin-A | FLNA_HUMAN | 3 &4 | |||
| 1.66 | 0.00078 | ||||
| Heat shock protein beta-1(hsp27) | HSPB1_HUMAN | 20 | 23 | ||
| 6.0 | 1.74 | 0.0028 | |||
| Programmed cell death 6 interacting protein | PDCD6IP_HUMAN | 3 &4 | 96 | ||
| 6.1 | 1.66 | 0.00078 | |||
| (also known as ALIX) | |||||
| Lamin-A/C, | LMNA_HUMAN | 11 | 74 | ||
| 6.6 | −1.3 | 0.019 | |||
| Dihydropyrimidinase-like 2 | DPYSL2_HUMAN | 11 | 62 | ||
| 5.9 | −1.3 | 0.019 | |||
| Vimentin | VIME_HUMAN | 16 | 54 | ||
| 5.1 | −4.5 | 0.0075 | |||
| F-actin capping protein subunit alpha-1 | CAPZA1_HUMAN | 18 | 33 | ||
| 5.4 | −2.27 | 3.30E-06 | |||
| Stathmin | STMN1_HUMAN | 22 | 17 | ||
| 5.7 | −1.53 | 0.002 | |||
| Translation | |||||
| Eukaryotic translation initiation factor 4 | IF4A1/2_HUMAN | 14 | 46 | ||
| 5.3 | 1.46 | 0.0013 | |||
| Ribosomal protein S3 | RPS3_HUMAN | 19 | 27 | ||
| 9.7 | 1.34 | 0.015 | |||
| Miscellaneous | |||||
| Dipeptidyl-peptidase 3 | DPP3_HUMAN | 8, 9 & 10 | 83 | ||
| 5.0 | 1.69 | 0.0037 | |||
| Trifunctional enzyme subunit beta | HADHB_HUMAN | 14 | 51.3 | ||
| 9.4 | 1.46 | 0.0013 | |||
| Protein CDV3 homolog | CDV3_HUMAN | 19 | 27 | ||
| 6.1 | 1.34 | 0.015 | |||
| Hsp90 | HS90A_HUMAN | 8, 9 & 10 | 85 | ||
| 4.9 | 1.69 | 0.0037 | |||
| Deoxyuridine triphosphatase | DUT_HUMAN | 21 | 27 | ||
| 9.6 | −1.36 | 0.031 | |||
Network and Functional Analysis
The relative levels of the vast majority of proteins were minimally altered by VJ115 treatment. As expected, no differential expression was observed in any of the Control-6 hr/Control-2 hr groups (P > 0.05, data not shown). Because a 2 hr exposure to VJ115 was insufficient for inducting a change in protein expression, we only considered proteins that showed at least a 20% differential expression at the 6 hr time point in our functional analysis. Exposure to VJ115 resulted in the differential expression of several endoplasmic reticulum-associated molecular chaperones (HYOU1, GRP78, PDIA1, ENPL, and GRP94) as well as several key cytoskeleton-remodeling proteins (Filamin-A, Hsp27, and ALIX) and the molecular chaperone Hsp90. The cytoskeleton-associated proteins lamin A/C, dihydropyrimidinase-like 2, (also known as CRMP2), vimentin, F-actin capping protein subunit alpha-1, and stathmin were found to be down-regulated.
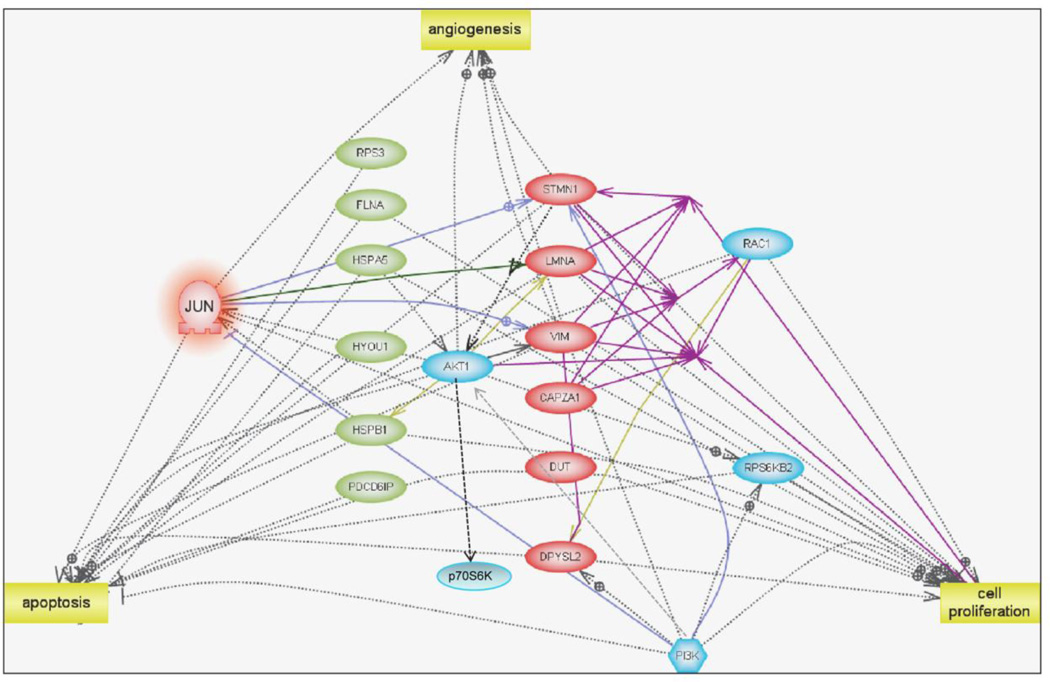
We used Pathway Studio software to determine if the differentially-expressed proteins at this time point displayed any relationship to each other. Of the 14 up-regulated proteins only six proteins exhibited direct interaction (RPS3, FLNA, GRP78, HYOU1, HSPB1, and PDCD6IP, Figure 5). Of these six proteins, four of them, filamin alpha (FLNA), GRP78 (BiP), hypoxia up-regulated 1 (HYOU1) and heat shock 27kDa protein 1 (HSPB1), are identified as negative regulators of apoptosis (Figure 5, green ovals). We found that all down-regulated proteins (red ovals) interacted with each other (represented by purple solid lines) and four of the six down-regulated proteins identified the GTPase Rac1 as a common binding target.
Figure 5.
Networks and Functional Analysis. An overview of significantly altered proteins and their interactions during a 6 hour treatment of HUVECs with VJ115 (50-µM). Pathway Studio 7.1 (Adriane Genomics) was used to assess the functional similarities between the differentially expressed proteins identified by the discovery proteomics analysis. Down-regulated proteins identified by proteomics are shown as pink ovals while up-regulated proteins are shown as green ovals. Down-regulation of the protein c-Jun (highlighted in orange) was predicted using network analysis. Proteins represented in blue color are other whose expression/activity is predicted to decrease upon VJ115 treatment by network analysis
Analysis of the down regulated proteins by Pathway Studio software predicted c-Jun (P < 10−5, highlighted in orange in Figure 4) as a positive regulator and so we hypothesized that exposure to VJ115 might down-regulate c-Jun’s expression or suppress its activity (reduction of phospho-cJun levels). The immunoblot shown in Supplemental Figure 2 confirms the inhibition of phospho-cJun levels by 61% following a 6 hr exposure to VJ115. Based upon network analysis it is predicted that the expression of the proteins (or their activity) represented in blue color will decrease upon VJ115 treatment (Figure 5). For example, stathmin has been shown to be a biomarker for PI3K/AKT signaling [15]. Down regulation of phospho p70S6K by a VJ115 exposure (Supplemental Figure 2) is consistent with this hypothesis. Congruent with previous phenotypic assays [5], cell proliferation and angiogenesis were identified as cellular processes (yellow squares) targeted by these differentially-regulated proteins.
Validation of discovery proteomics data by Reverse Transcriptase PCR (RT-PCR) and Western Blotting
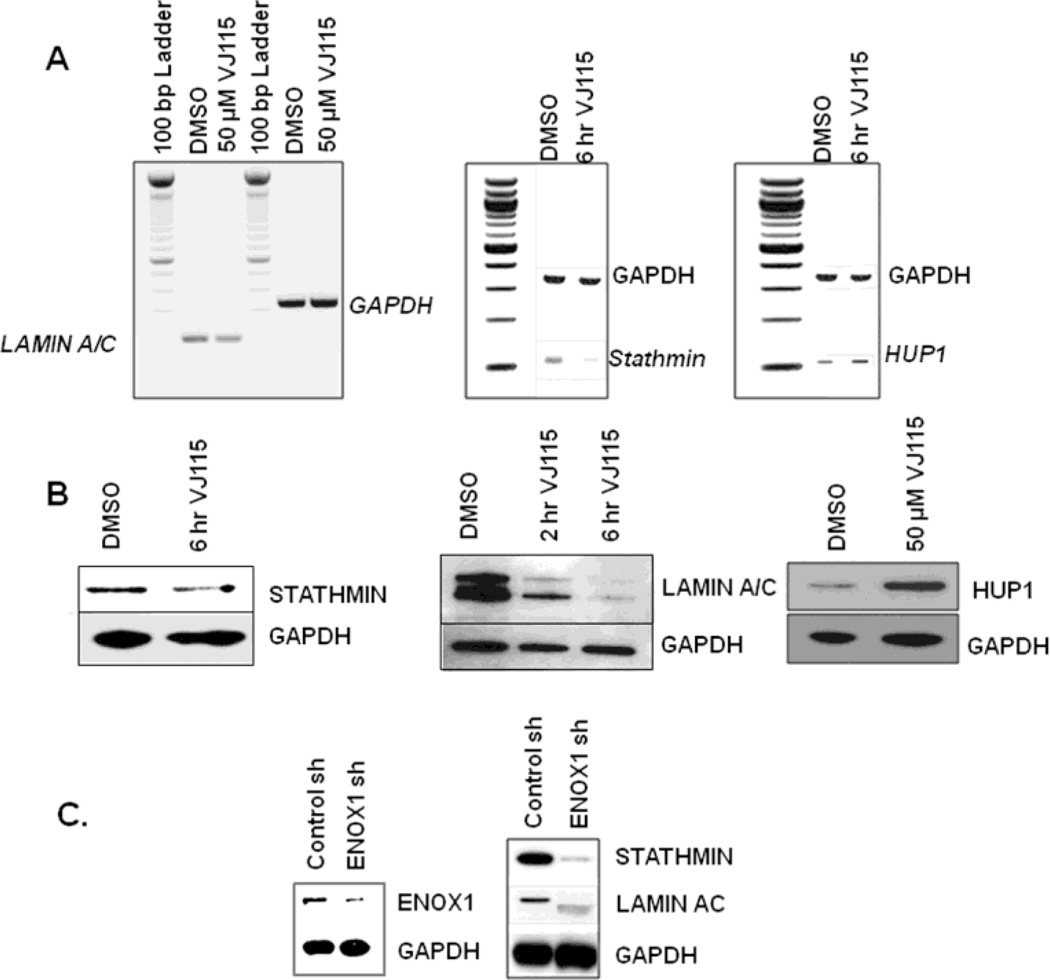
Total RNA was extracted from HUVECs exposed to 50 µM of VJ115 for 6 hrs. We observed a 37% decrease in lamin a/c (LMNA) mRNA, an 88% decrease in stathmin mRNA, and a 38% increase in hypoxia up-regulated protein (Hup1), as quantified by ImageJ analysis software (Figure 6A). Western blot analysis verified that exposure to VJ115 for 6 hrs resulted in down-regulation of stathmin (40%), lamin A/C (greater than 70%) and up-regulation of Hup1 (3.2 fold) (Figure 6B), as determined by ImageJ software analysis. Differential expression of these proteins is consistent with the discovery proteomic analysis described herein.
Figure 6.
Validation of discovery proteomics. A) RT-PCR showing down-regulation of lamin A/C and stathmin, as well as up-regulation of Hup1. B) Western blots confirming down-regulation of stathmin and lamin A/C and up-regulation of Hup1 upon a 6 hr exposure to VJ115. GAPDH was used as a loading control. C) Immunoblot evaluation of stathmin, and lamin A/C expression after siRNA based inhibition of ENOX1 in HUVECs
A pool of nonoverlaping retroviral-based shRNAs directed against ENOX1 was used to determine if suppression of ENOX1 affected the expression of stathmin and lamin A/C. As shown in Figure 6C, RNAi-mediated knockdown of ENOX1 resulted in suppression of stathmin and lamin A/C, consistent with the discovery proteomic analysis.
Discussion
There is now growing evidence from numerous preclinical and clinical studies that targeting tumor vasculature can significantly increase the effectiveness of radiation therapy [16–21]. VJ115 is a novel antiangiogenic targeting ENOX1 and a radiation sensitizer of tumor microvasculature [5]. However, the biochemical pathways impacted by this compound are not well characterized. We employed discovery proteomics together with biochemical approaches to study the effect of VJ115 on protein expression in HUVECs. Of the portion of the proteome queried by our analysis, 20 proteins exhibited differential expression that exceeded 20%, indicating that the phenotype produced by the sensitizer is relatively target-specific.
Several up-regulated proteins participate in protein processing in the endoplasmic reticulum (Table 1). Grp78, PDI, Hypoxia up-regulated protein-1/GRP170, and Grp94 are ER proteins and their up regulation can be considered a response to an ER stress. Of note, Alix is induced during the unfolded protein response [22]. Procollagen lysyl-hydroxylase (PLOD2) is a collagen processing enzyme that colocalizes with protein disulfide isomerase in the lumen during ER stress [23].
Eight of the differentially-regulated proteins are cytoskeleton-associated. Five downregulated proteins; vimentin, F-actin capping protein subunit alpha-1, lamin A/C, dihydropyrimidinase-like 2 (DPYSL2), and stathmin are directly associated with cytoskeleton reorganization. Dihydropyrimidinase-like 2/ Crmp2 interacts with tubulin dimers and kinesin [24]. F-actin capping protein regulates actin polymerization by capping the barbed ends during actin polymerization [25]. Vimentin and lamin A/C are part of major intermediate filaments; vimentin participates in critical cellular processes related to the organization and regulation of proteins involved in adhesion and migration [26, 27], and lamin A and C proteins are encoded by LMNA family of genes as splice variants [28]. The A and C proteins form a complex and are involved in maintenance of nuclear stability and chromatin structure during mitosis [28] and required for cellular spreading and proliferation [29]. Stathmin is a phosphoprotein that promotes cell motility by inducing tubulin catastrophe by binding to a microtubule plus-end thereby causing it to undergo a conformational change that favors depolymerization. This increases microtubule turnover and contributes to cytoskeletal reorganization, a process important for angiogenesis [30]. Three of the up-regulated proteins are associated with the cytoskeleton. Alix/PDCD6IP has recently been shown to assemble with ESCRT-III and participate in contractile filament helices during abscission at the completion of cytokinesis [31]. Heat shock protein beta 1 (Hsp27) is known to cause a decrease in endothelial cell migration by stabilization of the actin cytoskeleton [32] and has been found to be commonly up-regulated by angiogenesis inhibitors [32, 33]. Filamin alpha (FLNA) is an actin binding protein that is required for the F-actin-dependent arrest of caveolin-1 vesicles and stable anchorage to the plasma membrane [34]. Thus, an exposure to VJ115 alters the expression of several key proteins involved in cytoskeletal reorganization.
Our network analysis indicated that proteins impacted by exposure to VJ115 could be categorized into pathways regulating apoptosis (ie ER stress proteins) and cytosketetal motility. The analysis predicted suppression of phospho p70S6K1 and this was validated by immunoblotting. The pathway analysis of the down-regulated proteins identified c-Jun as a common regulator and immunoblotting confirmed that phosphorylation of c-Jun was suppressed by VJ115 (Supplemental Figure 2). Recently, De Luca et al [35] have shown that inhibition of ENOX activity and the subsequent increase in NADH resulted in sphingosine kinase inhibition. Su et al [36] found that sphingosine 1-phosphate positively regulates cJun. Therefore we hypothesize that VJ115 may inhibit an ENOX1/sphingosine kinase/cJun axis resulting in down-regulation of selected cytoskeleton proteins.
Supplementary Material
The relative change in protein expression over time for 20 polypeptides from 2D-DIGE study whose abundance changed by 20% or more (p <0.05) after VJ115 treatment.
A) Western blot analysis was employed to confirm the down-regulation of phosphor-c-Jun expression. B) Immunoblot of phosphorylation status of p70S6K and its down-stream target, ribosomal protein S6 in HUVECs exposed to VJ115.
Acknowledgements
Supported in part by grants from U.S. National Institutes of Health/National Cancer Institute grants RO1CA140409, T32CA093240, Vanderbilt-Ingram Cancer Center grant P30 CA68485, and the Vanderbilt Academic Venture Capital fund for proteomics. We are thankful to Drs. V.J. Sonar and Y.T.R. Reddy for the synthesis of VJ115.
Footnotes
Conflict of Interest Statement:
The authors declare that they have no conflict of interest
References
- 1.Folkman J. Tumor angiogenesis: therapeutic implications. The New England Journal of Medicine. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 2.Goel S, Duda DG, Xu L, Munn LL, Boucher Y, Fukumura D, et al. Normalization of the vasculature for treatment of cancer and other diseases. Physiol Rev. 2011;91:1071–1121. doi: 10.1152/physrev.00038.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sitohy B, Nagy JA, Dvorak HF. Anti-VEGF/VEGFR therapy for cancer: reassessing the target. Cancer Research. 2012;72:1909–1914. doi: 10.1158/0008-5472.CAN-11-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Z, Gorenstein NM, Morre DM, Morre DJ. Molecular cloning and characterization of a candidate human growth-related and time-keeping constitutive cell surface hydroquinone (NADH) oxidase. Biochemistry. 2008;47:14028–14038. doi: 10.1021/bi801073p. [DOI] [PubMed] [Google Scholar]
- 5.Geng L, Rachakonda G, Morre DJ, Morre DM, Crooks PA, Sonar VN, et al. Indolyl-quinuclidinols inhibit ENOX activity and endothelial cell morphogenesis while enhancing radiation-mediated control of tumor vasculature. FASEB J. 2009;23:2986–2995. doi: 10.1096/fj.09-130005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venkateswaran ASK, Melville DB, Levic DS, Rybski WM, Walsh AJ, Skala MC, Crooks PA, Freeman ML, Knapik EW. The NADH Oxidase Enox1 is required for vasculogenesis in Zebrafish. In Preparation. [Google Scholar]
- 7.Miyashita H, Kanemura M, Yamazaki T, Abe M, Sato Y. Vascular endothelial zinc finger 1 is involved in the regulation of angiogenesis: possible contribution of stathmin/OP18 as a downstream target gene. Arteriosclerosis, Thrombosis, and Vascular biology. 2004;24:878–884. doi: 10.1161/01.ATV.0000126373.52450.32. [DOI] [PubMed] [Google Scholar]
- 8.Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, et al. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophysical Journal. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skala MC, Riching KM, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, White JG, et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, et al. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- 11.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Analytical Biochemistry. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 12.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klaidman LK, Leung AC, Adams JD., Jr High-performance liquid chromatography analysis of oxidized and reduced pyridine dinucleotides in specific brain regions. Analytical Biochemistry. 1995;228:312–317. doi: 10.1006/abio.1995.1356. [DOI] [PubMed] [Google Scholar]
- 14.Avi-Dor Y, Olson JM, Doherty MD, Kaplan NO. Fluorescence of Pyridine Nucleotides in Mitochondria. J Biol Chem. 1962;237:2377–2383. [Google Scholar]
- 15.Andersen JN, Sathyanarayanan S, Di Bacco A, Chi A, Zhang T, Chen AH, et al. Pathway-based identification of biomarkers for targeted therapeutics: personalized oncology with PI3K pathway inhibitors. Sci Transl Med. 2010;2:43ra55. doi: 10.1126/scitranslmed.3001065. [DOI] [PubMed] [Google Scholar]
- 16.Chen HX. Expanding the clinical development of bevacizumab. Oncologist. 2004;9(Suppl 1):27–35. doi: 10.1634/theoncologist.9-suppl_1-27. [DOI] [PubMed] [Google Scholar]
- 17.Kozin SV, Boucher Y, Hicklin DJ, Bohlen P, Jain RK, Suit HD. Vascular endothelial growth factor receptor-2-blocking antibody potentiates radiation-induced long-term control of human tumor xenografts. Cancer Research. 2001;61:39–44. [PubMed] [Google Scholar]
- 18.Wachsberger P, Burd R, Dicker AP. Tumor response to ionizing radiation combined with antiangiogenesis or vascular targeting agents: exploring mechanisms of interaction. Clinical Cancer Research. 2003;9:1957–1971. [PubMed] [Google Scholar]
- 19.Willett CG, Boucher Y, di Tomaso E, Duda DG, Munn LL, Tong RT, et al. Direct evidence that the VEGF-specific antibody bevacizumab has antivascular effects in human rectal cancer. Nat Med. 2004;10:145–147. doi: 10.1038/nm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkler F, Kozin SV, Tong RT, Chae SS, Booth MF, Garkavtsev I, et al. Kinetics of vascular normalization by VEGFR2 blockade governs brain tumor response to radiation: role of oxygenation, angiopoietin-1, and matrix metalloproteinases. Cancer Cell. 2004;6:553–563. doi: 10.1016/j.ccr.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Yoon SS, Duda DG, Karl DL, Kim TM, Kambadakone AR, Chen YL, et al. Phase II Study of Neoadjuvant Bevacizumab and Radiotherapy for Resectable Soft Tissue Sarcomas. International Journal of Radiation Oncology, Biology, Physics. Int J Radiat Oncol Biol Phys. 2011;81:1081–1090. doi: 10.1016/j.ijrobp.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amodio G, Moltedo O, Monteleone F, D'Ambrosio C, Scaloni A, Remondelli P, et al. Proteomic Signatures in Thapsigargin-Treated Hepatoma Cells. Chem Res Toxicol. 2010;24:1215–1222. doi: 10.1021/tx200109y. [DOI] [PubMed] [Google Scholar]
- 23.Kellokumpu S, Sormunen R, Heikkinen J, Myllyla R. Lysyl hydroxylase, a collagen processing enzyme, exemplifies a novel class of luminally-oriented peripheral membrane proteins in the endoplasmic reticulum. J Biol Chem. 1994;269:30524–30529. [PubMed] [Google Scholar]
- 24.Rahajeng J, Giridharan SS, Naslavsky N, Caplan S. Collapsin response mediator protein-2(Crmp2) regulates trafficking by linking endocytic regulatory proteins to dynein motors. J Biol Chem. 2010;285:31918–31922. doi: 10.1074/jbc.C110.166066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SH, Dominguez R. Regulation of actin cytoskeleton dynamics in cells. Mol Cells. 29:311–325. doi: 10.1007/s10059-010-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eriksson JE, Dechat T, Grin B, Helfand B, Mendez M, Pallari HM, et al. Introducing intermediate filaments: from discovery to disease. J Clin Invest. 2009;119:1763–1771. doi: 10.1172/JCI38339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazarides E. Intermediate filaments as mechanical integrators of cellular space. Nature. 1980;283:249–256. doi: 10.1038/283249a0. [DOI] [PubMed] [Google Scholar]
- 28.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 29.Emerson LJ, Holt MR, Wheeler MA, Wehnert M, Parsons M, Ellis JA. Defects in cell spreading and ERK1/2 activation in fibroblasts with lamin A/C mutations. Biochim Biophys Acta. 2009;1792:810–821. doi: 10.1016/j.bbadis.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Yoshie M, Miyajima E, Kyo S, Tamura K. Stathmin, a microtubule regulatory protein, is associated with hypoxia-inducible factor-1alpha levels in human endometrial and endothelial cells. Endocrinology. 2009;150:2413–2418. doi: 10.1210/en.2008-1333. [DOI] [PubMed] [Google Scholar]
- 31.Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, et al. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 32.Dai S, Jia Y, Wu SL, Isenberg JS, Ridnour LA, Bandle RW, et al. Comprehensive characterization of heat shock protein 27 phosphorylation in human endothelial cells stimulated by the microbial dithiole thiolutin. J Proteome Res. 2008;7:4384–4395. doi: 10.1021/pr800376w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keezer SM, Ivie SE, Krutzsch HC, Tandle A, Libutti SK, Roberts DD. Angiogenesis inhibitors target the endothelial cell cytoskeleton through altered regulation of heat shock protein 27 and cofilin. Cancer Research. 2003;63:6405–6412. [PubMed] [Google Scholar]
- 34.Muriel O, Echarri A, Hellriegel C, Pavon DM, Beccari L, Del Pozo MA. Phosphorylated filamin A regulates actin-linked caveolae dynamics. J Cell Sci. 124:2763–2776. doi: 10.1242/jcs.080804. [DOI] [PubMed] [Google Scholar]
- 35.De Luca T, Morre DM, Morre DJ. Reciprocal relationship between cytosolic NADH and ENOX2 inhibition triggers sphingolipid-induced apoptosis in HeLa cells. J Cell Biochem. 2010;110:1504–1511. doi: 10.1002/jcb.22724. [DOI] [PubMed] [Google Scholar]
- 36.Su Y, Rosenthal D, Smulson M, Spiegel S. Sphingosine 1-phosphate, a novel signaling molecule, stimulates DNA binding activity of AP-1 in quiescent Swiss 3T3 fibroblasts. J Biol Chem. 1994;269:16512–16517. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The relative change in protein expression over time for 20 polypeptides from 2D-DIGE study whose abundance changed by 20% or more (p <0.05) after VJ115 treatment.
A) Western blot analysis was employed to confirm the down-regulation of phosphor-c-Jun expression. B) Immunoblot of phosphorylation status of p70S6K and its down-stream target, ribosomal protein S6 in HUVECs exposed to VJ115.