Abstract
Morning stiffness and increased symptoms of inflammatory arthritis are among the most common manifestations of rheumatoid arthritis (RA). Tumor necrosis alpha (TNF-α), an important mediator of inflammation in RA, regulates the circadian expression of clock proteins, and adenosine A2A receptors (A2AR) mediate many of the anti-inflammatory and antirheumatic actions of methotrexate, the cornerstone drug in the treatment of RA. We found that A2AR activation and TNF-α activated the clock core loop of the human monocytic THP-1 cell line. We further observed that interleukin (IL)-10, but not IL-12, mRNA expression fluctuates in a circadian fashion and that TNF-α and A2AR stimulation combined increased IL-10 expression. Interestingly, TNF-α, but not CGS21680, dramatically inhibited IL-12 mRNA expression. The demonstration that A2AR and TNF-α regulate the intrinsic circadian clock in immune cells provides an explanation for both the pathologic changes in circadian rhythms in RA and for the adverse circadian effects of methotrexate, such as fatigue.
Keywords: adenosine A2 receptor, TNF-α, IL10, IL-12, clock genes, methotrexate
INTRODUCTION
The daily rhythm of life follows from both the revolution of the earth on its axis and from the inherent circadian rhythms set by an internal clock apparatus. The internal clock in mammals and lower animals is organized by transcriptional machinery, namely the core clock genes Clock, Bmal1, Period, and Cryptochrome [1]. The master pacemaker apparatus resides in the hypothalamic suprachiasmatic nucleus, but virtually all cells in the body express genes related to periodicity [2–4]. In the immune system, many functions depend on the time of day, including variation in susceptibility to infection [5] and the course of such diseases as rheumatoid arthritis (RA [6]) or asthma [7]. In particular, recent observations indicate that periodicity genes alter inflammation in animal models of arthritis, and inflammation leads to changes in expression of the pacemaker apparatus [8].
One of the most striking characteristics of RA, a chronic polyarthritis of unknown etiology, is the circadian fluctuation in symptoms [9–12]. In fact, morning stiffness, included in the diagnostic criteria of RA, is one of the most common complaints of afflicted patients [13, 14] and methotrexate (MTX) administration, the cornerstone in the treatment of RA, also manifests in perturbations of the circadian rhythm, primarily in the form of severe fatigue on the day that the weekly dose is taken [15, 16]. Moreover, the expression of proinflammatory cytokines [interleukin (IL)-1β, IL-6, and tumor necrosis alpha (TNF-α)] and the classic disease specific marker rheumatoid factor exhibit characteristic rhythmic patterns [17–20].
The demonstration that low-dose, intermittent MTX is a potent and effective therapy for RA [21] was followed by investigations indicating that many of the therapeutic effects of MTX are mediated by adenosine [22, 23]. Adenosine potently diminishes the proinflammatory actions of inflammatory and immune cells via interaction with specific cell surface receptors [A1R, A2AR, A2BR, and A3R, reviewed in [24]]. In human and murine monocytes/macrophages, the activation of adenosine receptors, particularly A2AR, modulates the production of inflammatory cytokines including TNF-α, IL-10, and IL-12 [25–29]. We therefore analyzed the impact of A2AR activation by the specific agonist CGS26180, as well as with the pro-inflammatory mediator TNF-α, on the molecular clock machinery of the human monocytic THP-1 cell line. The effects described here are, to our knowledge, the first description of the role of adenosine and its receptors in the regulation of circadian pacemakers in the immune system and provide further insight into the effects of inflammation on circadian pacemakers.
MATERIALS AND METHODS
Cell Culture and Stimulation
THP-1 (TIB-202) cells were purchased from American Type Culture Collection (ATCC) and maintained in RPMI 1640 medium (30-2001; ATCC) supplemented with 10 % FBS (Gibco). For the stimulation, cells were thawed and split in every experiment as follows: a new vial from ATCC was thawed into 20 ml of RPMI 1640 medium with 10 % FBS; after 2 days of culture, cells had achieved a concentration of 2.5–4×05/ml and were then split into 120 ml of medium (passage 1); after 3 days, the cell concentration reached 1–2×105/ml, and the cells were then spun down and resupended into 120 ml of medium (passage 2). After 3 days, the confluence was 6–8×105/ml, and they were then split into 600 ml of medium (passage 3). Three days after (day of the stimulus), the concentration was 2.5–5×105/ml. Cells were discarded if the concentration was outside the specified range at any of the passages described above.
On the day of study, passage 4 of THP-1 cells was diluted into the appropriate volume of RPMI 1640 medium with 10 % serum to get 120×106 cells at 0.8×106/ml, which were then split into 25-cm2 flasks with 5 ml on each at 12pm. Stimulation with CGS21680 (TOCRIS) 100 nM (final) and/or TNF-α (rhTNF-α, R&D Systems) 100 U/ml (final) was begun at 12am, and samples were harvested every 4 h. For harvesting, the THP-1 cells were spun down, washed with PBS, and the resulting pellet was immediately frozen at –80 °C. The changes in gene expression were studied in four separate independent experiments, and all results reported represent the mean (±SEM) changes observed in these experiments.
RNA Extraction and Real-Time Reverse Transcription PCR
After stimulating the THP-1 cells with CGS21680, 100 nM, and/or TNF-α 100 U/ml at the indicated time points, the RNA was extracted and purified using the RNeasy Mini Kit columns (Qiagen) according to the manufacturer’s protocol, including sample homogenization with QIAshredder columns (Qiagen) and DNA digestion with RNAse-free DNAase set (Qiagen). Following this step, the RNA was reverse transcribed with MuLV Reverse transcriptase (Applied Biosystems) at 2.5 U/μl, including in the same reaction the following reagents: RNAase Inhibitor 1 U/μl (Applied Biosystems,), Random Hexamers 2.5 U/μl (Applied Biosystems), MgCl2 5 mM (Applied Biosystems), PCR buffer II 1× (Applied Biosystems), and dNTPs 1 mM (Applied Biosystems,).
For Clock, Bmal1, Cry1, IL-10, IL-12, and actin, relative quantification of gene expression was performed using real-time RT-PCR on Mx3005P Real-Time PCR System (Stratagene) with SYBR Green (Agilent Technologies, 600548) according to the manufacturer’s protocol. For Per1, Cry2, hrevErbA1, Per2, and GAPDH, Multiplex relative quantification of gene expression was performed using real-time RT-PCR on Mx3005P Real-Time PCR System (Stratagene) with Brilliant Multiplex QPCR Master Mix (Agilent Technologies). Fluorescence was measured during the annealing step of each cycle. In the initial tests, the PCR products were separated by electrophoresis on agarose gels to verify the size of the amplified product and to check for the presence of primer dimers or nonspecific bands. Single-plex real-time PCR assays were performed as described above with only one set of primers and probe in the amplification reaction. To rule out the possibility of discrepancies between SYBR Green and TaqMan Multiplex technologies, we analyzed one of the genes, Per1, with both technologies and found similar fold changes throughout the 24-h time course in control non-stimulated THP-1 cells, as well as cells stimulated with CGS21680 100 nM or TNF-α 100 U/ml (Supplemental Figure).
Primers and TaqMan Probes’ Design and Validation
Multiplex primers and probes were designed using the online software RealTimeDesign™ (Biosearch Technologies) with TaqMan technology. Next, specificity of the primers and probes was assessed by MFEprimer online software [30]. Primers and probe dimerization was next analyzed by PriDimerCheck online software, and melting temperatures (Tm) were calculated with the Applied Biosystems™ online calculator. For SYBR Green primer design and validation, the same online tools were used, but the initial design was addressed by Primer3 (v.0.4.0). Target genes, primers’ and probes’ sequences, Tm, and amplicon sizes are shown in Table 1.
Table 1.
Target Genes, Primers and Probes Sequences, Tm, and Amplicon Sizes
| Target (human) | Technology | Forward primer 5′−3′ | Reverse primer 5′−3′ | Probe | Probe labeling | Amplicon size (bp) |
|---|---|---|---|---|---|---|
| Actin | SYBRGreen | TCACCCACACTGTGCCCATCTACGA | CAGCGGAACCGCTCATTGCCAATGG | 295 | ||
| Clock | SYBRGreen | AAGTTAGGGCTGAAAGACGACG | GAACTCCGAGAAGAGGCAGAAG | 172 | ||
| Bmal1a | SYBRGreen | GTAACCTCAGCTGCCTCGTC | TAGCTGTTGCCCTCTGGTCT | 153 | ||
| Cry1 | SYBRGreen | TTGCTTGATGCAGATTGGAG | TTTTGCAGGGAAGCCTCTTA | 174 | ||
| Per1 | SYBRGreen | CACCCTGATGACCCACTCTT | GGTAAGGCTGGACTGGATGA | 209 | ||
| IL-10 | SYBRGreen | GACTTTAAGGGTTACCTGGGTTG | TCACATGCGCCTTGATGTCTG | 112 | ||
| IL-12 | SYBRGreen | CCTTGCACTTCTGAAGAGATTGA | TCCACTGTGCTGGTTTTATCTTT | 63 | ||
| GAPDH (1) | TaqMan (Multiplex 1) | GCCCTCAACGACCACTTTG | CCACCACCCTGTTGCTGTAG | CAAGCTCATTTCCTGGTATGACAACGA | FAM-BHQ1 | 73 |
| Cry2 | TaqMan (Multiplex 1) | GGTGTGCATTCAGAGCATTGG | GGCCCGATTCTCTTTCCTCTC | CTTCCCTGTTCCCTCAGCCCCAG | HEX-BHQ1 | 96 |
| Per1 | TaqMan (Multiplex 1) | GCACCAGCTAGACTCCATTC | GGGCAGTTTCCCACTGGTT | TGGGACCATCTCCAGGAGTCCAT | ROX-BHQ2 | 46 |
| hrevErbA1 | TaqMan (Multiplex 1) | GGGCTTCCGTGACCTTTC | TGCGGCTTAGGAACATCAC | TGACCAAGTCACCCTGCTTAAGGC | CY5-BHQ2 | 128 |
| GAPDH (2) | TaqMan (Multiplex 2) | GCCAAGGTCATCCATGACAACT | GGGCCATCCACAGTCTTCTG | TGGTATCGTGGAAGGACTCATGACCA | FAM-BHQ1 | 92 |
| Per2 | TaqMan (Multiplex 2) | ACCAGGCTGTGACCAACACA | GGCTGGGTGGAAGCAGAT | TGTGAGACCCAGTCCTGTTTGGTTT | HEX-BHQ1 | 86 |
Statistical Analysis
Statistical differences were determined using repeated measures ANOVA carried out using GraphPad software on a PC. The alpha nominal level was set at 0.05 in all cases. A P value of <0.05 was considered significant.
RESULTS
A2AR Stimulation Activates the Clock Core Loop of THP-1 Cells
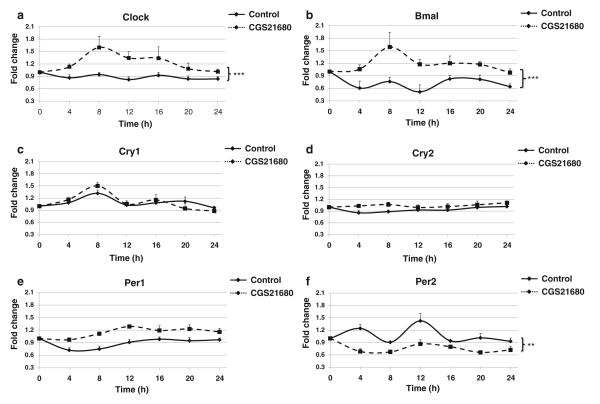
As noted above, the primary anti-inflammatory effects of adenosine are mediated via stimulation of adenosine A2A receptors, and we have previously demonstrated that adenosine A2A receptor stimulation of THP-1 cells suppresses TNF-α and stimulates IL-10 expression [31]. To determine whether stimulation of this receptor also regulates circadian fluctuations in clock proteins, we determined the effects of CGS21680 treatment on circadian gene expression over 24 h. As shown in Fig. 1a, b, A2AR activation promoted a significant increase of both Clock and Bmal, the main activators of the clock core loop, during the 24-h period studied. While maximal induction of these genes occurred after 8 h (Clock 1.6±0.1-fold increase and Bmal 1.6±0.4-fold increase of control), we found that even after A2AR activation, there were circadian fluctuations for these two genes throughout the entire 24-h period studied.
Fig. 1.
A2AR stimulation with CGS21680 activates the core loop machinery. THP-1 cells split at 12 pm were analyzed every 4 h starting at 12 am with quantitative real-time RT-PCR. Cells were kept in 10 % serum medium with CGS21680 100 nM, or without the agonist (control), under strict cell growth monitoring. Results show the x-fold variations from time 0 (t=0) for the mRNA of a clock, b Bmal, c Cry1, d Cry2, e Per1, and f Per2. Data represent mean±SEM of four independent experiments. Statistics was performed by ANOVA followed by Newman–Keuls posttest, control vs CGS21680 ***p<0.001, **p<0.01.
We next investigated the impact of A2AR activation on expression of the principal clock core loop repressor genes: Cry1, Cry2, Per1, and Per2 (Fig. 1c–f). We found that CGS21680 did not significantly alter either the fluctuations or the expression levels of Cry1, Cry2, or Per1, but A2AR activation did flatten the fluctuations and reduced the expression levels of Per2 throughout the period studied.
TNF-α Increases Bmal and Decreases Cry1 Expression in THP-1 Cells
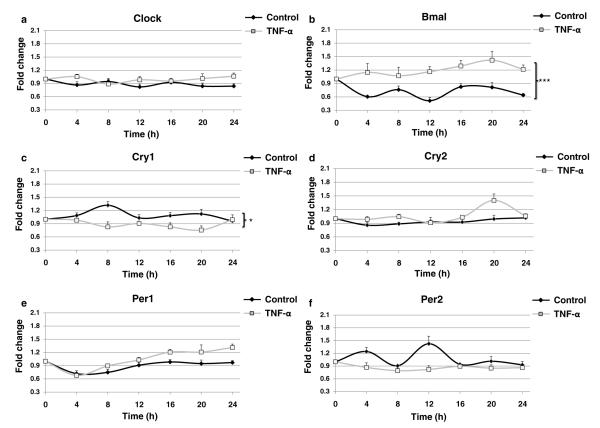
Previous studies have demonstrated that splenic macrophages secrete TNF-α in a rhythmic fashion governed by a circadian clock within the cell [32] and, conversely, it has also been demonstrated that TNF-α suppresses the expression of clock genes [33]. We therefore sought to analyze the impact of TNF-α on the expression of the clock core activators and inhibitors in the THP-1 cell line. As shown in Fig. 2a, b, TNF-α incubation did not promote significant modification of most clock gene fluctuations, but stimulated a strong and significant increase of Bmal expression with reduced fluctuation during the 24-h period studied. TNF-α incubation dramatically altered Cry1 fluctuations with inversions at 8 and 20 h. For Cry2, which has dampened periodicity when compared to the other clock genes, TNF-α promoted an increase on expression only after 20 h. TNF-α promoted a slight increase in Per1 expression from 16 to 24 h when compared to control cells and flattened the fluctuations of Per2, although these differences were not significant.
Fig. 2.
TNF-α activates the core loop machinery. THP-1 cells split at 12 pm were analyzed every 4 h starting at 12 am with quantitative real-time RT-PCR. Cells were kept in 10 % serum medium with TNF-α 100 U/ml, or without the cytokine (control), under strict cell growth monitoring. Results show the x-fold variations from time 0 (t=0) for the mRNA of a clock, b Bmal, c Cry1, d Cry2, e Per1, and f Per2. Data represent mean±SEM of four independent experiments. Statistics was performed by ANOVA followed by Newman–Keuls posttest, control vs TNF-α ***p<0.001, *p<0.05.
TNF-α+CGS21680 Impact on the Clock Core Loop of THP-1 Is Similar to the TNF-α Alone
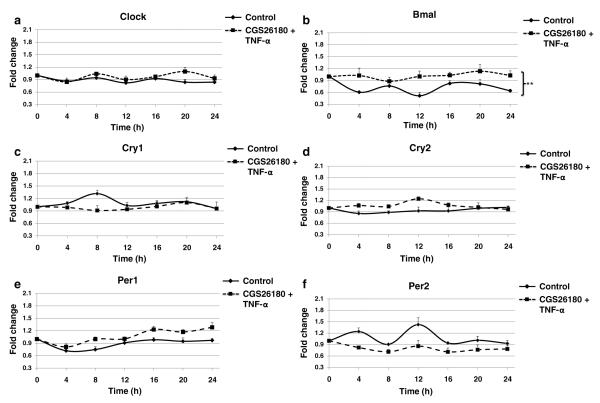
Since our research group has previously established that TNF-α treatment potentiates the effect of CGS21680 in THP-1 cells [31], we therefore stimulated the THP-1 cells with both TNF-α and CGS21680 and analyzed the fluctuations of the different components of the clock machinery. As shown in Fig. 3a, TNF-α+CGS21680 induced a similar circadian pattern for clock when compared to the non-stimulated control, and only after 20 h was there a slight increase in expression. On the other hand, TNF-α+CGS21680 stimulated a significant increase of Bmal over the 24-h period studied and, similar to the impact of TNF-α alone, the expression of Bmal was not only higher but also exhibited less fluctuations (Fig. 3b).
Fig. 3.
Co-treatment with CGS21680 and TNF-α impact on the core loop machinery. THP-1 cells split at 12 pm were analyzed every 4 h starting at 12 am with quantitative real-time RT-PCR. Cells were kept in 10 % serum medium with CGS21680 100 nM+TNF-α 100 U/ml, or without the agonist and the cytokine (control), under strict cell growth monitoring. Results show the x-fold variations from time 0 (t=0) for the mRNA of a clock, b Bmal, c Cry1, d Cry2, e Per1, and f Per2. Data represents mean±SEM of four independent experiments. Statistics was performed by ANOVA followed by Newman–Keuls posttest, control vs CGS21680+TNF-α **p<0.01.
When we examined the effect of TNF-α+CGS21680 on the clock core loop repressors (Fig. 3c–f), we observed that Cry1 expression was decreased only after 8 h, and that Cry2 was only increased after 12 h. For Per1 and Per2, we observed similar changes to those following TNF-α treatment alone with a slight increase in Per1 mRNA from 16 to 24 h, and Per2 expression was not only decreased but also it presented less fluctuations.
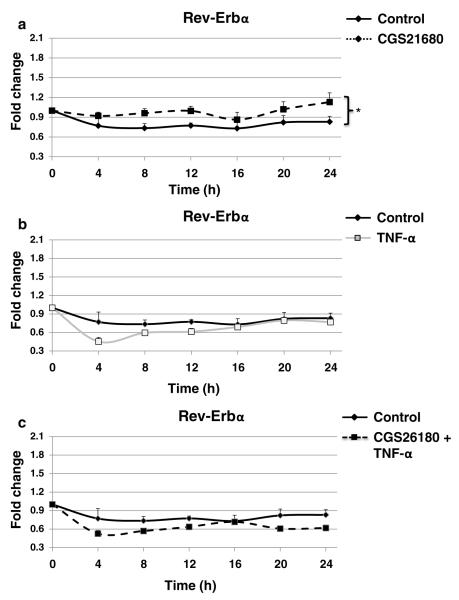
A2AR Activation with CGS21680, but Not TNF-α Incubation, Increases Rev-Erb α in THP-1 Cells
Increasing evidence has established the existence of a stabilizing secondary loop, which is not required for circadian rhythm generation, but reinforces the primary loop by regulating core components [1]. The nuclear orphan receptor Rev-Erbα, a key component of the secondary loop, has been repeatedly shown to repress Bmal1 transcription through direct binding to the Bmal1 promoter [34]. Because our results suggested that the only core component affected by both CGS21680 and TNF-α was Bmal1, we determined whether Rev-Erbα is regulated by A2AR activation with CGS21680 or TNF-α treatment alone, or in combination. As shown in Fig. 4a–c, CGS21680 stimulated a modest but significant increase of Rev-Erbα expression. On the other hand, TNF-α only decreased Rev-Erbα after 4 h and, similarly, CGS21680+TNF-α reduced the expression levels of Rev-Erbα after 4 h.
Fig. 4.
CGS21680, but not TNF-α or CGS21680+TNF-α, increases Rev-Erbα expression. THP-1 cells split at 12 pm were analyzed every 4 h starting at 12 am with quantitative real-time RT-PCR. Cells were kept in 10 % serum medium with a CGS21680 100 nM, b TNF-α 100 U/ml, and c CGS21680 100 nM+TNF-α 100 U/ml, or without the agonist and the cytokine (control), under strict cell growth monitoring. Results show the x-fold change for Rev-Erbα mRNA variations from time 0 (t=0). Data represent mean±SEM of four independent experiments. Statistics was performed by ANOVA followed by Newman–Keuls posttest, control vs CGS21680 *p<0.05.
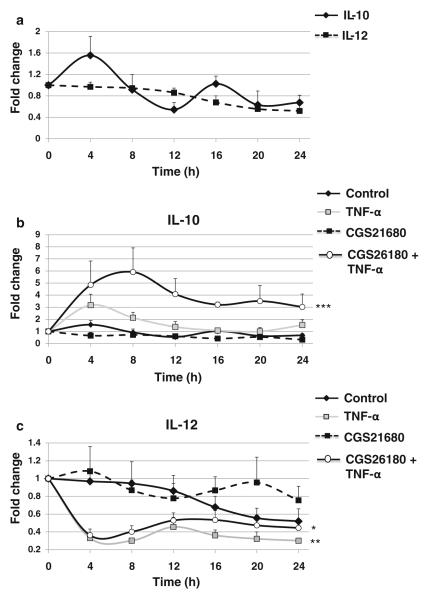
Impact of TNF-α and CGS26180 Stimulation on IL-10 and IL-12 Expression in THP-1 Cells
We have previously reported that stimulation of the A2AR with CGS21680 selectively inhibits IL-12 release and promotes IL-10 secretion [31], and prior work with murine splenic macrophages indicates that there is cyclic expression of cytokines [32]. Here, we observed that in non-preactivated THP-1 cells, IL-10 but not IL-12 expression fluctuated spontaneously (Fig. 5a). TNF-α treatment stimulated a slight but nonsignificant increase in IL-10 expression from 4 to 8 h (3.2±0.9-fold increase over control) while inducing a dramatic decrease of IL-12 at the same time points (0.33±0.02 of control; Fig. 5b, c). Stimulation of the A2AR with CGS21680 did not change IL-10 or IL-12 mRNA expression, but coestimulation with CGS21680 and TNF-α potentiated the effect of TNF-α eliciting a dramatic increase in IL-10 expression (5.9±2.0-fold increase over control). The effect of CGS21680+TNF-α coestimulation on IL-12 expression was identical to that of TNF-α alone.
Fig. 5.
TNF-α and A2AR impact on IL-10 and IL-12 expression. THP-1 cells split at 12 pm were analyzed every 4 h starting at 12 am with quantitative real-time RT-PCR. Cells were kept in 10 % serum medium under strict cell growth monitoring. a Results show the x-fold variations from time 0 (t=0) for the mRNA of IL-10 and IL-12. Impact of TNF-α 100 U/ml and CGS21680 100 nM on b IL-10 and c IL-12. Data represent mean±SEM of four independent experiments. Statistics was performed by ANOVA followed by Newman–Keuls posttest, ***p<0.001, **p<0.01 vs control.
DISCUSSION
The molecular mechanisms by which clock genes maintain circadian rhythm are becoming increasingly clear. In the immune system, many functions and parameters vary based on the time of day including variation in susceptibility to infection [5] and course of immunologically mediated diseases like RA or asthma [6, 7] highlighting the importance of the circadian clock for health and disease [35]. Consistent with these observations, fully operational autonomous circadian clockworks function in immunological tissues like spleen, lymph nodes, and resident peritoneal macrophages [32]. In fact, the strength of proinflammatory cytokine production by macrophages in response to bacterial endotoxin is determined by the circadian phase of the macrophage clock rather than by systemic circadian modulators such as rhythmic cortisol levels [36].
There are clear circadian manifestations of RA, a chronic polyarthritis of unknown etiology affecting ~1 % of the population worldwide, such as morning stiffness, a symptom which is included in the diagnostic criteria for RA [13]. Such circadian characteristics are also seen in the proinflammatory cytokines and disease-specific markers important in RA: IL-1β and IL-6 are elevated in sera of RA patients reaching peak levels in the early morning [6, 17, 19, 20]. In fact, TNF-α, a major cytokine implicated in RA [37], exhibits a delayed secretion rhythm in patients, with the highest levels measured at 6 am and remaining upregulated until 10 am [38] coinciding with morning stiffness in RA [9, 39, 40]. In our studies, we found that TNF-α or costimulation with CGS+TNF-α promotes dramatic changes in IL-10 and IL-12 from 4 am to 8 am, suggesting that signs and symptoms of RA can be modulated by the circadian clock and that, conversely, circadian rhythms modulate arthritis [8]. Thus, TNF-α interferes with the expression of clock genes, suggesting that the “TNF-α inflammatory clock gene response” may induce fatigue, a manifestation of CNS circadian dysregulation, diminishing the quality of life in autoimmune diseases [33] and that inhibition of TNF-α improves disabling fatigue in RA [41].
Methotrexate (MTX), the cornerstone drug in the treatment of RA and other rheumatic diseases, promotes the release of adenosine from cells and tissues, and adenosine mediates many of the anti-inflammatory effects of MTX [23]. Moreover, the beneficial effects of MTX have been reported to be reduced in RA patients who drink coffee (caffeine is a well-known adenosine receptor antagonist) [42]. When stimulated, the adenosine A2A receptor increases intracellular cAMP, and cAMP signaling oscillates in a circadian manner, which in turn sustains core oscillation machinery of the circadian clock [43], suggesting a potential link between adverse circadian effects of MTX, such as fatigue, and A2A receptor activation in the CNS after MTX-induced adenosine release [44].
The validation of TNF-α as a therapeutic target in RA encouraged the investigation of signaling pathways regulating its production [45]. The cAMP signaling pathway downregulates TNF-α production and upregulates production of the anti-inflammatory cytokine IL-10 [46, 47]. In the present work we found that, as previously described [26, 48–52], TNF-α stimulation promotes an increase of IL-10 and a marked decrease in IL-12, and these effects are potentiated by coincubation with the A2AR agonist CGS26180 consistent with prior observations [25, 28, 31, 52].
We analyzed the impact of TNF-α and A2AR activation on the clock genes of the human monocytic cell line THP-1. Although circadian genes’ expression has not been extensively studied in cultured macrophages or monocytes, it has been previously suggested that altering the fetal bovine serum content from 10 % to either 50 [53] or 0 % [33, 54] serum synchronizes circadian gene expression in cultured mammalian cells [53]. In our preliminary studies, we found that there was no impact of varying the serum content of medium to 0 or 50 % serum changed the periodicity of expression of clock gene in THP-1 cells (not shown) and did not, therefore, shock cells for the experiments described here.
Circadian alterations in symptoms of patients with RA and other inflammatory arthritides correlate with aberrant circadian alterations in levels of TNF-α; levels of this inflammatory cytokine are upregulated from 6 am to 10 am [38] coinciding with the sign of morning stiffness [9, 39, 40]. It was therefore interesting to note that TNF-α increases Bmal1, but reduces Cry1 and Per2 expression in THP-1 cells, and the greatest effects of cytokine treatment were noted over a short period of time. Activation of A2AR also regulates expression of circadian clock genes with increases in the clock core loop (clock and Bmal1) and downregulation of Per2, the core loop repressor. Prior experiments have shown that TNF-α potentiates the A2AR impact on IL-10 and IL-12 [31], and similar potentiation was observed for circadian clock genes as well.
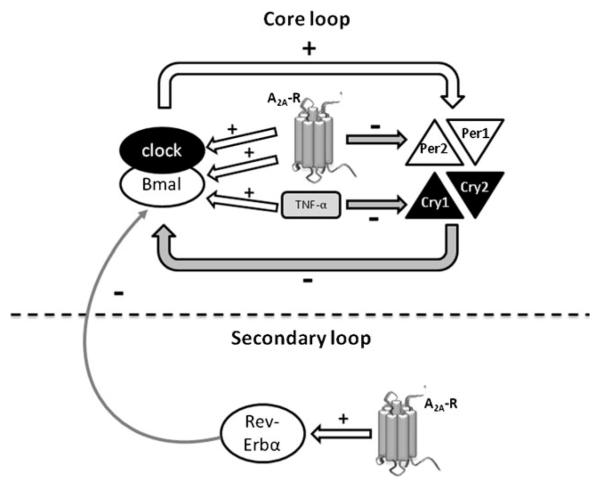
The nuclear orphan receptor Rev-Erbα, a key component of the secondary loop, is the major regulator of cyclic Bmal1 transcription and constitutes a molecular link through which components of the negative limb drive antiphasic expression of the positive limb [55]. Stimulation of A2AR slightly, but significantly, increased its expression. Since the secondary loop stabilizes, but is not required for circadian rhythm generation [1], our findings suggest that adenosine activation of the A2AR confers additional robustness to the core loop via Rev-Erbα upregulation. On the other hand, neither TNF-α nor CGS21680+TNF-α significantly affected Rev-Erbα levels.
Our results demonstrate, for the first time to our knowledge, that A2AR stimulation activates the intrinsic clockwork machinery of the human monocytic THP-1 cell line by increasing the expression of the core loop activators, clock and Bmal1, while decreasing the expression of the core loop repressor Per2 and confers greater robustness to the circadian phase by increasing the secondary loop repressor Rev-Erbα (summarized in Fig. 6). TNF-α also activates the core loop by increasing Bmal1 expression and decreasing Cry1. On the other hand, A2AR activation does not alter the impact of TNF-α neither on the core nor on the secondary loops here studied. The demonstration that A2AR activation and TNF-α concomitantly regulate the intrinsic circadian clock in immune cells provides an attractive explanation for the observation that methotrexate, which promotes adenosine release, is associated with such adverse circadian affects as fatigue and may therefore shed further light on the anti-inflammatory actions of MTX therapy and may provide new insights to improve the quality of life for RA patients undergoing therapy.
Fig. 6.
Diagram depicting the A2AR and TNF-α roles on the intrinsic molecular clockwork of THP-1 cells.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (AR56672, AR56672S1 and AR54897).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10753-012-9530-x) contains supplementary material, which is available to authorized users.
REFERENCES
- 1.Hogenesch JB, Herzog ED. Intracellular and intercellular processes determine robustness of the circadian clock. FEBS Letters. 2011;585(10):1427–1434. doi: 10.1016/j.febslet.2011.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288(5466):682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 4.Yoo SH, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(15):5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shackelford PG, Feigin RD. Periodicity of susceptibility to pneumococcal infection: Influence of light and adrenocortical secretions. Science. 1973;182(4109):285–287. doi: 10.1126/science.182.4109.285. [DOI] [PubMed] [Google Scholar]
- 6.Cutolo M, et al. Circadian rhythms in RA. Annals of the Rheumatic Diseases. 2003;62(7):593–596. doi: 10.1136/ard.62.7.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutherland ER. Nocturnal asthma. The Journal of Allergy and Clinical Immunology. 2005;116(6):1179–1186. doi: 10.1016/j.jaci.2005.09.028. quiz 1187. [DOI] [PubMed] [Google Scholar]
- 8.Hashiramoto A, et al. Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. Journal of Immunology. 2010;184(3):1560–1565. doi: 10.4049/jimmunol.0903284. [DOI] [PubMed] [Google Scholar]
- 9.Harkness JA, et al. Circadian variation in disease activity in rheumatoid arthritis. British Medical Journal. (Clinical Research Ed) 1982;284(6315):551–554. doi: 10.1136/bmj.284.6315.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowanko IC, et al. Circadian variations in the signs and symptoms of rheumatoid arthritis and in the therapeutic effectiveness of flurbiprofen at different times of day. British Journal of Clinical Pharmacology. 1981;11(5):477–484. doi: 10.1111/j.1365-2125.1981.tb01153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowanko IC, et al. Time of day of prednisolone administration in rheumatoid arthritis. Annals of the Rheumatic Diseases. 1982;41(5):447–452. doi: 10.1136/ard.41.5.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowanko IC, et al. Domiciliary self-measurement in the rheumatoid arthritis and the demonstration of circadian rhythmicity. Annals of the Rheumatic Diseases. 1982;41(5):453–455. doi: 10.1136/ard.41.5.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnett FC, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis and Rheumatism. 1988;31(3):315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 14.Pinals RS, Masi AT, Larsen RA. Preliminary criteria for clinical remission in rheumatoid arthritis. Arthritis and Rheumatism. 1981;24(10):1308–1315. doi: 10.1002/art.1780241012. [DOI] [PubMed] [Google Scholar]
- 15.Bijlsma JW, Jacobs JW. Methotrexate: Still the anchor drug in RA treatment. Joint, Bone, Spine. 2009;76(5):452–454. doi: 10.1016/j.jbspin.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Furst DE. The rational use of methotrexate in rheumatoid arthritis and other rheumatic diseases. British Journal of Rheumatology. 1997;36(11):1196–1204. doi: 10.1093/rheumatology/36.11.1196. [DOI] [PubMed] [Google Scholar]
- 17.Choy EH, Panayi GS. Cytokine pathways and joint inflammation in rheumatoid arthritis. The New England Journal of Medicine. 2001;344(12):907–916. doi: 10.1056/NEJM200103223441207. [DOI] [PubMed] [Google Scholar]
- 18.Cutolo M, Straub RH, Buttgereit F. Circadian rhythms of nocturnal hormones in rheumatoid arthritis: Translation from bench to bedside. Annals of the Rheumatic Diseases. 2008;67(7):905–908. doi: 10.1136/ard.2008.088955. [DOI] [PubMed] [Google Scholar]
- 19.Danis VA, et al. Circulating cytokine levels in patients with rheumatoid arthritis: Results of a double blind trial with sulphasalazine. Annals of the Rheumatic Diseases. 1992;51(8):946–950. doi: 10.1136/ard.51.8.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dasgupta B, et al. Serial estimation of interleukin 6 as a measure of systemic disease in rheumatoid arthritis. Journal of Rheumatology. 1992;19(1):22–25. [PubMed] [Google Scholar]
- 21.Weinblatt ME, et al. Efficacy of low-dose methotrexate in rheumatoid arthritis. The New England Journal of Medicine. 1985;312(13):818–822. doi: 10.1056/NEJM198503283121303. [DOI] [PubMed] [Google Scholar]
- 22.Chan ES, Cronstein BN. Molecular action of methotrexate in inflammatory diseases. Arthritis Research. 2002;4(4):266–273. doi: 10.1186/ar419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan ES, Cronstein BN. Methotrexate—how does it really work? Nature Reviews. Rheumatology. 2010;6(3):175–178. doi: 10.1038/nrrheum.2010.5. [DOI] [PubMed] [Google Scholar]
- 24.Gessi S, et al. Adenosine receptors in health and disease. Advances in Pharmacology. 2011;61:41–75. doi: 10.1016/B978-0-12-385526-8.00002-3. [DOI] [PubMed] [Google Scholar]
- 25.Hasko G, et al. Adenosine inhibits IL-12 and TNF-[alpha] production via adenosine A2a receptor-dependent and independent mechanisms. The FASEB Journal. 2000;14(13):2065–2074. doi: 10.1096/fj.99-0508com. [DOI] [PubMed] [Google Scholar]
- 26.Le Moine O, et al. Adenosine enhances IL-10 secretion by human monocytes. Journal of Immunology. 1996;156(11):4408–4414. [PubMed] [Google Scholar]
- 27.Le Vraux V, et al. Inhibition of human monocyte TNF production by adenosine receptor agonists. Life Sciences. 1993;52(24):1917–1924. doi: 10.1016/0024-3205(93)90632-d. [DOI] [PubMed] [Google Scholar]
- 28.Link AA, et al. Ligand-activation of the adenosine A2a receptors inhibits IL-12 production by human monocytes. Journal of Immunology. 2000;164(1):436–442. doi: 10.4049/jimmunol.164.1.436. [DOI] [PubMed] [Google Scholar]
- 29.Thiel M, Chouker A. Acting via A2 receptors, adenosine inhibits the production of tumor necrosis factor-alpha of endotoxin-stimulated human polymorphonuclear leukocytes. The Journal of Laboratory and Clinical Medicine. 1995;126(3):275–282. [PubMed] [Google Scholar]
- 30.Qu W, et al. MFEprimer: Multiple factor evaluation of the specificity of PCR primers. Bioinformatics. 2009;25(2):276–278. doi: 10.1093/bioinformatics/btn614. [DOI] [PubMed] [Google Scholar]
- 31.Khoa ND, et al. Inflammatory cytokines regulate function and expression of adenosine A(2A) receptors in human monocytic THP-1 cells. Journal of Immunology. 2001;167(7):4026–4032. doi: 10.4049/jimmunol.167.7.4026. [DOI] [PubMed] [Google Scholar]
- 32.Keller M, et al. A circadian clock in macrophages controls inflammatory immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(50):21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavadini G, et al. TNF-alpha suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(31):12843–12848. doi: 10.1073/pnas.0701466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin L, Lazar MA. The orphan nuclear receptor Reverbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Molecular Endocrinology. 2005;19(6):1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi JS, et al. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nature Reviews Genetics. 2008;9(10):764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krieger DT. Rhythms of ACTH and corticosteroid secretion in health and disease, and their experimental modification. Journal of Steroid Biochemistry. 1975;6(5):785–791. doi: 10.1016/0022-4731(75)90068-0. [DOI] [PubMed] [Google Scholar]
- 37.Semaan N, et al. miR-346 controls release of TNF-alpha protein and stability of its mRNA in rheumatoid arthritis via tristetraprolin stabilization. PLoS One. 2011;6(5):e19827. doi: 10.1371/journal.pone.0019827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straub RH, Cutolo M. Circadian rhythms in rheumatoid arthritis: Implications for pathophysiology and therapeutic management. Arthritis and Rheumatism. 2007;56(2):399–408. doi: 10.1002/art.22368. [DOI] [PubMed] [Google Scholar]
- 39.Drewes AM, et al. Sleep in rheumatoid arthritis: A comparison with healthy subjects and studies of sleep/wake interactions. British Journal of Rheumatology. 1998;37(1):71–81. doi: 10.1093/rheumatology/37.1.71. [DOI] [PubMed] [Google Scholar]
- 40.Latman NS. Relation of menstrual cycle phase to symptoms of rheumatoid arthritis. American Journal of Medicine. 1983;74(6):957–960. doi: 10.1016/0002-9343(83)90789-1. [DOI] [PubMed] [Google Scholar]
- 41.Pollard LC, et al. Fatigue in rheumatoid arthritis reflects pain, not disease activity. Rheumatology (Oxford, England) 2006;45(7):885–889. doi: 10.1093/rheumatology/kel021. [DOI] [PubMed] [Google Scholar]
- 42.Nesher G, Mates M, Zevin S. Effect of caffeine consumption on efficacy of methotrexate in rheumatoid arthritis. Arthritis and Rheumatism. 2003;48(2):571–572. doi: 10.1002/art.10766. [DOI] [PubMed] [Google Scholar]
- 43.Koyanagi S, et al. cAMP-response element (CRE)-mediated transcription by activating transcription factor-4 (ATF4) is essential for circadian expression of the Period2 gene. Journal of Biological Chemistry. 2011;286(37):32416–32423. doi: 10.1074/jbc.M111.258970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernini JC, et al. Aminophylline for methotrexate-induced neurotoxicity. The Lancet. 1995;345(8949):544–547. doi: 10.1016/s0140-6736(95)90464-6. [DOI] [PubMed] [Google Scholar]
- 45.Foey AD, et al. Impact of VIP and cAMP on the regulation of TNF-alpha and IL-10 production: Implications for rheumatoid arthritis. Arthritis Research & Therapy. 2003;5(6):R317–R328. doi: 10.1186/ar999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kambayashi T, et al. Cyclic nucleotide phosphodiesterase type IV participates in the regulation of IL-10 and in the subsequent inhibition of TNF-alpha and IL-6 release by endotox-in-stimulated macrophages. Journal of Immunology. 1995;155(10):4909–4916. [PubMed] [Google Scholar]
- 47.Meisel C, et al. Differential regulation of monocytic tumor necrosis factor-alpha and interleukin-10 expression. European Journal of Immunology. 1996;26(7):1580–1586. doi: 10.1002/eji.1830260726. [DOI] [PubMed] [Google Scholar]
- 48.Elbim C, et al. Intracellular pool of IL-10 receptors in specific granules of human neutrophils: Differential mobilization by proinflammatory mediators. Journal of Immunology. 2001;166(8):5201–5207. doi: 10.4049/jimmunol.166.8.5201. [DOI] [PubMed] [Google Scholar]
- 49.Hodge-Dufour J, et al. Inhibition of interferon gamma induced interleukin 12 production: A potential mechanism for the anti-inflammatory activities of tumor necrosis factor. Proc Natl Acad Sci USA. 1998;95(23):13806–13811. doi: 10.1073/pnas.95.23.13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keller CC, et al. Acquisition of hemozoin by monocytes down-regulates interleukin-12 p40 (IL-12p40) transcripts and circulating IL-12p70 through an IL-10-dependent mechanism: In vivo and in vitro findings in severe malarial anemia. Infection and Immunity. 2006;74(9):5249–5260. doi: 10.1128/IAI.00843-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma X, et al. Inhibition of IL-12 production in human monocyte-derived macrophages by TNF. Journal of Immunology. 2000;164(4):1722–1729. doi: 10.4049/jimmunol.164.4.1722. [DOI] [PubMed] [Google Scholar]
- 52.Platzer C, et al. Up-regulation of monocytic IL-10 by tumor necrosis factor-alpha and cAMP elevating drugs. International Immunology. 1995;7(4):517–523. doi: 10.1093/intimm/7.4.517. [DOI] [PubMed] [Google Scholar]
- 53.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93(6):929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 54.Cardone L, et al. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309(5739):1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 55.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.