Using a newly developed method that measures nucleosome density-normalized chromatin accessibility, Tsukiyama and colleague find a Mec1-dependent increase in chromatin accessibility specifically at and ahead of replication forks under conditions of replication stress. Mec1 is also found to be required for normal replication fork progress rate. The study provides valuable insight into how chromatin structure is regulated to facilitate replication fork progression through the chromatin template.
Keywords: ATR, DNA replication, Mec1, S-phase checkpoint, chromatin, chromatin accessibility, replication fork progression
Abstract
Faithful DNA replication is essential for normal cell division and differentiation. In eukaryotic cells, DNA replication takes place on chromatin. This poses the critical question as to how DNA replication can progress through chromatin, which is inhibitory to all DNA-dependent processes. Here, we developed a novel genome-wide method to measure chromatin accessibility to micrococcal nuclease (MNase) that is normalized for nucleosome density, the NCAM (normalized chromatin accessibility to MNase) assay. This method enabled us to discover that chromatin accessibility increases specifically at and ahead of DNA replication forks in normal S phase and during replication stress. We further found that Mec1, a key regulatory ATR-like kinase in the S-phase checkpoint, is required for both normal chromatin accessibility around replication forks and replication fork rate during replication stress, revealing novel functions for the kinase in replication stress response. These results suggest a possibility that Mec1 may facilitate DNA replication fork progression during replication stress by increasing chromatin accessibility around replication forks.
Eukaryotic DNA replication is a highly regulated process that must be completed faithfully once and only once in every S phase of the cell cycle to ensure that two genetically identical daughter cells are produced. In the budding yeast Saccharomyces cerevisiae, DNA replication initiates at multiple discrete genomic locations known as origins of replication (ORIs). A well-defined temporal pattern of ORI activation (firing) exists in yeast such that some ORIs fire early and others fire late during S phase (Raghuraman et al. 2001). ORI firing is followed by the formation of bidirectional DNA replication forks (forks hereafter) that move away from ORIs as DNA replication progresses (Bell and Dutta 2002). One major complication during initiation and progression of DNA replication arises from the fact that eukaryotic genomes are tightly packaged into chromatin (Tabancay and Forsburg 2006). The basic structural and functional unit of chromatin is the nucleosome, which consists of ∼147 base pairs (bp) of DNA wrapped around an octamer of the highly conserved core histones (H3, H4, H2A, and H2B) (Luger et al. 1997; Richmond and Davey 2003). In addition to their genome-packaging function, nucleosomes exhibit strong inhibitory effects on DNA-dependent processes such as transcription and DNA repair, recombination (Kouzarides 2007), and replication (Tabancay and Forsburg 2006).
During DNA replication, the replisome, the >2-MDa protein assembly that replicates DNA, has to travel through nucleosome arrays. In yeast cells under normal growth conditions, replication forks move at an average rate of 3 kb per minute (Raghuraman et al. 2001; Yabuki et al. 2002; Vincent et al. 2008), which means that the replisome, on average, moves past a nucleosome every 3 sec. Despite the importance of the issue, very little is known about the mechanisms by which cells overcome strong inhibitory effects of nucleosomes during replication fork progression. In contrast to DNA replication, effects of the presence of nucleosomes have been extensively studied for transcription elongation, which, like DNA replication fork progression, involves a large protein complex progressing through nucleosome arrays. In this case, there is clear evidence that the presence of nucleosome arrays strongly inhibits RNA polymerase II (Pol II) progression in vitro (Izban and Luse 1991; Orphanides et al. 1998). To alleviate this strong inhibitory effect, eukaryotic cells have evolved multiple mechanisms, such as removal of H2A–H2B dimers by the FACT complex as well as covalent modification of specific histone residues by histone-modifying enzymes traveling with the elongating Pol II (Selth et al. 2010). These results suggest that similar to transcription, chromatin changes may take place during replication to accommodate replisome progression. However, to date, these changes have remained elusive, perhaps because many of the studies to date were done using asynchronously growing cell populations.
The intra-S-phase checkpoint (checkpoint hereafter) plays key roles in both ORI firing and replication progression. Checkpoint protein functions have been best characterized in the presence of replication stress, during which replication forks slow down and/or stall (Friedel et al. 2009). Replication stress leads to targeting of multiple proteins to stalled replication forks, which initiates a cascade of events to activate the checkpoint response. In S. cerevisiae, a central player in this reaction is the protein kinase Mec1 (ATR in metazoans), which directs a number of events to ensure cell survival (Friedel et al. 2009). These include delaying the time of late ORI firing (Santocanale and Diffley 1998; Shirahige et al. 1998; Alvino et al. 2007) and prevention of replication fork collapse (Lopes et al. 2001; Tercero and Diffley 2001). Understanding the molecular mechanisms by which the checkpoint regulates multiple aspects of DNA replication and genome stability is a major challenge in the field.
In this study, we sought to determine the properties of replicating chromatin under replication stress conditions in cells synchronously undergoing DNA replication. Surprisingly, our results indicate that the positions of nucleosomes change very little, if at all, during DNA replication. We extended our analysis of chromatin structure by developing a novel method to measure accessibility of chromatin to micrococcal nuclease (MNase hereafter) that is normalized for histone density, which we named NCAM (normalized chromatin accessibility to MNase) assay. This approach has led to the discovery that nucleosomes at and around DNA replication forks increase accessibility to MNase as compared with nonreplicating chromatin both in normal S phase and during replication stress conditions. We further found that the checkpoint kinase Mec1 is required for normal chromatin accessibility around forks under replication stress, revealing a previously unknown function of this kinase. In addition, using the mec1-100 allele, we provide strong evidence that Mec1 promotes replication fork progression during replication stress. These results suggest an interesting possibility that Mec1 may facilitate replication fork progression by increasing chromatin accessibility.
Results
Chromosome-wide mapping of genomic regions undergoing DNA replication
DNA replication is a very fast and dynamic process that progresses through the entire genome. Therefore, in order to map the regions of active DNA replication for the analysis of chromatin structure, we required the cell populations to proceed synchronously through DNA replication. We therefore performed the first sets of experiments by arresting cells in G1 phase with α-factor, then releasing them into rich medium containing 200 mM hydroxyurea (HU), a drug that decreases the overall rate of fork progression through the reduction of the intracellular dNTP pools (see the Materials and Methods; Koc et al. 2004). Cells were harvested during G1 arrest (nonreplicating control) and at 60 min post-release into S phase. Both arrest and release steps were monitored by flow cytometry analysis (data not shown). High-density custom tiling microarrays covering chromosomes III, VI, and XII (Yadon et al. 2010) were used for the subsequent mapping of DNA replication sites and chromatin features.
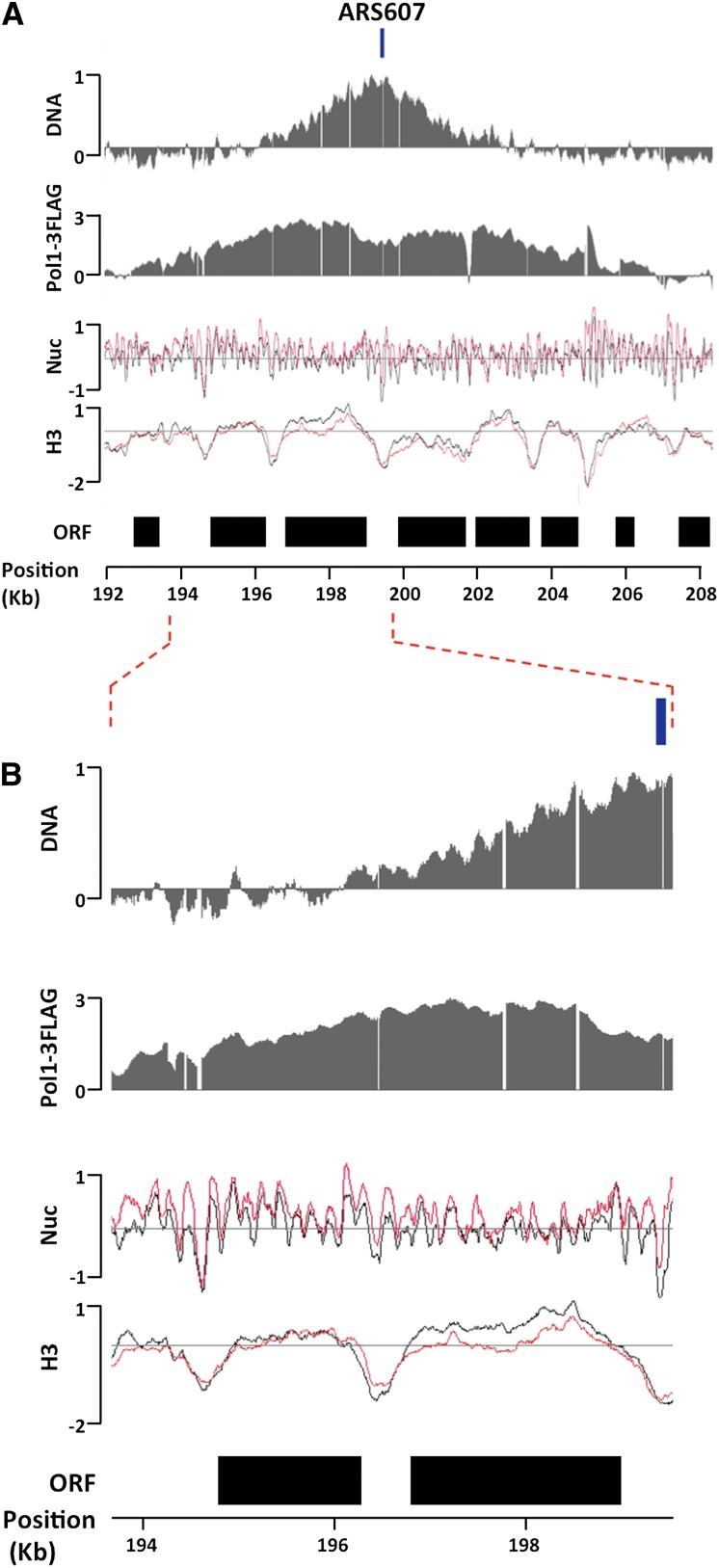
Two complementary approaches were taken in order to map the sites of active DNA replication. First, we mapped the sites of active DNA synthesis by generating DNA copy number profiles (DNA profiles hereafter) (Yabuki et al. 2002). By 60 min in HU, firing activity was detected at 23 ORIs across chromosomes III, VI, and XII (Supplemental Table S1). We refer to these ORIs as early firing, as previously reported (Supplemental Table S1). As shown in Figure 1, for the early-firing ORI ARS607 (see Supplemental Fig. S1 for additional early ORIs), the maximal signal in DNA profiles coincided with previously published ORI positions (Supplemental Table S1). ORIs that had not fired by 60 min in HU were considered as either late firing or inactive, as previously reported (Supplemental Table S1), and were used as nonreplicating control regions (Supplemental Fig. S2). A total of 18 late-firing ORIs were found across our data set (Supplemental Table S1). Second, we mapped fork positions by chromatin immunoprecipitation (ChIP) of a Flag-tagged DNA Pol I, a replication fork component (Burgers 1998). As expected, Pol I-enriched regions were only detected in S-phase samples and precisely mapped around the same early-firing ORIs where we detected DNA synthesis (Fig. 1; Supplemental Figs. S1, S2). Our chromatin preparation protocol consistently yielded samples sonicated to an average fragment size of 250 bp (Supplemental Fig. S3A), which, together with the use of high-density tiling arrays, provided sufficient resolution to clearly distinguish between left and right forks moving away from ORIs (Fig. 1; Supplemental Fig. S1).
Figure 1.
Analysis of chromatin structure around replication forks. (A) DNA profiles (DNA), DNA Pol I ChIP signals (Pol1-3Flag), nucleosome mapping (Nuc), and H3 ChIP (H3) signals around the efficient early-firing ORI ARS607, whose previously annotated position is delimited by a box above the DNA panel. All Y-axes are on the log2 scale. DNA profile analysis and Pol I ChIP were done in S phase (60 min in HU). Black and red lines in nucleosome mapping and H3 ChIP data correspond to G1 and S phase (60 min in HU) samples, respectively. Black boxes denote the position of ORFs. (B) The same as A, except that the region between the dotted lines is expanded.
Changes in chromatin structure during DNA replication
We next investigated the properties of chromatin in the regions undergoing DNA replication by first mapping nucleosome positions. Cells for nucleosome mapping experiments were harvested simultaneously with those for DNA profile and replication fork mapping (see the Materials and Methods). Mononucleosomal DNA and genomic DNA from the same pool of cells were competitively hybridized onto our arrays. Surprisingly, at the levels of synchrony and resolution achieved in our experiments, we did not detect notable changes in nucleosome positions at and around replication forks (Fig. 1; Supplemental Fig. S1, Nuc panels). Interestingly, the most prominent change in chromatin structure in S phase was that nucleosome signals at and around replication forks were higher than in the G1 sample (Fig. 1; cf. Supplemental Figs. S1 [Nuc panels] and S2 [nonreplicating regions]).
There are two parameters that could affect the nucleosome signal intensity: nucleosome occupancy (fraction of cells within a population that have a nucleosome formed at a given locus at a given time) and accessibility of nucleosome to MNase (Supplemental Fig. S4). For example, it is possible that nucleosome occupancy is generally higher immediately after nucleosomes are newly deposited behind replication forks and gradually decreases as the cell cycle progresses. If this were the case, even if the sensitivity of each nucleosome to MNase stayed constant between G1 and S phases, higher nucleosome signals would be expected from S-phase chromatin. To test whether higher nucleosome occupancy caused the increase in nucleosome signals at forks, we measured histone density independently from MNase by histone H3 ChIP following extensive chromatin sonication (Lee et al. 2004; Fan et al. 2008, 2010). Interestingly, histone H3 density at and around forks in S phase was not higher, but similar to or lower, than in G1 phase (Fig. 1; Supplemental Fig. S1). No significant changes in H3 density were detected between G1 and S phase at nonreplicating regions (Supplemental Fig. S2). Similar results were obtained by ChIP using anti-histone H2B antibodies (data not shown). These results demonstrated that increases in nucleosome density are not the cause for the increases in nucleosome signals around replication forks. Rather, the higher nucleosome signals were obtained from regions that have similar or lower nucleosome occupancy.
Nucleosome accessibility increases at and around DNA replication forks
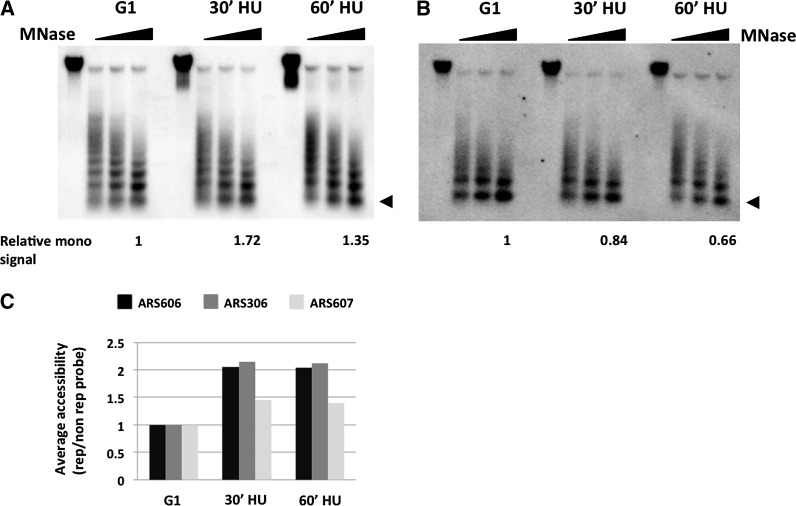
The results above suggested that nucleosomes around replication forks might be more accessible to MNase (Supplemental Fig. S4). Consistent with this, it was recently reported that elevated MNase accessibility leads to higher nucleosome signals at or around transcription start sites of Pol II genes (Weiner et al. 2010). To address this hypothesis, we directly measured the sensitivity of chromatin to digestion by MNase. G1- and S-phase chromatin samples were digested with MNase and analyzed by Southern blot hybridization with probes corresponding to replicating and nonreplicating regions. If our hypothesis is correct, mononucleosomes should be produced more readily from chromatin around replication forks than that from nonreplicating control regions upon MNase digestion. Indeed, larger fractions of mononucleosome signals were obtained from S-phase samples compared with G1 samples when hybridized with a probe from a replicating region adjacent to ARS606 (Fig. 2A). To confirm that the shift in nucleosome signal was replication dependent, the same membrane was rehybridized with a probe corresponding to a nonreplicating control region in chromosome VI. In this case, we did not detect any significant increase in the relative amount of mononucleosomal signals between G1- and S-phase chromatin (Fig. 2B). Quantification and normalization of these results showed that chromatin around replication forks is more readily digested by MNase (Fig. 2C). Similar results were obtained by using a probe adjacent to the early-firing ORIs ARS306 and ARS607 (Fig. 2C). Taken together, these results establish that chromatin is more accessible to MNase around replication forks.
Figure 2.
Evidence that chromatin accessibility to MNase increases around replication forks. (A) MNase digestion kinetics at a replicating region. Chromatin samples from cells in G1 and S phase (30 min and 60 min in HU) were digested by increasing amounts of MNase, followed by Southern blotting using a probe adjacent to an efficient early-firing ORI, ARS606. The fraction of mononucleosome signal from the strongest MNase digestion relative to the total signal in the same lane is shown below. The triangle denotes the mononucleosome signal. (B) MNase digestion kinetics at a control locus. The membrane used in A was stripped and hybridized to a probe corresponding to a nonreplicating region on the same chromosome. The relative fraction of mononucleosome signal from the strongest MNase digestion is shown as in A. The triangle denotes the mononucleosome signal. (C) Quantification of the fraction of mononucleosome signals from the highest MNase concentration at each time point around three efficient early-firing ORIs: ARS606, ARS306, and ARS607. The relative mononucleosome signal around each ORI is normalized to that at a nonreplicating region on the same chromosome, and the relative value in G1 was set at 1.0.
Establishment of a normalized measure of chromatin accessibility to MNase
The finding that chromatin around replication forks exhibits elevated accessibility to MNase prompted us to determine chromatin accessibility during DNA replication in a global fashion using tiled microarrays. Supporting this approach, a recent study demonstrated that more accessible nucleosomes yield higher signals when mapped on microarrays (Weiner et al. 2010). However, as discussed in Supplemental Figure S4, nucleosome signal strengths are affected by both histone density and MNase accessibility of chromatin at any given locus. Therefore, the intensity of nucleosome signals alone cannot be used to assess MNase accessibility. Indeed, currently, there is no available method to determine MNase accessibility of chromatin independently from nucleosome density in a genome-wide fashion. We therefore sought to measure MNase accessibility of chromatin by normalizing nucleosome signals by histone H3 ChIP signal. This method, the NCAM assay, calculates the degree of chromatin digestion by MNase per amount of histone (see the Materials and Methods for details). We validated the utility of this method by measuring NCAM at gene promoters. As shown in Supplemental Figure S5, NCAM at promoters correlated well with the transcriptional levels of genes at a global level.
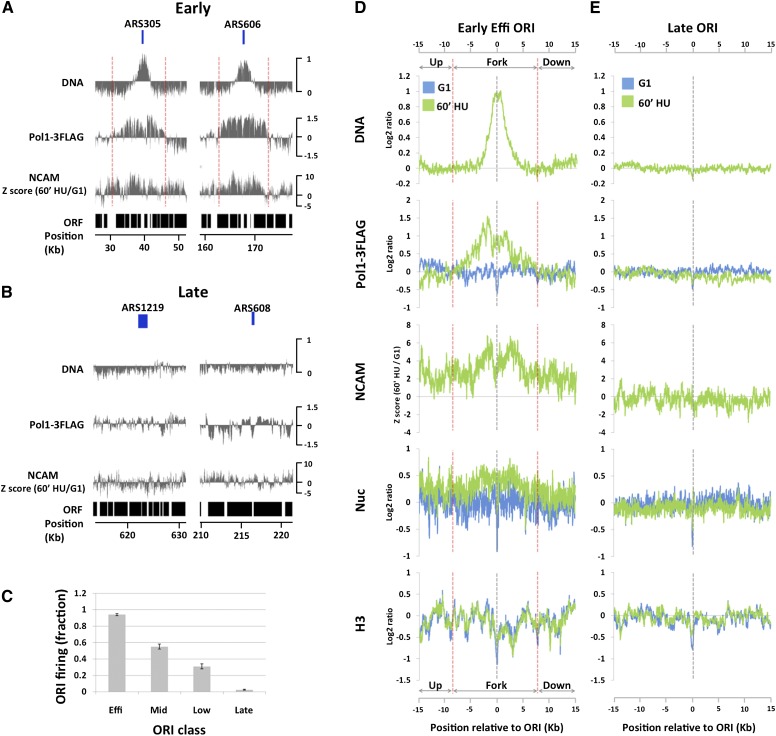
We then performed the NCAM assay during DNA replication. As shown in Figure 3A and consistent with results in Figure 1, cells in S phase displayed an increase in NCAM at early efficient ORIs ARS305 and ARS606 but not at late ORIs, where neither DNA synthesis nor Pol I enrichment was detected (Fig. 3B). Importantly, the replication-dependent increase in NCAM does not peak at the sites of maximal DNA signal and extends ahead of Pol I and DNA synthesis regions—in some cases, for several thousands of base pairs. We then averaged data for the most efficient early ORIs in our data set, which, based on their DNA profiles, had fired in ∼90% of cells, on average (Fig. 3C, early efficient ORIs). To facilitate visualization and quantification of the averaged data, we defined a fork region around ORI midpoints, where the level of Pol I enrichment is above the genome average. The regions upstream of and downstream from the fork region were considered ahead of forks. In agreement with single ORI analysis (Fig. 3A), NCAM increased by 60 min in HU at and ahead of the fork region, with a distribution similar to that of Pol I ChIP signals and clearly different from DNA copy number profiles (Fig. 3D). At forks, the increase in NCAM is mainly driven by an increase in nucleosome signal and, to a much lesser extent, by a decrease in histone H3 occupancy (Fig. 3D, nucleosome and H3 panels). On the other hand, the increase in NCAM ahead of forks, which extends a minimum of 7 kb (see Supplemental Fig. S6 for broader chromosomal regions), is exclusively driven by an increase in nucleosome signal, as no significant H3 loss is detected (Fig. 3D, nuc and H3 panels, up and down regions). Similar results were obtained for less efficient early ORIs, except that the decreasing levels of ORI activity (Fig. 3C, early mid and low columns) were associated with decreasing NCAM levels both at the forks and ahead of them (Supplemental Fig. S7, NCAM panels). In contrast to early ORIs, no NCAM increase was detected at late ORIs, where no replication activity was detected by 60 min in HU, as shown by complete absence of Pol I and DNA signals (Fig. 3E). In summary, the results above clearly establish that a previously unknown increase in chromatin accessibility occurs at and ahead of replication forks in the presence of replication stress.
Figure 3.
Global analysis of chromatin accessibility around replication forks. (A) DNA profiles (DNA) and Pol I ChIP signals (Pol1-3Flag) in S phase (60 min in HU) and difference in NCAM between S and G1 phases (NCAM) around two efficient early-firing ORIs: ARS305 and ARS606. Previously annotated positions of ARSs are delimited by the boxes above the DNA panels. The Y-axes are on the log2 scale, except for the NCAM panel. The dotted red lines denote replication fork regions where Pol I ChIP signals are above the genome average. (B) The same as A, except that the analyses were done around two late-firing origins: ARS1219 and ARS608. (C) Origin firing efficiencies (mean ± standard error of the mean from two biological replicates), as judged by DNA profile analysis, of different classes of ORIs at the time of the experiment (60 min in HU). (D) DNA profiles (DNA), Pol I ChIP signals (Pol1-3Flag), NCAM signals (NCAM), nucleosome mapping (Nuc), and H3 ChIP signals (H3) averaged from all eight efficient early-firing ORIs on chromosomes III, VI, and XII. G1 data are in blue, and S-phase data (60 min in HU) are in green, except for NCAM signals, which show the differences between S and G1. Data sets are aligned at the peak of DNA profiles, which precisely coincide with the previously published ORI midpoints. The dotted red lines denote replication fork regions where Pol I ChIP signals are above the genome average. The Y-axes are on the log2 scale, except for the NCAM panel. (E) The same as D, except that the data are averaged from 18 late-firing ORIs on chromosomes III, VI, and XII.
Chromatin accessibility increases around replication forks during normal S phase
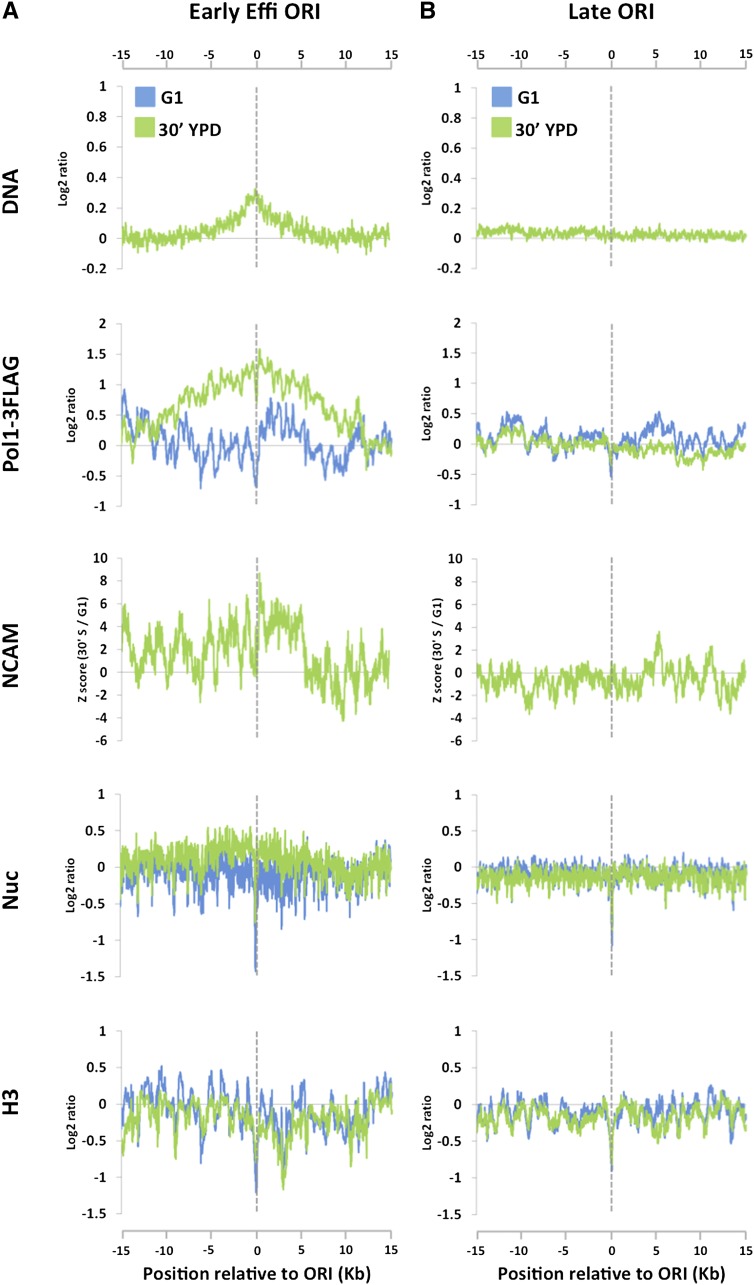
The results above prompted us to ask whether chromatin accessibility also increases around replication forks during normal S phase (in the absence of replication stress). To address this question, cells were released from α-factor arrest into rich medium at a temperature of 24°C to slow down forks. To compensate for the higher replication fork asynchrony later in S phase, which is caused by heterogeneous ORI firing and faster fork progression, cells were harvested at an earlier time point (30 min) for NCAM assay. Despite the fact that a much smaller fraction of cells within a population fired early ORIs and that replication forks progress less synchronously compared with the experiment in the presence of HU (cf. Figs. 3D and 4A [DNA and Pol1-3Flag panels]), we observed a sharp increase in NCAM around replication forks (Fig. 4A). The increase in NCAM was not detected in nonreplicating control regions (Fig. 4B). These results clearly demonstrate that an increase in chromatin accessibility around replication forks takes place during normal S phase.
Figure 4.
NCAM signals increase around replication forks during normal S phase. Cells were released from α-factor arrest into rich medium at 24°C, and the NCAM assay was performed during S phase as in Figure 3. (A) DNA profiles (DNA), Pol I ChIP signals (Pol1-3Flag), NCAM signals (NCAM), nucleosome mapping (Nuc), and H3 ChIP signals (H3) averaged from all eight efficient early-firing ORIs on chromosomes III, VI, and XII. G1 data are in blue, and S-phase data (30 min in S phase) are in green, except for NCAM signals, which show the differences between S and G1. Data sets are aligned at the peak of DNA profiles, which precisely coincide with the previously published ORI midpoints. The Y-axes are on the log2 scale, except for the NCAM panel. (B) The same as A, except that the data are averaged from 18 late-firing ORIs on chromosomes III, VI, and XII.
Increased chromatin accessibility at DNA replication forks during replication stress is Mec1-dependent
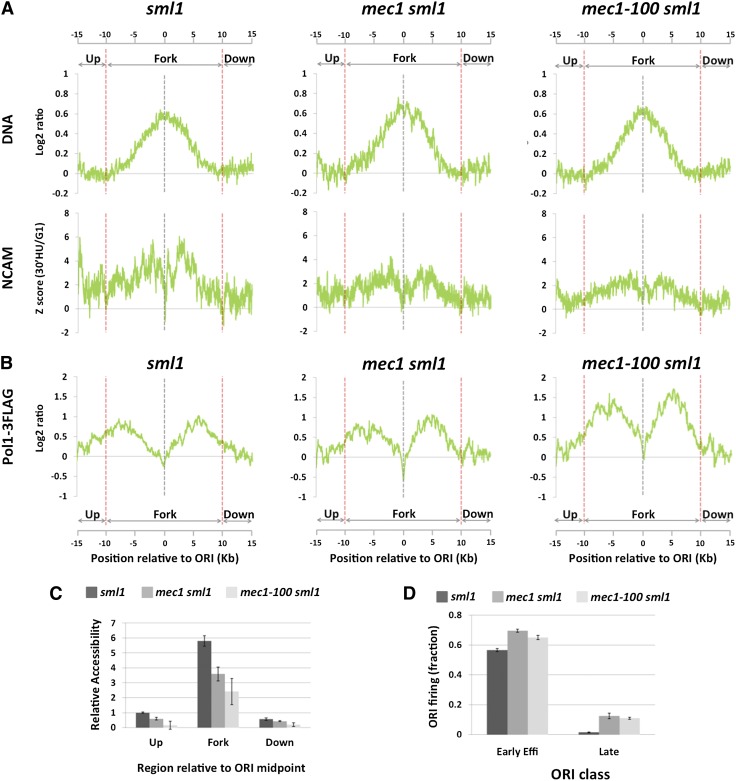
Because the S-phase checkpoint controls many aspects of DNA replication under replication stress (Friedel et al. 2009), we considered the possibility that it might affect chromatin accessibility around forks in the presence of replication stress. To test this hypothesis, we deleted MEC1, a key kinase in this checkpoint pathway. SML1 was simultaneously deleted to allow viability of the mec1-null mutants (Zhao et al. 2001). The sml1 strain was used as a control. Similar to experiments with wild-type cells, mec1 sml1 and sml1 cells were arrested in G1 and released into S phase in the presence of HU. Cells were harvested at an earlier time point in S phase (30 min post-release from α-factor) to compensate for the faster replication progression of the sml1 background (Poli et al. 2012). Both sml1 and mec1 sml1 mutants showed increases in NCAM at and around forks (Fig. 5A, NCAM panels). However, quantification by signal integration of the increase in NCAM at early efficient ORIs showed that mec1 sml1 double mutants have a 45% reduction in accessibility compared with sml1 cells (Fig. 5C). This is likely an underestimation, as firing activity at early efficient ORIs was, on average, 13% higher in mec1 sml1 cells compared with sml1 cells at the time point at which they were harvested (Fig. 5D).
Figure 5.
MEC1-dependent increase in chromatin accessibility around replication forks. (A) DNA (DNA, top panels) and NCAM (NCAM, bottom panels) profiles at early efficient ORIs (n = 8) by 30 min in S phase in the presence of 200 mM HU in sml1, mec1-100 sml1, and mec1-100 sml1 mutant strains. (B) Same as in A for Pol1-3Flag signal profiles. (C) Quantification of the relative increase in NCAM between G1 and S phases from A in sml1, mec1 sml1, and mec1-100 sml1 cells upstream of, within, and downstream from replication fork regions. The value upstream of the replication fork region in sml1 cells is set as 1.0. (D) Origin firing efficiencies, as judged by DNA profile analysis, of early efficient and late-firing ORIs in sml1, mec1 sml1, and mec1-100 sml1 cells at the time of the experiment (30 min in HU). Values in panels C and D express the mean ± standard error of the mean from two independent biological replicates.
We next tested whether increased NCAM around replication forks during normal S phase is also Mec1-dependent. As shown in Supplemental Figure S8, NCAM around replication forks in mec1 sml1 and sml1 mutants is very similar, showing that the roles of Mec1 in chromatin accessibility control are a part of the replication stress response. Together, these results suggest that Mec1 has a previously unknown role in promoting an increase in chromatin accessibility at and around stalled DNA replication forks in response to replication stress.
mec1-null and mec1-100 mutants display similar losses in chromatin accessibility at forks
We next sought to identify potential physiological roles for Mec1-dependent chromatin accessibility at replication forks. We hypothesized that the increased chromatin accessibility around replication forks may facilitate replication fork progression. However, analysis of the fork progression rate in mec1-null strains is difficult due to the fact that replication forks in these mutants degrade after prolonged exposure to replication stress (Supplemental Fig. S9; Lopes et al. 2001; Tercero and Diffley 2001; Cotta-Ramusino et al. 2005). To circumvent this issue, we took advantage of the mec1-100 mutant allele, which contains two amino acid substitutions outside of the kinase domain (Paciotti et al. 2001). mec1-100 mutants are partially defective for S-phase checkpoint activation. However, unlike mec1-null mutants, the mec1-100 mutants maintain a significant fraction of the replication fork protection function (Tercero et al. 2003), making them suitable for the analysis of fork progression. Indeed, viability of mec1-100 cells after 30 min of treatment in 200 mM HU is ∼63%, ∼30% lower than that of wild-type cells (∼90%). This is still much higher than an ∼3% viability of mec1-null mutants (data not shown). Consistent with this notion, DNA and Pol I profiles of mec1-100 sml1 were very similar to those in sml1 control cells at 30 min in HU (Fig. 5A,B), showing no detectable sign of fork degradation at this time point. We therefore performed NCAM assay at 30 min in HU in mec1-100 sml1 cells. This analysis revealed that mec1-100- and mec1-null mutations cause very similar loss of NCAM (Fig. 5A [NCAM panels], C). This loss in accessibility is not caused by forks moving less synchronously in mec1 sml1 and mec1-100 sml1 than sml1 mutants, as indicated by the very similar distribution of the Pol I ChIP signals among the three mutant strains (Fig. 5B). In addition, the fact that mec1-100- and mec1-null mutations cause a very similar loss of chromatin accessibility despite a very large difference in viability argues against the possibility that the loss of chromatin accessibility in mec1-100 is caused by compromised replication fork integrity.
Mec1-dependent increase in chromatin accessibility at forks is associated with faster fork progression
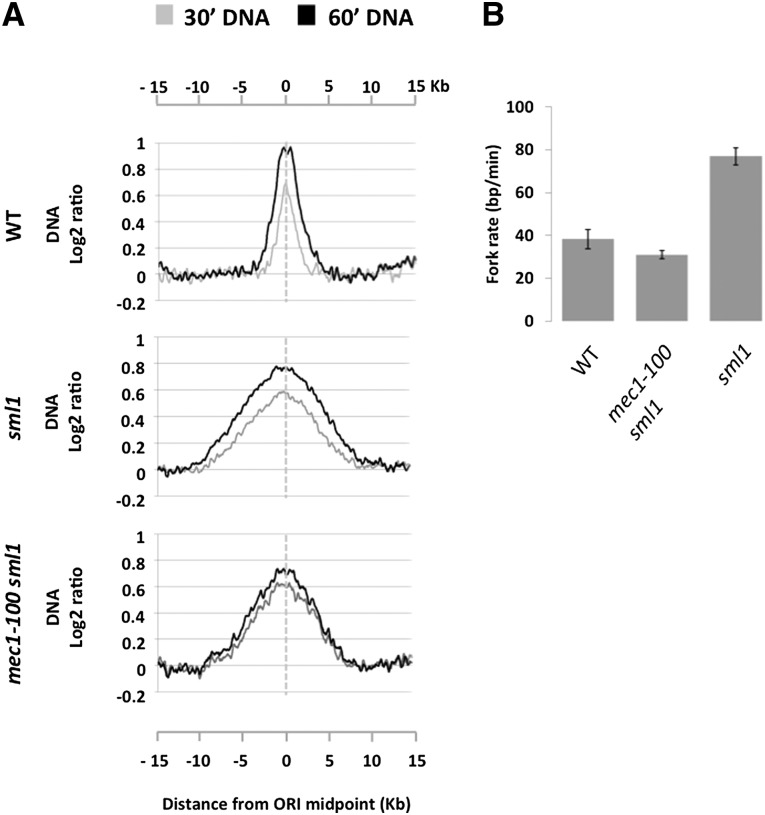
Importantly, the results above also meant that we could use the mec1-100 mutant to test whether Mec1 affects the fork progression rate. We used DNA content analysis to estimate the progression rate of forks originating from early efficient ORIs in the presence of 200 mM HU. The fork rate was estimated measuring the distance traveled by forks between 30 and 60 min post-release into S phase (see the Materials and Methods; Supplemental Fig. S10 for complete details). In wild-type cells, the fork rate from the average early efficient ORI was 38 ± 4 bp min−1 (Fig. 6A,B). Deletion of SML1, which increases dNTP levels (Zhao et al. 1998), resulted in a higher fork rate of 76 ± 4 bp min−1 (P < 0.05, two-tailed t-test) (Fig. 6A,B). This is in agreement with a recent report (Poli et al. 2012) that dNTP levels regulate fork progression. We next measured the fork rate in the mec1-100 sml1 mutant. Surprisingly, double-mutant cells displayed an average fork rate of 31 ± 2 bp min−1, much slower than that of sml1 cells (P < 0.005, two-tailed t-test) (Fig. 6A,B). The decrease in fork rate in mec1-100 sml1 compared with sml1 is a general phenomenon, as every single fork from early efficient ORIs was significantly slower in the double mutant compared with the single mutant (Supplemental Figs. S11, S12). These results revealed that Mec1 facilitates replication fork progression during replication stress conditions.
Figure 6.
Replication fork rate is compromised in mec1-100 cells in HU. (A) Average DNA profiles at 30 min (gray line) and 60 min (black line) from early efficient ORIs (n = 8) in wild type (WT), sml1, and mec1-100 sml1 cells. Data were smoothed using a moving average window of 800 bp. (B) Average replication fork rates from two independent biological replicates measured from 30 to 60 min in 200 mM HU in the same strains as in A. Error bars indicate the standard error of the mean.
We next asked whether previously described defects in mec1 mutants could explain the slow fork rate in the mec-100 sml1 strain. It has been shown that Mec1 is required for up-regulating dNTP levels during replication stress conditions (Huang and Elledge 1997), which, as shown above, can impact fork rates. Recently, the intracellular dNTP levels in several checkpoint mutants have been determined (Poli et al. 2012), which revealed that the dNTP levels are higher in mec1-100 sml1 than in wild-type cells both in G1 and at 60 min in S phase in the presence of 200 mM HU. Therefore, if Mec1 facilitates fork rate exclusively through increasing dNTP levels, we would expect fork rates to be higher in mec1-100 sml1 than in wild-type cells. However, as shown in Figure 6B, this is clearly not the case. Another possibility is that mec1-100 mutation affects the fork rate indirectly because it allows late ORIs to fire during replication stress (Tercero et al. 2003), which could lead to less available dNTPs and other resources for fork progression. We did not believe that this was very likely, as replication from late-firing ORIs is low even at 60 min in HU in mec1-100 sml1 cells (Supplemental Fig. S13A). To test this possibility directly, we measured fork rates in wild type and mec1-100 sml1 between earlier time points (29 and 40 min after release from G1). During earlier time points in S phase, we expected fork rates to be higher in both strains, as the available dNTP levels are higher earlier in S phase in the presence of HU (Poli et al. 2012). More importantly, if late ORI firing is responsible for slower forks from early ORIs in the mec1-100 sml1 mutant, we would expect forks from early ORIs to be even faster between 29 and 40 min in this mutant, as late ORI firing is lower at this time point (Supplemental Fig. S13B). In wild-type cells, in agreement with the description above, we measured a faster fork rate of 51 ± 10 bp min−1 (compared with 38 ± 4 bp min−1 between 30 and 60 min). In contrast, the fork rate in mec1-100 sml1 between 29 and 40 min in S phase was 30 ± 18 bp min−1, almost identical to that measured between 30 and 60 min (31 ± 2 bp min−1). These results strongly argue against the possibility that the slower fork rate in the mec1-100 mutant is due to its inability to delay late ORI firing. We therefore concluded that Mec1 facilitates replication fork progression under replication stress through a mechanism other than regulation of dNTP levels or late ORI firing. These results are consistent with the possibility that Mec1 facilitates replication fork progression in the presence of replication stress by up-regulating chromatin accessibility at and ahead of replication forks.
Discussion
NCAM assay around replication forks
In this study, we described the NCAM assay, a new method to measure chromatin accessibility to MNase in a way normalized for histone density. Although it has been shown that MNase accessibility strongly affects the strength of nucleosome signals upon nucleosome mapping (Weiner et al. 2010), a genome-wide measurement of chromatin accessibility to MNase has not been reported. The utility of the NCAM assay as a biologically meaningful parameter of chromatin structure was demonstrated by applying it to chromatin around replication forks and gene promoters. DNase I, the most commonly used endonuclease to measure chromatin accessibility, preferentially digests DNA in the long linkers between nucleosomes, such as nucleosome-depleted regions (NDR) at gene promoters (Bell et al. 2011), as well as sites where DNA structure is distorted (Zhu and Thiele 1996). In contrast, the NCAM assay can be applied to larger regions where regular arrays of nucleosomes are present to determine how chromatin accessibility changes under different conditions. We anticipate that the NCAM assay will be a useful tool in the study of chromatin structure in a wide variety of contexts in which access of protein factors to chromatin plays significant roles.
By measuring NCAM at different genomic locations as they undergo replication, we found that accessibility of chromatin to MNase significantly increases around replication forks during normal S phase as well as in the presence of replication stress caused by HU. How does chromatin accessibility increase around replication forks? This change is mainly driven by increased nucleosome signals and, to a much lesser extent, by decreased nucleosome occupancy. This means that the increased MNase sensitivity of nucleosomes is mainly responsible for the increased NCAM. It has been proposed that nucleosomes newly deposited after replication are hypersensitive to nucleases (Seale 1975, 1978; Hildebrand and Walters 1976). However, the increase in normalized chromatin accessibility coincides with the presence of the replisome (DNA Pol I ChIP signals), not with newly synthesized DNA on which nucleosomes are newly deposited. Moreover, increased accessibility extends several kilobases ahead of the replisomes. Therefore, it is highly unlikely that the nuclease hypersensitivity of newly deposited nucleosomes is the major cause for the increased chromatin accessibility that we observed. We envision that changes in the properties of nucleosomes, such as altered histone–DNA contacts or topological constraints on nucleosomal DNA, may be responsible for the increased accessibility of chromatin to MNase around replication forks. Besides changes in nucleosome accessibility, we also detected a reduced histone density at the regions where Pol I ChIP signals peak. The peak of Pol I marks the regions where the replisome is present in the highest fraction of cells within the population. The fact that the same decrease in histone density was detected using antibodies against both histones H3 and H2B likely reflects transient loss of entire nucleosomes at replication forks, as previously suggested (Tabancay and Forsburg 2006). This result strongly argues against the possibility that the increase in chromatin accessibility at the forks is due to the presence of nucleosomes lacking specific histones.
Mec1 promotes chromatin accessibility and replication fork progression
The S-phase checkpoint regulates diverse aspects of DNA replication under stress conditions that are directed to ensure cell survival, among which are the stabilization of replication forks and the delay in late ORI firing. The highly conserved Mec1 kinase plays central roles in the S-phase checkpoint (Friedel et al. 2009). Here, we present evidence that Mec1 function is required for the maintenance of high levels of both chromatin accessibility around replication forks and replication fork progression rates under replication stress conditions. One exciting possibility is that Mec1 facilitates the replication fork rate by increasing chromatin accessibility at and ahead of replication forks, although we cannot exclude other possibilities. For example, because Mec1 phosphorylates a large number of replisome components (Friedel et al. 2009), it may also facilitate replication fork progression by activating some of them. Our results suggest that, besides generally promoting a slower S-phase progression through the inhibition of late ORI firing (Santocanale and Diffley 1998; Shirahige et al. 1998), the S-phase checkpoint specifically helps forks maintain their progression rate. We hypothesize that both functions contribute to alleviate the deleterious effects of the nucleotide depletion imposed by HU: While the delay in late ORI firing helps maintain higher dNTP levels and other resources for replication forks originated from early ORIs, the checkpoint facilitates the progression of the large replication complex, possibly by making chromatin more accessible, such that S phase would not be unnecessarily long during this vulnerable state.
How does Mec1 promote chromatin accessibility? One possibility is that Mec1 increases chromatin accessibility through modification of replisome components. Alternatively, Mec1 may function through activation of chromatin remodeling factors, some of which are targeted to stalled replication forks (Papamichos-Chronakis and Peterson 2008; Shimada et al. 2008; Vincent et al. 2008). It is also possible that Mec1 activates and/or recruits complexes that covalently modify histones. The fact that the two amino acid substitutions in the mec1-100 allele reside outside of its kinase domain (Paciotti et al. 2001) leads us to speculate that these mutations impair the ability of Mec1 to interact directly with the regulators of chromatin accessibility. In any case, it is worth noting that NCAM does not completely diminish in mec1 and mec1-100 mutant cells, thus suggesting the presence of unidentified mechanisms that function in a MEC1-independent manner to promote chromatin accessibility around replication forks. In addition, the fact that the mec1-100- and mec1-null mutations cause a very similar degree of loss in chromatin accessibility indicates that the functions of Mec1 to promote chromatin accessibility and protect replication forks are genetically separable.
It has been suggested that chromatin structure negatively influences DNA replication fork progression, as the replisome has to progress through nucleosome arrays for this process (Groth et al. 2007). However, compared with other DNA-dependent processes such as transcription and DNA repair, much less is known about how chromatin structure is regulated for specific steps in DNA replication. Here we showed that chromatin accessibility increases around replication forks during normal S phase and replication stress conditions. We also found that this chromatin regulation is partially dependent on the checkpoint kinase Mec1 during replication stress, revealing a novel aspect of replication stress response. Finally, our work revealed that Mec1 facilitates replication fork progression during replication stress. Based on these results, we suggest a model in which Mec1 facilitates replication fork progression through increased chromatin accessibility. Together, these results provide a step toward elucidating the molecular mechanisms by which chromatin is regulated to control DNA replication and replication stress response.
Materials and methods
Yeast strains, cell culture synchronization, and sample cross-linking
The yeast strains used in this study (Supplemental Table S2) are congenic to W303-1a in which the weak rad5 mutant allele in the original W303 has been corrected (Thomas and Rothstein 1989; Zhao et al. 1998). At least two independent biological replicates were used for each experiment. Cells were grown in YPD medium at 30°C, arrested in G1 with 5 μg/mL α-factor, and released into YPD containing 200 mM HU, as previously described (Unnikrishnan et al. 2010). Cells for genomic DNA, nucleosome harvest, ChIP, and flow cytometry analysis were simultaneously collected and cross-linked at the time of harvest in G1 and S phase (30 and 60 min after α-factor release). Samples were cross-linked with 0.5% (genomic DNA and nucleosome harvest samples) or 1% (ChIP samples) formaldehyde for 20 min at room temperature. Cross-linking reactions were stopped with 0.125 M glycine.
Genomic DNA copy number profiles
For each S-phase time point, DNA copy number profiles were obtained by competitive hybridization of S-phase DNA against G1 DNA as previously described (Yabuki et al. 2002).
Nucleosome harvest
Mononucleosomal DNA was prepared as previously described (Whitehouse et al. 2007), typically obtaining 60%–80% mononucleosomal DNA (Supplemental Fig. S3B,C). Nucleosome maps were obtained by competitive hybridization of labeled nucleosomal DNA against labeled genomic DNA from the same time point.
ChIP
Chromatin was sonicated using a Bioruptor sonicator bath (Diagenode) to an average fragment size of 250 bp (Supplemental Fig. S3A), as extensive sonication has been previously shown to increase ChIP resolution (Fan et al. 2008). Soluble chromatin was immunoprecipitated as previously described (Gelbart et al. 2005; Lindstrom et al. 2006) using antibodies against Flag (Sigma), H3 C terminus (Active Motif), or H2B (Active Motif). Immunoprecipitation efficiency was routinely checked by Western blot. Libraries for array hybridization of both immunoprecipitated and input DNA were obtained using ligation-mediated PCR (LM-PCR) or whole-genome amplification (WGA; Sigma) strategies.
Tiling array hybridization and data processing
Samples were fragmented and labeled as previously described (Whitehouse et al. 2007). Labeled samples were competitively hybridized to custom tiling microarrays (Roche Nimblegen), as previously described (Yadon et al. 2010). Both strands of chromosomes III, VI, and XII were tiled with 50-mer probes overlapping by an average of 42 bp, covering 14% of the yeast genome. Only data corresponding to the forward strand were used in this analysis. Nimblescan software was used to obtain Tukey by-weight mean-adjusted log2 Cy5/Cy3 ratio files. Both raw and analyzed hybridization files are available for download at http://labs.fhcrc.org/tsukiyama. A pseudomedian smoothing method was applied to all ratio files to eliminate outliers (Royce et al. 2007) using a 25-bp window for smoothing nucleosome data and a 50-bp window for the rest of the data files. A set of custom-made Perl scripts was used to align data according to multiple genomic features and generate all of the averaged profiles.
NCAM assay
Nucleosome mapping data were normalized against histone H3 ChIP data by subtraction of the log2 values. The fact that whole nucleosome signals, not just peak heights or valleys, are integrated in this analysis makes the NCAM assay insensitive to differences in translational nucleosome positioning, as such differences do not alter the overall nucleosome signal across large genomic regions. To normalize for sample-to-sample differences in NCAM signals due to differences in MNase digestion, which have been shown to significantly affect nucleosome signals (Weiner et al. 2010), we performed Z-score transformation (Cheadle et al. 2003). In the Z-score transformation, values are expressed as a unit of standard deviation (SD) from a normalized mean of 0, therefore allowing for direct sample-to-sample comparisons. Briefly, for each NCAM data file, the mean and SD were calculated from a region encompassing 1 kb upstream of and 2 kb downstream from 673 ORFs aligned at their transcription start sites. None of the ORFs included in this analysis were replicated during the course of the experiments in any of the samples in order to avoid any biases in the Z-score transformation due to replication-dependent increases in NCAM. For each strain, the average increase in NCAM from G1 to S phase was calculated at early efficient ORIs (n = 8) as a Z-score difference between S phase and G1. The average increase in NCAM was then measured by integration of the area below each spline-smoothed Z-score difference profile (smoothing parameter 0.1) using R (http://www.r-project.org).
ORI classification based on firing activity
A list of all ORI positions included in this analysis as well as their firing activity in HU and ACS locations can be found in Supplemental Table S1. The ORI midpoints used in this study were derived from the maxima of the spline-smoothed DNA copy number profiles, similar to previously described (Raghuraman et al. 2001). ORIs were initially classified as early (n = 23) and late (n = 18) firing based on their activity in 200 mM HU-containing medium such that late ORIs were those exclusively firing in S-phase checkpoint mutants in the presence of HU, as previously reported (Santocanale and Diffley 1998; Shirahige et al. 1998). ORIs were further classified based on their firing activity in a wild-type strain by 60 min in S phase in the presence of 200 mM HU (Supplemental Table S1). For each ORI, firing activity was calculated as the fraction of cells that had fired the ORI, measured as the maxima of the spline-fitted DNA profile. Thus, firing activity ranged from 0 (nonreplicating) to 1 (fully replicated). ORIs were classified as being early efficient (firing activity >0.7; n = 8), mid (between 0.7 and 0.40; n = 8), or low (<0.4; n = 7). ORIs were aligned based on the orientation of the T-rich strand, as recently published (Xu et al. 2006; Berbenetz et al. 2010; Eaton et al. 2010).
Southern blot analysis of MNase chromatin accessibility
Following a described method (Fazzio and Tsukiyama 2003), cells from G1 and S phase (30 and 60 min) were treated with increasing amounts of MNase (100, 200, and 400 U) for 5 min at 37°C. For each sample, ∼1.5 μg of MNase-digested DNA was loaded onto a 1% agarose gel, transferred to a nylon membrane, and hybridized with radioactive probes. Signals from exposed membranes were quantified using Image Quant software (Amersham), and the fraction of mononucleosome signal versus the total signal in the lane was calculated. We first used a probe corresponding to a region upstream of ARS606 that replicates by 30 min in HU, as seen in DNA profiles. This blot was then stripped and rehybridized with a probe corresponding to a region in chromosome VI that does not replicate during the course of the experiment (negative control). The mononucleosome signal fraction from the replicating probe was normalized to that of the nonreplicating probe. Two additional membranes were similarly probed against two other replicating regions adjacent to ARS306 and ARS607 and their nonreplicating control probes in chromosomes III and VI, respectively. The PCR primers used to generate the probes are listed in Supplemental Table S3.
Fork progression rate
The rate of fork progression was measured as the distance traveled between DNA peak edges in successive time points, divided by the time difference in minutes between the two time points, as previously described (see Supplemental Fig. S10 for further details; Sekedat et al. 2010). Data were normalized before the analysis so that nonreplicating regions had an average signal intensity equal to 0. DNA synthesis rates are expressed as mean base pairs per minute ± standard deviation.
Supplementary Material
Acknowledgments
We thank Martin Morgan and Jeff Delrow for their advice on data analysis, Stephen Bell for helpful discussions, and Maria Pia Longhese for providing the mec1-100 mutant strain. We thank Sue Biggins and Susan Parkhurst and the members of the Tsukiyama laboratory for helpful discussions and critical reading of the manuscript. This research was supported by grants GM058465 and GM078259 from NIGMS. J.R. was supported in part by a fellowship from the Spanish Ministerio de Educacion y Ciencia.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.202978.112.
References
- Alvino GM, Collingwood D, Murphy JM, Delrow J, Brewer BJ, Raghuraman MK 2007. Replication in hydroxyurea: It's a matter of time. Mol Cell Biol 27: 6396–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Bell O, Tiwari VK, Thoma NH, Schubeler D 2011. Determinants and dynamics of genome accessibility. Nat Rev Genet 12: 554–564 [DOI] [PubMed] [Google Scholar]
- Berbenetz NM, Nislow C, Brown GW 2010. Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet 6: e1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM 1998. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 107: 218–227 [DOI] [PubMed] [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, Becker KG 2003. Analysis of microarray data using Z score transformation. J Mol Diagn 5: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M 2005. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17: 153–159 [DOI] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM 2010. Conserved nucleosome positioning defines replication origins. Genes Dev 24: 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Lamarre-Vincent N, Wang Q, Struhl K 2008. Extensive chromatin fragmentation improves enrichment of protein binding sites in chromatin immunoprecipitation experiments. Nucleic Acids Res 36: e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Moqtaderi Z, Jin Y, Zhang Y, Liu XS, Struhl K 2010. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3′-end formation. Proc Natl Acad Sci 107: 17945–17950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Tsukiyama T 2003. Chromatin remodeling in vivo: Evidence for a nucleosome sliding mechanism. Mol Cell 12: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Friedel AM, Pike BL, Gasser SM 2009. ATR/Mec1: Coordinating fork stability and repair. Curr Opin Cell Biol 21: 237–244 [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T 2005. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev 19: 942–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G 2007. Chromatin challenges during DNA replication and repair. Cell 128: 721–733 [DOI] [PubMed] [Google Scholar]
- Hildebrand CE, Walters RA 1976. Rapid assembly of newly synthesized DNA into chromatin subunits prior to joining to small DNA replication intermediates. Biochem Biophys Res Commun 73: 157–163 [DOI] [PubMed] [Google Scholar]
- Huang M, Elledge SJ 1997. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol Cell Biol 17: 6105–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS 1991. Transcription on nucleosomal templates by RNA polymerase II in vitro: Inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev 5: 683–696 [DOI] [PubMed] [Google Scholar]
- Koc A, Wheeler LJ, Mathews CK, Merrill GF 2004. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J Biol Chem 279: 223–230 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lindstrom KC, Vary JC Jr, Parthun MR, Delrow J, Tsukiyama T 2006. Isw1 functions in parallel with the NuA4 and Swr1 complexes in stress-induced gene repression. Mol Cell Biol 26: 6117–6129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116 [DOI] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Scotti M, Lucchini G, Longhese MP 2001. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol 21: 3913–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL 2008. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol 15: 338–345 [DOI] [PubMed] [Google Scholar]
- Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P 2012. dNTP pools determine fork progression and origin usage under replication stress. EMBO J 31: 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL 2001. Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Richmond TJ, Davey CA 2003. The structure of DNA in the nucleosome core. Nature 423: 145–150 [DOI] [PubMed] [Google Scholar]
- Royce TE, Carriero NJ, Gerstein MB 2007. An efficient pseudomedian filter for tiling microrrays. BMC Bioinformatics 8: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Seale RL 1975. Assembly of DNA and protein during replication in HeLa cells. Nature 255: 247–249 [DOI] [PubMed] [Google Scholar]
- Seale RL 1978. Nucleosomes associated with newly replicated DNA have an altered conformation. Proc Natl Acad Sci 75: 2717–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekedat MD, Fenyo D, Rogers RS, Tackett AJ, Aitchison JD, Chait BT 2010. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol Syst Biol 6: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth LA, Sigurdsson S, Svejstrup JQ 2010. Transcript elongation by RNA polymerase II. Annu Rev Biochem 79: 271–293 [DOI] [PubMed] [Google Scholar]
- Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol 18: 566–575 [DOI] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621 [DOI] [PubMed] [Google Scholar]
- Tabancay AP Jr, Forsburg SL 2006. Eukaryotic DNA replication in a chromatin context. Curr Top Dev Biol 76: 129–184 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Longhese MP, Diffley JF 2003. A central role for DNA replication forks in checkpoint activation and response. Mol Cell 11: 1323–1336 [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R 1989. The genetic control of direct-repeat recombination in Saccharomyces: The effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics 123: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnikrishnan A, Gafken PR, Tsukiyama T 2010. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol 17: 430–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JA, Kwong TJ, Tsukiyama T 2008. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat Struct Mol Biol 15: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20: 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035 [DOI] [PubMed] [Google Scholar]
- Xu W, Aparicio JG, Aparicio OM, Tavare S 2006. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K 2002. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7: 781–789 [DOI] [PubMed] [Google Scholar]
- Yadon AN, Van de Mark D, Basom R, Delrow J, Whitehouse I, Tsukiyama T 2010. Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol Cell Biol 30: 5110–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 2: 329–340 [DOI] [PubMed] [Google Scholar]
- Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J 20: 3544–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Thiele DJ 1996. A specialized nucleosome modulates transcription factor access to a C. glabrata metal responsive promoter. Cell 87: 459–470 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.