Abstract
Cell-free protein synthesis harnesses the synthetic power of biology, programming the ribosomal translational machinery of the cell to create macromolecular products. Like PCR, which uses cellular replication machinery to create a DNA amplifier, cell-free protein synthesis is emerging as a transformative technology with broad applications in protein engineering, biopharmaceutical development, and post-genomic research. By breaking free from the constraints of cell-based systems, it takes the next step towards synthetic biology. Recent advances in reconstituted cell-free protein synthesis (Protein synthesis Using Recombinant Elements (PURE) expression systems) are creating new opportunities to tailor the reactions for specialized applications including in vitro protein evolution, printing protein microarrays, isotopic labeling, and incorporating nonnatural amino acids.
Keywords: cell-free translation, protein engineering, protein synthesis, PURE expression, synthetic biology
Introduction
Proteins represent the ultimate expression of biochemical function: as enzymes, they perform exquisitely selective chemical reactions; as receptors, they recognize specific partners in signaling pathways that integrate and regulate metabolism; as transporters, they control trafficking across cell membranes; and as macromolecular building blocks, they form the scaffolding of cell architecture. In biotechnology, proteins are important not only for their remarkable catalytic power, but also for their unique physical properties, and as tools of discovery in the development of new therapeutics. As a result, the production of pure proteins has itself become an important goal, driving the evolution of both chemical and biochemical methods for protein synthesis. Conventional solid phase chemical synthesis provides the ultimate control over molecular structure, but is generally restricted to smaller peptides (<40 residues) (Nilsson et al. 2005), limiting its ability to meet the need for a broad range of proteins for industrial and research applications. Alternatively, several decades of biochemical research has expanded and refined methods for recombinant protein production based on ribosomal protein synthesis. The protein expression field has traditionally been dominated by cell-based expression systems, including both conventional (Escherichia coli, Saccharomyces cerevisiae) and non-conventional (e.g., Pichia pastoris, Thermus thermophilus) host organisms that have been developed as ‘cell factories’. However, cell-free protein synthesis is rapidly emerging as an important complementary approach, rivaling cell-based protein production in terms of both convenience and scaleability, and becoming widely recognized as a valuable tool for protein engineering (Hodgman and Jewett 2012, Kai et al. 2012, Swartz 2012).
Cell-free protein synthesis is not a new concept. Protein synthesis in cell-free extracts helped to crack the genetic code and has been extensively used to dissect ribosomal protein biosynthesis, providing a specialized niche for the development of these methods. However, advances in cell-free protein synthesis in recent years, particularly the ability to reconstitute protein synthesis from well-defined, purified components, has transformed the approach from a specialized analytical tool to a powerful preparative method with broad applicability. Cell extract - or lysate-derived systems for protein synthesis - continue to be exploited and optimized, with the traditional wheat germ, rabbit reticulocyte and E. coli lysates being extended with Leischmanii (Kovtun et al. 2011), Thermus (Zhou et al. 2012) and even human (Mikami et al. 2008) cell-free systems. Systems based on reconstituted highly purified components (PURE expression systems) have significantly extended the range of applications and will be the primary focus of this mini-review, which surveys recent advances and emerging applications of cell-free protein synthesis.
PURE expression systems
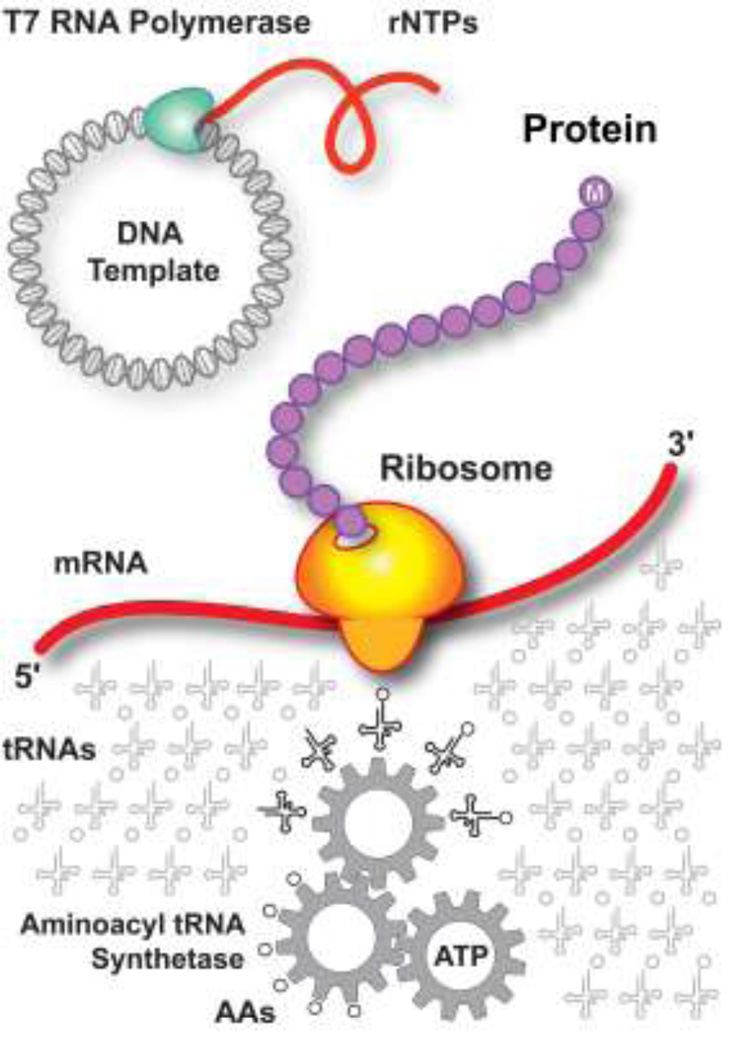
The PURE (Protein synthesis Using Recombinant Elements) approach to cell-free protein synthesis is based on modular reconstitution of the translational machinery of the cell from affinity-purified protein components (Shimuzu et al. 2005, Shimuzu and Ueda 2010, Wang et al. 2012), including initiation factors (IF1, IF2, IF3), elongation factors (EF-Tu, EF-Ts, EF-G), release factors (RF1, RF2, RF3), ribosome recycling factors, 20 aminoacyl tRNA synthetases, methionyl tRNA formyltransferase and pyrophosphatase, all bearing a 6×His tag. These recombinant components are combined with ribosomes and tRNAs isolated from specially engineered E. coli strains, together with all of the necessary NTPs and amino acids, an ATP-generating catalytic module and recombinant T7 RNA polymerase, creating a self-contained reaction system that can be programmed for protein synthesis using a variety of DNA templates (Fig. 1). The PURE cell-free protein synthesis system has been commercialized (e.g., PURESYSTEM, Cosmo Bio, Tokyo, Japan; PURExpress, New England Biolabs, Beverly, MA, USA), making it available for a variety of laboratory research applications.
Figure 1.
PURE protein translation system. The PURE expression system is programmed by a DNA template and includes a transcription module, an energy-coupling module for NTP regeneration, aminoacyl tRNA synthetases and tRNAs, and a translation module comprised of ribosomes, elongation factors, and release factors that are integrated to perform peptide elongation and macromolecular synthesis.
The advantages of the PURE systems include reduced levels of contaminating proteases, nucleases, and phosphatases, greater reproducibility resulting from more defined chemistry, and the flexibility of a modular system. Metabolic side reactions that deplete the amino acid pool in cell extracts can be entirely avoided. In addition, the 6×His affinity tags associated with the PURE components can be utilized in ‘reverse purification’ of products, extracting the tagged proteins by metal affinity chromatography. Because it is modular, the PURE system supports a variety of modifications for specialized applications, including ribosome display and site-selective incorporation of nonnatural amino acids, described in more detail below. Both cell extracts and the components of the PURE system can be modified to mimic the macromolecular crowding of the interior of a cell (Ge et als. 2011).
ATP-regeneration module
Ribosomal protein synthesis is an energy-intensive process, requiring approximately 4 ATP equivalents/peptide bond (including 2 ATP equivalents/residue for amino acid activation, 1 GTP/residue for transfer of the charged aminoacyl-tRNA to the A site of the ribosome, and 1 GTP/residue for translocation of the ribosome through coupling to EF-G)(Calhoun and Swartz 2007, Kim and Kim 2009). Consequently, the choice of ATP-regenerating system is a critical factor for cell-free protein synthesis. A variety of ATP-regenerating reactions have been explored to achieve sustained, stable production of ATP while avoiding accumulation of inorganic phosphate, which inhibits translation by binding Mg2+ that serve as essential cofactor in many nucleotide-dependent reactions, including protein synthesis. The choice of ATP-regeneration module determines both the rate and duration of active protein synthesis.
In the PURE systems, all enzymes required to form a complete in vitro catalytic pathway must be included to regenerate ATP in one or more substrate-level phosphorylation steps. High phosphoryl-group transfer potential substrates (creatine phosphate, phosphoenolpyruvate, 3-phosphoglycerate) can be used directly as phosphate donors, although they are relatively expensive. Alternatively, non-phosphorylated substrates may be used, with the phosphate entering from ATP (lowering the overall yield) or as inorganic phosphate. Adenylate kinase and nonspecific nucleotide diphosphate kinase must also be included to catalyze phosphate exchange within the pool of ribonucleotides. These two enzymes equilibrate the phosphorylation state of all four nucleotides, and couple ATP regeneration to formation of GTP, which is required for delivery of charged tRNA and for translocation of the ribosome during protein synthesis.
Phosphate donors
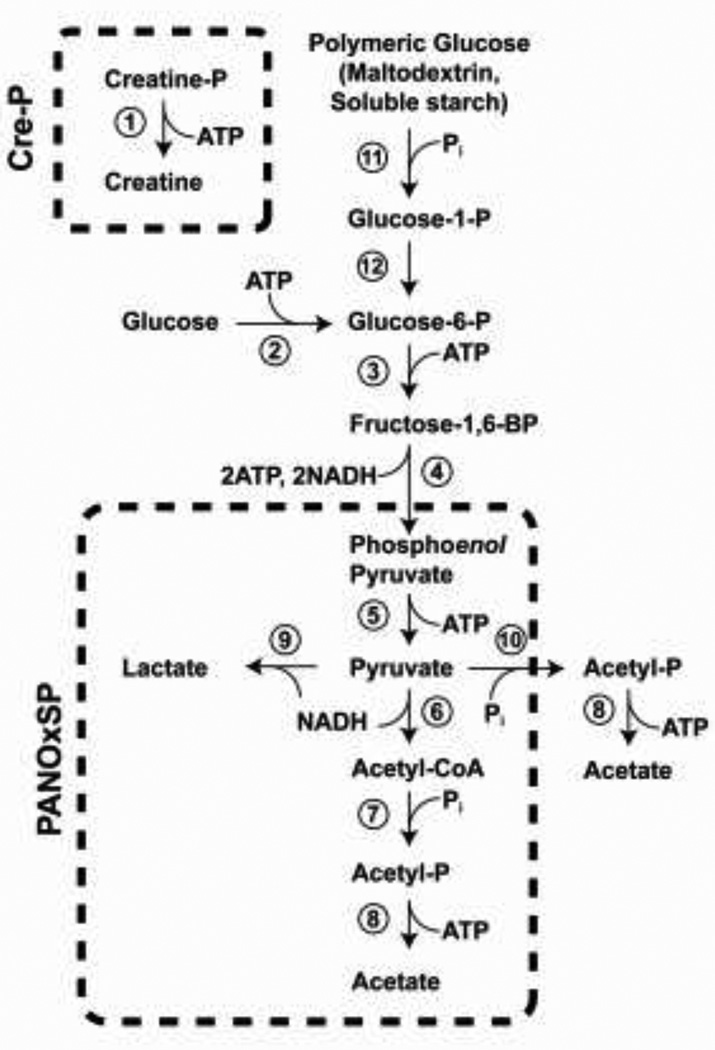
The simplest ATP regeneration system uses high group transfer potential phosphorylated compounds to power the translational machinery. While its simplicity is an advantage, this type of system invariably leads to accumulation of inorganic phosphate as an end product. In the original formulation of the PURE translation system, creatine phosphate was added as a relatively stable source of high group transfer potential phosphate, and the enzyme creatine kinase was used to catalytically regenerate ATP from ADP (Fig. 2). The organic end product, creatine, is relatively innocuous and does not appear to interfere with protein synthesis. However, because of the 1:1 stoichiometry of the phosphoryl transfer reaction, relatively large amounts of creatine phosphate substrate are required, and one equivalent of inorganic phosphate is formed in each reaction cycle. The accumulation of phosphate can be delayed by including glucose as a secondary energy source (Kim et al. 2007).
Figure 2.
ATP regeneration pathways for cell-free protein synthesis. Commonly used reaction sequences (Cre-P (Creatine-phosphate); PANOxSP (Phosphoenolpyruvate (PEP), Amino acids, NAD+, Oxalic acid, Spermidine and Putrescine)) are indicated. The enzymes required for these reactions are: (1) Creatine kinase; (2) hexokinase; (3) phosphofructokinase; (4) aldolase + triose phosphate isomerase + glyceraldehydes-3-phosphate dehydrogenase + 3-phosphoglycerate mutase + enolase; (5) pyruvate kinase; (6) pyruvate dehydrogenase; (7) phosphate acetyltransferase ; (8) acetate kinase; (9) lactate dehydrogenase; (10) pyruvate oxidase; (11) phosphorylase; (12) phosphoglucomutase.
The glycolytic intermediate, 3-phosphoglycerate, supports substrate-level phosphorylation in the presence of three enzymes of the glycolytic pathway (phosphoglyceromutase, enolase and pyruvate kinase). Once again the 1:1 stoichiometry for ATP production is a limiting factor. Another simple ATP regeneration module is based on acetate kinase-catalyzed formation of ATP from ADP and acetyl phosphate, which is readily available (Ryabova et al. 1995), although formation of an organic acid (acetic acid) as an end product may interfere with protein synthesis (Wang and Zhang 2009). Polyphosphate, a linear polymer of phosphoric acid that occurs naturally in cells, can also serve as a phosphate donor for ATP regeneration. However, as in the other examples described above, chelation of divalent metal ions by inorganic phosphate is a complicating factor.
Reconstituted pathways
Some of the problems associated with ATP-regeneration based on simple phosphate donors can be resolved by reconstituting extended metabolic pathways, increasing the yield of ATP and recycling the inorganic phosphate. So far, approaches based on well-characterized cellular metabolism (including glycolysis and oxidative phosphorylation) have mainly been applied in cell extracts, although reconstitution of extended metabolic pathways in vitro from purified components is also possible (Stevenson et al. 2012).
The glycolytic reaction sequence is well-suited to ATP regeneration in the cell-free system, since the reactions do not require O2 and the presence of two substrate-level phosphorylation steps enhances the yield of ATP. When glucose is used, two ATP equivalents are required for substrate activation but four ATP are formed, and two equivalents of inorganic phosphate are recycled. One drawback of this scheme is that the redox stoichiometry of glycolysis results in a net production of NADH as an end product (Kim et al. 2008).
A number of variations on this theme have been described. The PANOxSP [Phosphoenolpyruvate (PEP), Amino acids, NAD+, Oxalic acid, Spermidine and Putrescine] system utilizes PEP as a simple phosphate donor but extends the biochemical processing through the pyruvate dehydrogenase complex, and recycles one equivalent of inorganic phosphate, with a net yield of two ATP for each PEP (Calhoun and Swartz 2007) (Fig. 2). Another system, called cytomim (cytosolic mimicry), effectively reconstitutes oxidative phosphorylation, consuming simple organic acids (e.g., succinate) and using inverted respiratory membranes to generate ATP (Jewett and Swartz 2004, Jewett et al. 2008).
A particularly interesting method of ATP-regeneration has recently been described that involves the use of glucose storage polymers (maltodextrin or soluble starch) as a reservoir of chemical free energy (Wang and Zhang 2009, Kim et al 2011). This approach is related to the reconstituted glycolysis system described above but is extended to include phosphorolytic cleavage of the storage polymer. There are four major advantages of this scheme: (1) it allows use of an inert and inexpensive maltodextrin or starch substrate, (2) it provides a ‘pacekeeper’ reaction that moderates ATP production, (3) it generates a high phosphoryl group transfer potential intermediate (glucose 1-phosphate) in situ, and (4) it completely recycles inorganic phosphate. When combined with the reactions of glycolysis and the PANOx pathway, the reactions have a theoretical yield of four ATP per glucose. This appears to be the most efficient energy coupling module currently available. Development of reaction modules to drive cell-free protein synthesis is an active area of research, and the possibilities are far from being exhausted.
Continuous exchange
Some of the limitations inherent in closed system (batch) reaction scheme described above can be overcome in a continuous exchange open-system format allowing substrates to be replenished during the reaction (Yin et al. 2012). For example, filtration or dialysis can be used to continuously supply ATP as it is consumed and to remove inorganic phosphate as it is formed. This strategy is very effective at extending the duration of active translation in small scale cell-free systems, but is not as scaleable. Other strategies, in which the open-system character is restricted to pH control, have been used to extend cell-free protein synthesis to the industrial scale resulting in a high yield of a disulfide-bonded protein product (700 mg/l) (Zawada et al. 2011).
Transcription module
Cell-free protein synthesis is programmed by addition of a DNA template, formed from either closed circular vector DNA or a linear PCR product. Transcription is performed by recombinant phage T7 RNA polymerase (RNAP), generating the mRNA upon which the ribosomal translation machinery acts (Beckert and Masquida 2011). T7 RNAP is a relatively simple, single-subunit polymerase with high promoter specificity and transcriptional fidelity (Sousa and Mukherjee 2003).
The minimum requirements for the DNA template include a 5´-untranslated region (UTR) comprising a strong T7 promoter sequence to support a high transcription rate, a Shine-Delgarno sequence that serves as the ribosome entry point, and a 3´-UTR that includes an efficient translation termination codon (e.g., TAA), followed by six or more nucleotides (Shimuzu and Ueda 2010). An epsilon (Enhancer of Protein Synthesis Initiation) sequence may be added to the 5´-UTR to improve translation, and a T7 terminator in the 3´-UTR improves efficient release and recycling of the ribosome. This simple transcription unit is easily prepared by two-step PCR, allowing virtually any coding sequence to be assembled together with promoter and terminator elements for cell-free synthesis.
Transcription and translation processes can be arranged as either linked or coupled reactions. Linked transcription/translation implies a two-step sequential process, where the transcript is formed first. In contrast, coupled transcription/translation implies simultaneous synthesis of mRNA and protein within an extended polysome complex. Cell-free protein synthesis based on prokaryotic components generally involves a coupled reaction system (Fig. 1).
It is possible to extend cell-free synthesis to simultaneous production of multiple polypeptides in a single reaction mixture, since addition of multiple templates results in the parallel synthesis of distinct proteins. This approach can be used to assemble complex multicomponent proteins, as demonstrated by the successful cell-free synthesis of the heterotrimeric core of Paracoccus denitrificans cytochrome c oxidase in an E. coli extract (Katayama et al. 2010). An alternative strategy for multigene expression from polycistronic constructs has been demonstrated for production of up to five distinct protein products from a single ‘BioBrick’ plasmid template (Du et al. 2009). Sequential synthesis is also possible: by immobilizing template DNA on magnetic microbeads, cell-free protein synthesis can be arbitrarily reset and reprogrammed, an example of artificial gene circuits (Lee et al. 2012).
Typical protein synthesis reactions are driven by transcription from sub-pmol quantities of template, and increasing improvements in the efficiency of transcriptional processing has brought the technology near the single-molecule limit of template sensitivity. In a recent example, green fluorescent protein was produced at quantized expression levels from 1–2 copies of template in picoliter volume reaction microchambers (Okano et al. 2012). This level of sensitivity is important for a variety of nanoscale processing applications, including printing protein microarrays.
Addition of a species-independent translation sequences (SITS) in the 5´-UTR of the template eliminates species barriers to cell-free translation. This universal adapter element relaxes secondary structure in the transcript and facilitates assembly of the translation complex in yeast, wheat germ, insect cell, rabbit reticulocyte and E. coli translation systems, opening up new possibilities for cell-free protein expression (Mureev et al. 2009).
tRNA aminoacylation module
Amino acids are activated for peptide bond formation by amino acyl tRNA synthetases that are specific for the amino acid substrate and the cognate set of tRNAs, covalently linking the residue to the 3´-terminal adenosine of the tRNA (Ling et al. 2009). Formation of inorganic pyrophosphate as a by-product during amino acid activation requires inorganic pyrophosphatase and adenylate kinase to be present to drive the reaction and recycle AMP.
Protein synthesis requires that all 20 amino acids are present in amounts super-stoichiometric with the amount of protein to be formed, in addition to catalytic amounts of all 20 amino acid tRNA synthetases and the full complement of tRNAs. In the PURE system the synthetases are affinity purified recombinant products, and tRNAs are isolated from special E. coli strains (Shimuzu and Ueda 2010). The modular design of the PURE translation system makes it possible to supplement with rare tRNAs to compensate for codon bias in the template.
Modifying the composition of the amino acid mixture can be used to control the translation process. For example, using a drop-out mixture lacking one amino acid will result in ribosome stalling or pausing, allowing expression to be synchronized. Substitution with isotopomers facilitates isotopic labeling (with stable isotopes or radionuclides) while avoiding isotope dilution and scrambling resulting from unavoidable metabolic processing in cell-based systems (Ozawa et al. 2005, Su et al. 2011, Yokoyama et al. 2011). These approaches also permit efficient incorporation of Se-methionine into proteins without the toxic side effects that can compromise in vivo selenium labeling (Kigawa et al. 2002). Insertion of photochemically active analogs that are recognized by the native E. coli tRNA aminoacylation machinery (e.g., photo-leucine and photo-methionine (Suchanek et al. 2005)) is also expected to be straightforward. Alternatively, bioorthogonal pairs of suppressor tRNA/cognate aminoacyl tRNA synthetase can be added to the system to direct the incorporation of nonnatural amino acids into the protein product (Goerke and Swartz 2009, Ozawa et al. 2012), or aminoacylating ribozymes (flexizymes) can be used to expand the genetic code (Goto et al. 2011, Goto and Suga 2012) (see below).
Peptide synthesis module
Peptide synthesis is catalyzed by ribosomes, large (megadalton) ribonucleoprotein complexes comprised of more than 50 proteins and three RNAs that represents about 30% of the mass of rapidly growing bacterial cells (Bremer and Dennis 2008). Although in principle a small, catalytic amount of ribosomes could be continuously recycled during cell-free protein synthesis, in practice the yield of protein tends to be roughly proportional to the quantity of ribosomes, which therefore represent one of the main components of the reaction mixture. Because of their abundance, unmodified native ribosomes may be easily isolated from E. coli. However, an engineered strain of E. coli producing His-tagged ribosomes is now available thereby allowing a one-step purification of active ribosomes (Ederth et al. 2009) that lend themselves to the ‘reverse purification’ strategy described above. The PURE systems also provides control over the availability of release factors, facilitating applications like ribosome display (see below) by promoting stalling of the ribosome on the mRNA, or incorporation of nonnatural amino acids by suppressor tRNAs.
Posttranslational processing
The modular design of the cell-free expression system provides a high degree of control over posttranslational processing events. One of the most widely appreciated advantages of the cell-free system is the possibility of directing insertion of an integral membrane protein product into a lipid structure, avoiding problems that are often encountered in high-level expression of membrane proteins in cellular systems (Schneider et al. 2010). Expression of integral membrane proteins in living cells can be problematic as a result of toxic side-effects relating to membrane disruption. Cell-free synthesis of integral membrane proteins has been accomplished utilizing a variety of lipid structures, including liposomes, micelles, bicelles and nanodiscs (Lyukmanova et al. 2012). Bacteriorhodopsin synthesized in a cell-free system has been cotranslationally inserted into giant lipsomes, where its photochemical proton pumping function could be demonstrated (Kalmbach et al. 2007). Nanodisc technology appears to be particularly well-suited to marrying cell-free synthesis with protein structural studies, since the nanodisc is structurally well-defined (Bayburt and Sligar 2010).
Correct folding of the recombinant protein can be assisted by supplementing the basic PURE system with molecular chaperones to enhance the efficiency of protein folding, helping polypeptides navigate the folding funnel and reach their native conformational state (Shimizu et al. 2005, Ueda 2008). Interestingly, even when the cell-free synthesis protein product is insoluble, it tends to be more readily solubilized and re-folded than proteins that have been recovered from inclusion bodies in cell-based expression systems (Swartz 2012). The formation of disulfide bonds is another important posttranslational processing step that can be problematic in prokaryotic expression systems. In cell-free protein synthesis, disulfide bond formation has been shown to be enhanced by addition of a glutathione redox buffer that facilitates disulfide exchange (Goerke and Swartz 2008, Knapp et al. 2007).
Applications of cell-free protein synthesis
Cell-free protein synthesis is an enabling technology that has the potential to transform many aspects of biotechnology. Its value has already been demonstrated in production of ‘difficult’ proteins that may be toxic to conventional cell protein factories, and new applications are emerging in protein engineering, drug discovery and synthetic biology
Printing protein arrays
Protein microarrays have emerged as a promising platform for high-throughput screening in postgenomic biomedical research, permitting massively parallel functional analysis of complete proteomes and providing an important tool for vaccine development and personalized medicine (Berrade et al. 2011). Cell-free protein synthesis is a crucial link between the established technology of DNA microarrays and protein arrays allowing multiplexed, robotic processing to be used to print protein arrays from gene chips. Cell-free expression based in situ protein microarrays can be produced by a variety of methods that take advantage of the programmability of ribosomal protein synthesis by transcripts formed from either linear DNA (generated by PCR) or circular plasmid vectors: PISA (protein in situ array), DAPA (DNA array to protein array), NAPPA (nucleic acid programmable protein array) and TUS-TER mediated affinity labeling differ in the details of template construction and affinity capture strategy (Nand et al. 2012).
Ribosome display
Ribosome display is an approach to protein functional analysis that can be used as a platform for protein engineering that is based on the stability of the ribosomal translation complex (comprised of mRNA, ribosome, and nascent polypeptide) in the absence of release factors. This translation complex provides a physical link between gene and protein that can be utilized in a recursive cycle of functional selection and mutagenesis to drive protein evolution (Plückthun 2012). The modular composition of the PURE cell-free system is especially well-suited to this application (Ohashi et al. 2007, Ueda et al. 2010), because it allows release factors to be omitted as a rational choice in the preparation and can accommodate incorporation of nonnatural amino acids (Watts and Forster 2012).
Isotopic labeling
Labeling of recombinant proteins with stable isotopes or radionuclides is important for a variety of applications, including biological NMR and tracer experiments. When proteins are expressed in living cells or cell extracts, labeling is complicated by metabolic processes that dilute isotope or scramble labeling patterns as a result of molecular transformations, although strategies have been devised to suppress these side-reactions (Su et al. 2011, Yokoyama et al. 2011, Matsuda et al. 2007). In contrast, the severely edited metabolic map in PURE cell-free protein synthesis may be expected to eliminate many of these problems, because the interfering enzymes and metabolites are not present (Ozawa et al. 2005).
Incorporation of nonnatural amino acids
Expansion of the genetic code is perhaps the most significant development in ribosomal protein synthesis since biochemistry based on the canonical set of 20 amino acids evolved 3 billion years ago, adding new dimensions to protein engineering (Lin and Schultz 2010). The approach is conceptually simple, using a bioorthogonal cognate pair of tRNA and aminoacyl tRNA synthetase to suppress nonsense or frameshift mutations, incorporating nonnatural amino acids into the growing polypeptide and thereby creating a chemical toolbox for protein engineering. By systematic evolution of cognate tRNA/aminoacyl tRNA sythetases, more than 50 distinct nonnatural amino acids have been incorporated into proteins in this way, adding fluorescent tags or reactive groups that contribute novel chemistry or functionality in the protein product (de Graaf et al. 2009, Hartman et al. 2007).
While the majority of this work has been performed in vivo, cell-free expression systems offer some important advantages (Goerke and Swartz 2009, Ozawa et al. 2012). For example, cellular systems are susceptible to toxic side-effects of non-natural amino acids in cellular proteins or recombinant products. In contrast, cell-free systems avoid toxicity issues. Further, the cell-free system can be more easily tailored to accommodate unusual side chain structures by modifying the components of the translation machinery. Mutation of EF-Tu has already been shown to improve the efficiency of incorporating nonnatural amino acids with bulky side chains in the PURE system (Doi et al. 2007). The well-defined chemistry of the PURE system also lends itself to customization in terms of the choice of tRNAs and aminoacyl tRNA synthetases. Non-natural amino acid chemistry can be introduced into the protein product simply by adding the bioorthogonal components. The effectiveness of this approach has already been demonstrated in cell-free extracts by preparation of p-propargyloxyphenylalanine-containing proteins that can be crosslinked to form bioconjugates via click chemistry (Bundy and Swartz 2010). Selective omission of the RF1 release factor can enhance suppression by the bioorthogonal tRNA, allowing incorporation of nonnatural amino acids at multiple sites (Johnson et al. 2011, Losha et al. 2012).
Ribozyme technology has recently been developed as an alternative strategy for expanding the genetic code. Flexizymes are small catalytic RNAs that support acylation of tRNA with virtually any amino acid analog (Goto et al. 2011, Goto and Suga 2012). Flexizymes can be subjected to rapid in vitro molecular evolution to select for optimized performance, and the lack of membrane barriers in the cell-free system makes it the ideal platform for both selection and application of flexizyme technology.
Comparison of platforms for protein synthesis
Ribosomal protein synthesis (both cellular and cell-free) is characterized by exceptionally high fidelity, with an amino acid misincorporation frequency of approx. 0.01%, even at high elongation rates (20 residues/s)(Yadavalli and Ibba 2012). This makes it possible to accurately produce extremely large proteins (up to 3.5 MDa)(LeWinter and Granzier 2010) by these biochemical methods. ‘Cell factories’ are able to perform the complete metabolic transformation of simple nutrients (inorganic salts and carbon fuels) into protein products. Cell-free systems require input of pre-formed amino acids, but provide other advantages described above. Solid phase chemical synthesis of proteins can in principle be used to produce polypeptides with a virtually unlimited range of building blocks and stereochemistry, but requires protected amino acid derivatives, is relatively slow (approx. 15 min per coupling cycle) and the relatively high error rate (maximum coupling efficiency approximately 99% per cycle) (Merrifield 1997) places a practical limit on the size of the protein product that can be formed (Table 1).
Table 1.
Comparison of platforms for protein synthesis
Conclusions
Cell-free protein synthesis has emerged as an important and effective alternative to both cell-based expression systems and solid-phase protein synthesis (Table 1). Advances in cell-free methods, including commercialization of PURE expression technology, are creating new possibilities for applications in biotechnology, ranging from microscale to industrial scale and in diverse areas including protein microarrays, biotherapeutics and biomaterials.
Acknowledgements
The author would like to thank Corinna Tuckey (New England Biolabs) for helpful discussions. This work was supported by the National Institutes of Health (GM42680 to J. W. W.).
References
- Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckert B, Masquida B. Synthesis of RNA by in vitro transcription. Methods Mol Biol. 2011;703:29–41. doi: 10.1007/978-1-59745-248-9_3. [DOI] [PubMed] [Google Scholar]
- Berrade L, Garcia AE, Camarero JA. Protein microarrays: novel developments and applications. Pharm Res. 2011;28:1480–1499. doi: 10.1007/s11095-010-0325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer H, Dennis PP. Chapter 5.2.3, Modulation of chemical composition and other parameters of the cell at different exponential growth rates. In: Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, Ussery D, editors. EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 2008. http://www.ecosal.org. [DOI] [PubMed] [Google Scholar]
- Bundy BC, Swartz JR. Site-specific incorporation of p-propargyloxyphenylalanine in a cell-free environment for direct protein-protein click conjugation. Bioconjug Chem. 2010;21:255–263. doi: 10.1021/bc9002844. [DOI] [PubMed] [Google Scholar]
- Calhoun KA, Swartz JR. Energy Systems for ATP Regeneration in Cell-Free Protein Synthesis Reactions. Methods Mol Biol. 2007;375:3–17. doi: 10.1007/978-1-59745-388-2_1. [DOI] [PubMed] [Google Scholar]
- Cenatiempo Y, Twardowski T, Redfield B, Reid BR, Dauerman H, Weissbach H, Brot N. Simplified in vitro system for study of eukaryotic mRNA translation by measuring di- and tripeptide formation. Proc Natl Acad Sci U S A. 1983;80:3223–3226. doi: 10.1073/pnas.80.11.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf AJ, Kooijman M, Hennink WE, Mastrobattista E. Nonnatural amino acids for site-specific protein conjugation. Bioconjug Chem. 2009;20:1281–1295. doi: 10.1021/bc800294a. [DOI] [PubMed] [Google Scholar]
- Doi Y, Ohtsuki T, Shimizu Y, Ueda T, Sisido M. Elongation factor Tu mutants expand amino acid tolerance of protein biosynthesis system. J Am Chem Soc. 2007;129:14458–14462. doi: 10.1021/ja075557u. [DOI] [PubMed] [Google Scholar]
- Du L, Gao R, Forster AC. Engineering multigene expression in vitro and in vivo with small terminators for T7 RNA polymerase. Biotechnol Bioeng. 2009;104:1189–1196. doi: 10.1002/bit.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederth J, Mandava CS, Dasgupta S, Sanyal S. A single-step method for purification of active His-tagged ribosomes from a genetically engineered Escherichia coli. Nucleic Acids Res. 2009;37:e15. doi: 10.1093/nar/gkn992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Luo D, Xu J. Cell-free protein expression under macromolecular crowding conditions. PLoS One. 2011;6:e28707. doi: 10.1371/journal.pone.0028707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke AR, Swartz JR. Development of cell-free protein synthesis platforms for disulfide bonded proteins. Biotechnol Bioeng. 2008;99:351–367. doi: 10.1002/bit.21567. [DOI] [PubMed] [Google Scholar]
- Goerke AR, Swartz JR. High-level cell-free synthesis yields of proteins containing site-specific non-natural amino acids. Biotechnol Bioeng. 2009;102:400–416. doi: 10.1002/bit.22070. [DOI] [PubMed] [Google Scholar]
- Goto Y, Katoh T, Suga H. Flexizymes for genetic code reprogramming. Nat Protoc. 2011;6:779–790. doi: 10.1038/nprot.2011.331. [DOI] [PubMed] [Google Scholar]
- Goto Y, Suga H. Flexizymes as a tRNA acylation tool facilitating genetic code reprogramming. Methods Mol Biol. 2012;848:465–478. doi: 10.1007/978-1-61779-545-9_29. [DOI] [PubMed] [Google Scholar]
- Hartman MC, Josephson K, Lin CW, Szostak JW. An expanded set of amino acid analogs for the ribosomal translation of unnatural peptides. PLoS One. 2007;2:e972. doi: 10.1371/journal.pone.0000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgman CE, Jewett MC. Cell-free synthetic biology: thinking outside the cell. Metab Eng. 2012;14:261–269. doi: 10.1016/j.ymben.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett MC, Calhoun KA, Voloshin A, Wuu JJ, Swartz JR. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol Syst Biol. 2008;4:220. doi: 10.1038/msb.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett MC, Swartz JR. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol Bioeng. 2004;86:19–26. doi: 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
- Johansson M, Bouakaz E, Lovmar M, Ehrenberg M. The kinetics of ribosomal peptidyl transfer revisited. Mol Cell. 2008;30:589–598. doi: 10.1016/j.molcel.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Johnson DB, Xu J, Shen Z, Takimoto JK, Schultz MD, Schmitz RJ, Xiang Z, Ecker JR, Briggs SP, Wang L. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol. 2011;7:779–786. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai L, Roos C, Haberstock S, Proverbio D, Ma Y, Junge F, Karbyshev M, Dötsch V, Bernhard F. Systems for the cell-free synthesis of proteins. Methods Mol Biol. 2012;800:201–225. doi: 10.1007/978-1-61779-349-3_14. [DOI] [PubMed] [Google Scholar]
- Kalmbach R, Chizhov I, Schumacher MC, Friedrich T, Bamberg E, Engelhard M. Functional cell-free synthesis of a seven helix membrane protein: in situ insertion of bacteriorhodopsin into liposomes. Mol Biol. 2007;371:639–648. doi: 10.1016/j.jmb.2007.05.087. [DOI] [PubMed] [Google Scholar]
- Katayama Y, Shimokata K, Suematsu M, Ogura T, Tsukihara T, Yoshikawa S, Shimada H. Cell-free synthesis of cytochrome c oxidase, a multicomponent membrane protein. J Bioenerg Biomembr. 2010;42:235–240. doi: 10.1007/s10863-010-9285-8. [DOI] [PubMed] [Google Scholar]
- Kigawa T, Yamaguchi-Nunokawa E, Kodama K, Matsuda T, Yabuki T, Matsuda N, Ishitani R, Nureki O, Yokoyama S. Selenomethionine incorporation into a protein by cell-free synthesis. Struct Funct Genomics. 2002;2:29–35. doi: 10.1023/a:1013203532303. [DOI] [PubMed] [Google Scholar]
- Kim HC, Kim DM. Methods for energizing cell-free protein synthesis. J Biosci Bioeng. 2009;108:1–4. doi: 10.1016/j.jbiosc.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Kim HC, Kim TW, Kim DM. Prolonged production of proteins in a cell-free protein synthesis system using polymeric carbohydrates as an energy source. Process Biochem. 2011;46:1366–1369. [Google Scholar]
- Kim HC, Kim TW, Park CG, Oh IS, Park K, Kim DM. Continuous cell-free protein synthesis using glycolytic intermediates as energy sources. J Microbiol Biotechnol. 2008;18:885–888. [PubMed] [Google Scholar]
- Kim TW, Oh IS, Keum JW, Kwon YC, Byun JY, Lee KH, Choi CY, Kim DM. Prolonged cell-free protein synthesis using dual energy sources: Combined use of creatine phosphate and glucose for the efficient supply of ATP and retarded accumulation of phosphate. Biotechnol Bioeng. 2007;96:1510–1515. doi: 10.1002/bit.21337. [DOI] [PubMed] [Google Scholar]
- Knapp KG, Goerke AR, Swartz JR. Cell-free synthesis of proteins that require disulfide bonds using glucose as an energy source. Biotechnol Bioeng. 2007;97:901–908. doi: 10.1002/bit.21296. [DOI] [PubMed] [Google Scholar]
- Kovtun O, Mureev S, Jung W, Kubala MH, Johnston W, Alexandrov K. Leishmania cell-free protein expression system. Methods. 2011;55:58–64. doi: 10.1016/j.ymeth.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Lee KY, Lee KH, Park JW, Kim DM. Flexible programming of cell-free protein synthesis using magnetic bead-immobilized plasmids. PLoS One. 2012;7:e34429. doi: 10.1371/journal.pone.0034429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeWinter MM, Granzier H. Cardiac titin: a multifunctional giant. Circulation. 2010;121:2137–2145. doi: 10.1161/CIRCULATIONAHA.109.860171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- Loscha KV, Herlt AJ, Qi R, Huber T, Ozawa K, Otting G. Multiple-site labeling of proteins with unnatural amino acids. Angew Chem Int Ed Engl. 2012;51:2243–2246. doi: 10.1002/anie.201108275. [DOI] [PubMed] [Google Scholar]
- Lyukmanova EN, Shenkarev ZO, Khabibullina NF, Kopeina GS, Shulepko MA, Paramonov AS, Mineev KS, Tikhonov RV, Shingarova LN, Petrovskaya LE, Dolgikh DA, Arseniev AS, Kirpichnikov MP. Lipid-protein nanodiscs for cell-free production of integral membrane proteins in a soluble and folded state: comparison with detergent micelles, bicelles and liposomes. Biochim Biophys Acta. 2012;1818:349–358. doi: 10.1016/j.bbamem.2011.10.020. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Koshiba S, Tochio N, Seki E, Iwasaki N, Yabuki T, Inoue M, Yokoyama S, Kigawa T. Improving cell-free protein synthesis for stable-isotope labeling. J Biomol NMR. 2007;37:225–229. doi: 10.1007/s10858-006-9127-5. [DOI] [PubMed] [Google Scholar]
- Merrifield B. Concept and early development of solid-phase peptide synthesis. Methods Enzymol. 1997;289:3–13. doi: 10.1016/s0076-6879(97)89040-4. [DOI] [PubMed] [Google Scholar]
- Mikami S, Kobayashi T, Masutani M, Yokoyama S, Imataka H. A human cell-derived in vitro coupled transcription/translation system optimized for production of recombinant proteins. Protein Expr Purif. 2008;62:190–198. doi: 10.1016/j.pep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Mureev S, Kovtun O, Nguyen UT, Alexandrov K. Species-independent translational leaders facilitate cell-free expression. Nat Biotechnol. 2009;27:747–752. doi: 10.1038/nbt.1556. [DOI] [PubMed] [Google Scholar]
- Nand A, Gautam A, Pérez JB, Merino A, Zhu J. Emerging technology of in situ cell free expression protein microarrays. Protein Cell. 2012;3:84–88. doi: 10.1007/s13238-012-2012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson BL, Soellner MB, Raines RT. Chemical synthesis of proteins. Annu Rev Biophys Biomol Struct. 2005;34:91–118. doi: 10.1146/annurev.biophys.34.040204.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi H, Shimizu Y, Ying BW, Ueda T. Efficient protein selection based on ribosome display system with purified components. Biochem Biophys Res Commun. 2007;352:270–276. doi: 10.1016/j.bbrc.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Okano T, Matsuura T, Kazuta Y, Suzuki H, Yomo T. Cell-free protein synthesis from a single copy of DNA in a glass microchamber. Lab Chip. 2012;12:2704–2711. doi: 10.1039/c2lc40098g. [DOI] [PubMed] [Google Scholar]
- Ozawa K, Dixon NE, Otting G. Cell-free synthesis of 15N-labeled proteins for NMR studies. IUBMB Life. 2005;57:615–622. doi: 10.1080/15216540500217859. [DOI] [PubMed] [Google Scholar]
- Ozawa K, Loscha KV, Kuppan KV, Loh CT, Dixon NE, Otting G. High-yield cell-free protein synthesis for site-specific incorporation of unnatural amino acids at two sites. Biochem Biophys Res Commun. 2012;418:652–656. doi: 10.1016/j.bbrc.2012.01.069. [DOI] [PubMed] [Google Scholar]
- Plückthun A. Ribosome display: a perspective. Methods Mol Biol. 2012;805:3–28. doi: 10.1007/978-1-61779-379-0_1. [DOI] [PubMed] [Google Scholar]
- Ryabova LA, Vinokurov LM, Shekhovtsova EA, Alakhov YB, Spirin AS. Acetyl phosphate as an energy source for bacterial cell-free translation systems. Anal Biochem. 1995;226:184–186. doi: 10.1006/abio.1995.1208. [DOI] [PubMed] [Google Scholar]
- Schneider B, Junge F, Shirokov VA, Durst F, Schwarz D, Dötsch V, Bernhard F. Membrane protein expression in cell-free systems. Methods Mol Biol. 2010;601:165–186. doi: 10.1007/978-1-60761-344-2_11. [DOI] [PubMed] [Google Scholar]
- See YP, Burrow GN. Synthesis of high molecular weight thyroglobulin peptides by thyroid polysome in vitro. Biochim Biophys Acta. 1979;561:215–222. doi: 10.1016/0005-2787(79)90504-5. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Kanamori T, Ueda T. Protein synthesis by pure translation systems. Methods. 2005;36:299–304. doi: 10.1016/j.ymeth.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Ueda T. PURE Technology. Methods in Mol Biol. 2010;607:11–21. doi: 10.1007/978-1-60327-331-2_2. [DOI] [PubMed] [Google Scholar]
- Sousa R, Mukherjee S. T7 RNA Polymerase. Prog Nucleic Acid Res Mol Biol. 2003;73:1–41. doi: 10.1016/s0079-6603(03)01001-8. [DOI] [PubMed] [Google Scholar]
- Stevenson BJ, Liu JW, Kuchel PW, Ollis DL. Fermentative glycolysis with purified Escherichia coli enzymes for in vitro ATP production and evaluating an engineered enzyme. J Biotechnol. 2012;157:113–123. doi: 10.1016/j.jbiotec.2011.09.019. [DOI] [PubMed] [Google Scholar]
- Su XC, Loh CT, Qi R, Otting G. Suppression of isotope scrambling in cell-free protein synthesis by broadband inhibition of PLP enzymes for selective 15N-labelling and production of perdeuterated proteins in H2O. J Biomol NMR. 2011;50:35–42. doi: 10.1007/s10858-011-9477-5. [DOI] [PubMed] [Google Scholar]
- Suchanek M, Radzikowska A, Thiele C. Photo-leucine and photo-methionine allow identification of protein-protein interactions. Nat Methods. 2005;2:261–267. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- Swartz JR. Transforming biochemical engineering with cell-free biology. AIChE Journal. 2012;58:5–13. [Google Scholar]
- Ueda T. Chapter 2, The Constructive Approach for Cell-Free Translation. In: Spirin AS, Swartz JR, editors. Cell-Free Protein Synthesis: Methods and Protocols. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2008. [Google Scholar]
- Ueda T, Kanamori T, Ohashi H. Ribosome display with the PURE technology. Methods Mol Biol. 2010;607:219–225. doi: 10.1007/978-1-60327-331-2_18. [DOI] [PubMed] [Google Scholar]
- Wang HH, Huang PY, Xu G, Haas W, Marblestone A, Li J, Gygi S, Forster A, Jewett MC, Church GM. Multiplexed in vivo His-tagging of enzyme pathways for in vitro single-pot multi-enzyme catalysis. ACS Synth Biol. 2012;1:43–52. doi: 10.1021/sb3000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang YH. Cell-free protein synthesis energized by slowly-metabolized maltodextrin. BMC Biotechnol. 2009;9:58. doi: 10.1186/1472-6750-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts RE, Forster AC. Update on pure translation display with unnatural amino acid incorporation. Methods Mol Biol. 2012;805:349–365. doi: 10.1007/978-1-61779-379-0_20. [DOI] [PubMed] [Google Scholar]
- Yadavalli SS, Ibba M. Quality control in aminoacyl-tRNA synthesis its role in translational fidelity. Adv Protein Chem Struct Biol. 2012;86:1–43. doi: 10.1016/B978-0-12-386497-0.00001-3. [DOI] [PubMed] [Google Scholar]
- Yin G, Garces ED, Yang J, Zhang J, Tran C, Steiner AR, Roos C, Bajad S, Hudak S, Penta K, Zawada J, Pollitt S, Murray CJ. Aglycosylated antibodies and antibody fragments produced in a scalable in vitro transcription-translation system. mAbs. 2012;4:217–225. doi: 10.4161/mabs.4.2.19202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama J, Matsuda T, Koshiba S, Tochio N, Kigawa T. A practical method for cell-free protein synthesis to avoid stable isotope scrambling and dilution. Anal Biochem. 2011;411:223–229. doi: 10.1016/j.ab.2011.01.017. [DOI] [PubMed] [Google Scholar]
- Zawada JF, Yin G, Steiner AR, Yang J, Naresh A, Roy SM, Gold DS, Heinsohn HG, Murray CJ. Microscale to manufacturing scale-up of cell-free cytokine production--a new approach for shortening protein production development timelines. Biotechnol Bioeng. 2011;108:1570–1578. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Asahara H, Gaucher EA, Chong S. Reconstitution of translation from Thermus thermophilus reveals a minimal set of components sufficient for protein synthesis at high temperatures and functional conservation of modern and ancient translation components. Nucleic Acids Res. 2012;40:7932–7945. doi: 10.1093/nar/gks568. [DOI] [PMC free article] [PubMed] [Google Scholar]