Abstract
In this work, false positive rate of an arrayCGH platform for its use in day-3 single-blastomere analysis was calculated. For this purpose, 38 embryos diagnosed as abnormal on day-3 by FISH were re-biopsied on day-4. Single-cell day-4 arrayCGH diagnosis was then performed. A successful amplification was obtained in 97.4 % (37/38) of the day-4 cells analysed by arrayCGH. Day-3 FISH and day-4 arrayCGH diagnosis were concordant in 35/37 cases. The two discordant embryos were spread and all the cells from each embryo were re-analysed by FISH on day 5. The same error rate (2.7 %) for day-3 FISH and day-4 arrayCGH was obtained when comparing day-5 FISH re-analysis. After this pre-clinical phase, the platform was used for day-3 arrayCGH clinical application in 320 patients (1,760 embryos). Day-3 amplification rate was 98.6 %. An optimal reproductive outcome was obtained when applying arrayCGH to a clinical program: clinical pregnancy rate per cycle of 38.4 % and 60.3 % per transference were obtained, with an implantation rate of 53.5 %. Overall miscarriage rate was 10.6 %. Additionally, day-5 FISH re-analysis was performed in 42 of the embryos from the clinical phase, obtaining a concordance rate of 97.6 % with day-3 arrayCGH.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-012-9918-4) contains supplementary material, which is available to authorized users.
Keywords: arrayCGH, Blastomere biopsy, Day-3 PGS, Day-5 FISH re-analysis
Introduction
Preimplantation Genetic Aneuploidy Screening (PGS) is offered in many IVF centres to improve the reproductive outcome of specific groups of patients. In particular, PGS programs have widely employed fluorescence in situ hybridisation (FISH) for screening numerical chromosome anomalies in preimplantation embryos. Using this technique on single day-3 blastomeres has led to reported improvements in implantation and pregnancy rates in retrospective studies of cases of advanced maternal age (AMA), severe male factor (SMF) infertility, and recurrent miscarriage (RM); the benefit of this analysis in couples with repetitive implantation failure (RIF) has been more controversial [14]. Interestingly, a meta-analysis of the randomized control trials (RCT) conducted in PGS indicated that FISH screening does not improve live birth rates in IVF patients and, indeed, lowers live birth rates in AMA patients [11]. However, the results reported in these RCTs were attributed to several methodological flaws [2, 16, 20, 22] and, more recently, to the poor predictive value of day-3 FISH [18].
Another complication of PGS is the time of biopsy. The most common option has been day-3 embryo biopsy, but this approach has been criticized arguing a reduction in embryo viability due to the biopsy; in fact, a defective biopsy could damage the embryo [22]. Further, embryo mosaicism at cleavage stage and self-correction of aneuploidies between cleavage stage and blastocyst stage have been noted [6]. Recent studies, however, suggest that both phenomena may be overestimated by day-3 FISH analysis [23, 29]. To avoid any kind of misdiagnosis due to embryo mosaicism, polar bodies biopsies have been used [27], but in those cases, only maternal genetic information is obtained, thus all the paternal contribution as well as the mitotic errors are missed. The third option is to biopsy trophectoderm cells from blastocysts, considered less invasive and with a high concordance between inner cell mass and trophectoderm cells [1, 18].
Previous RCTs have been limited by the technique available for chromosomal analysis. Using a sequential FISH protocol, only a selected panel of chromosomes, usually between 9 and 12, could be analyzed simultaneously. Additionally, the technique required cell fixation on a microscope slide. Poor spreading could result in low-quality nuclei or even loss of chromatin with impairment on the accuracy of the diagnosis [17, 26]. Indeed, misdiagnosis can also result from overlapping signals, split signals, cross-hybridisation, or polymorphisms [5]. However, some of these limitations might be overcome by using different strategies. For example, to improve FISH accuracy, additional probes could be incorporated to double-check for dubious signals and false monosomies for certain chromosomes [3, 5, 10, 15, 19, 25]. Indeed, higher diagnosis accuracy was obtained when performing these additional hybridisation rounds comparing with embryos in which no re-hybrisation rounds were used (95 % vs. 82.7 %; p = 0.0443) [15]. Despite this improvement, the limitation on the number of tested chromosomes remained.
Recently, several approaches toward 24-chromosome analysis have been developed to improve clinical results by incorporating comprehensive chromosomal screening. These approaches require whole-genome amplification (WGA) to generate enough DNA for analysis. The first of these approaches applied to PGS was comparative genomic hybridisation (CGH) [28], in which the genome of interest is hybridized against a reference genome in a slide containing a spreading of euploid human metaphases. More recently, arrayCGH has emerged, providing higher resolution and more rapid and automated diagnosis. This approach uses microarray slides containing DNA spots representative of the human genome. Two types of array platforms have been described in PGS: arrayCGH and SNParray. In arrayCGH platforms the spots can be clones of bacterial artificial chromosomes (BAC-arrayCGH) or synthetic oligonucleotides (oligo-arrayCGH). In SNParray platforms, spots include SNPs (Single Nucleotide Polymorphisms) from all the genome and add to the 24-chrosomsome analysis the possibility of haplotyping the sample [8]. These advances based on array platforms, offer the opportunity to increase the reliability and standardisation of diagnosis for PGS, but the approaches need to be optimized for clinical use.
In this study, we sought to evaluate the potential of a single-cell adapted BAC-based arrayCGH platform for the detection of aneuploidies in cleavage stage embryos. A single blastomere from embryos previously diagnosed as abnormal by day-3 FISH in our routine PGS program was analysed by arrayCGH on day 4. Both diagnoses were then compared and, in cases of discordant results, the remaining cells of the embryo were fixed on day 5 and re-analysed by FISH. After this pre-clinical study, the same platform was applied to an initial set of patients who underwent day-3 arrayCGH diagnosis. Clinical results of these PGS cycles are presented.
Materials and methods
This prospective study was performed from October 2009 to March 2012. The pre-clinical phase of the study was performed from October 2009 to February 2010; subsequently, arrayCGH was applied clinically.
Clinical protocol for PGS cycles by FISH
In our routine PGS program, all patients with a specific clinical indication for PGS signed a specific informed consent before undergoing PGS. Standardized ovarian stimulation protocols were used and intracytoplasmic sperm injection was performed in all cases. Fertilization was assessed 17–20 h after microinjection, and embryo cleavage was recorded every 24 h. Embryos were grown in IVF/CCM medium (1/1) (Vitrolife, Göteborg, Sweden) until day 3, when embryo biopsy was performed. Subsequently, embyos were cultured in CCM medium with a monolayer of endometrial epithelial cells until day 5 [12].
For biopsy, embryos were placed on a droplet containing a Ca2+/Mg2+-free medium (G-PGD, Vitrolife, Göteborg, Sweden), and the zona pellucida was perforated using acid Tyrode’s solution or laser technology (OCTAX, Herbron, Germany). A single blastomere was removed in embryos with ≥5 nucleated blastomeres and ≤25 % of fragmentation degree. Individual blastomeres were fixed on glass slides under an inverted microscope, using a slightly modified Tarkowski’s protocol with methanol-acetic acid (3:1) solution. FISH diagnosis included the analysis of nine chromosomes in two consecutive rounds of hybridisation: in the first round chromosomes 13, 16, 18, 21, and 22 were analysed using MultiVysionTM PB panel; in the second round chromosomes 15, 17, X, and Y were analysed using MultiVysionTM 4 Colour Custom panel (Vysis, Inc., Downers Grove, IL, USA). Additionally, re-hybridisation rounds using additional probes were conducted to rescue non-informative or monosomic results as previously described [15]. Chromosomally normal embryos were transferred on day 5, and the surplus euploid embryos cryopreserved.
Pre-clinical phase
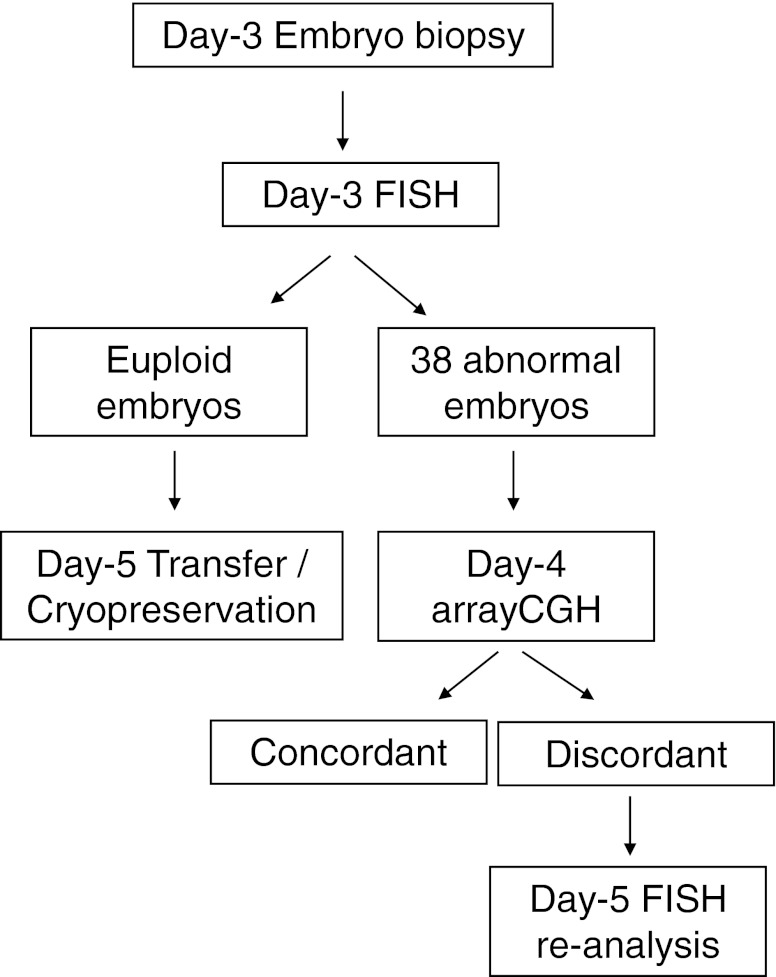
In the pre-clinical phase, 38 embryos diagnosed on day 3 as chromosomally abnormal by FISH were re-biopsied on day 4. A second cell from each one of these embryos was biopsied and analysed by arrayCGH on day 4. It was known prior to arrayCGH analysis that the sample came from an embryo known to have been given an aneuploid diagnosis by FISH. Then, comparison of both diagnoses was then performed. In cases of discordant results, day-5 re-analysis of the remaining cells of the embryo was performed by FISH using probes for the discordant chromosomes (Fig. 1).
Fig. 1.
Workflow of the validation phase. After performing our routine day-3 FISH program, a second cell was biopsied on day 4 in 38 embryos diagnosed as chromosomally abnormal. Single-cell arrayCGH diagnosis was performed in those samples. Day-3 FISH and day-4 arrayCGH diagnoses were compared, and, in cases of discordant results, the whole day-5 embryo was fixed to be re-analysed by FISH using probes directed to the discordant chromosomes
To perform day-4 arrayCGH analysis, a single cell from each embryo was amplified using SureplexTM DNA amplification system (BlueGnome, Cambridge, UK). Amplification quality was ensured by gel electrophoresis (Lonza, Rockland, USA). Then, sample and control DNA were labelled with Cy3 and Cy5 fluorophores, respectively. Labelling mixes were combined and hybridised on a 24sure array (BlueGnome, Cambridge, UK) for 6 to 12 h. Final results were obtained on early day 5 using a laser scanner (710 Innoscan, Innopsys, Carbonne, France; and Powerscanner, TECAN, Männedorf, Switzerland). BlueFuse software was used to analyse data (BlueGnome, Cambridge, UK). Analysis by arrayCGH was completed in a 24-h protocol.
In cases of discordant results between day-3 FISH and day-4 arrayCGH, the remaining cells of the day-5 embryo were fixed and re-analysed by FISH using probes directed to the discordant chromosomes. Due to the limited number of chromosomes that can be analysed simultaneously, day-5 FISH diagnoses were performed with knowledge of the chromosomes involved in the aneuploidies previously seen on day-3 and day-4, but without knowing what aneuploidies for these chromosomes were. Day-5 embryos were fixed using a slightly modified fixation method described in [9]. After removing zona pellucida with Tyrode’s acid, day-5 embryos were placed in a Ca2+/Mg2+ free medium (G-PGD, Vitrolife, Göteborg, Sweden). Then, the whole embryo was fixed using a spreading solution with 0.010 N HCl and Tween 20; when cytoplasm was dissolved, nuclei were re-fixed with methanol-acetic acid.
Clinical application of arrayCGH
After the pre-clinical evaluation of the platform, patients coming to our PGS program were offered the possibility of undergoing day-3 PGS by arrayCGH instead of by FISH. A total of 320 patients decided to undergo PGS by day-3 arrayCGH in these two-year clinical experience (March 2010–March 2012). The number of patients for each PGS indication was: 106 for AMA, 96 for RM, 62 for RIF, 28 for severe male factor (SMF), and 28 patients with a previous chromosomally abnormal gestation as a principal indication for PGS. RM indication included patients with at least 2 miscarriages, RIF patients with three or more previous IVF attempts failed, and SMF patients with seminal parameters severely affected. PGS was indicated for AMA, in patients over 38 years old until May 2010. After that, the AMA group was re-defined as patients over 40 years old due to a retrospective analysis of the results from our lab [13]. Also mixed indications were found in 47 of those 320 couples, in which AMA was the secondary indication for 22 RM couples, 14 RIF couples and 7 couples with a previous chromosomally abnormal gestation.
On day 3, a single blastomere was biopsied in the 1,760 embryos, and single-cell arrayCGH was performed using the same 24-h protocol used in the validation phase. ArrayCGH protocol started in the following morning, and results were obtained on early day-5. Chromosomally normal embryos were transferred on day 5 of the corresponding cycle and the surplus chromosomally normal embryos were cryopreserved.
Re-analysis of the results of the clinical application phase
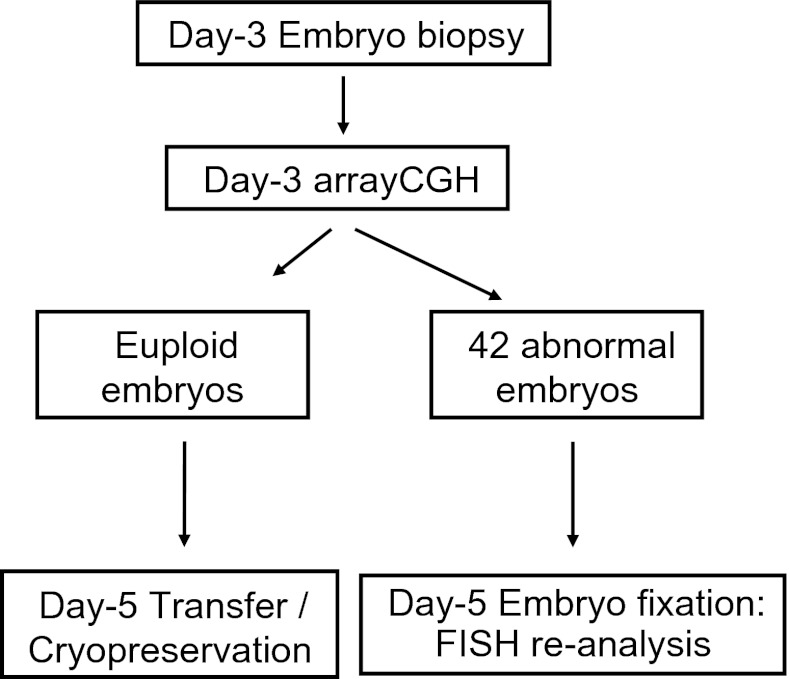
No special criteria in the selection of day-5 embryos for re-analysis were followed. The only limitation to include abnormal embryos in this study was their availability in the corresponding IVF units. We requested aneuploid embryos independtly of the affected chromosome, since we dispose FISH-probes for all the chromosomes. After all, 42 embryos diagnosed as chromosomally abnormal by arrayCGH in the clinical phase were spread and fixed to be re-analysed by FISH on day 5, using probes directed to the abnormal chromosomes. In these embryos, the same fixation protocol described in the validation phase was applied (Figs. 2 and 3).
Fig. 2.
Workflow of the clinical phase. Day-3 arrayCGH diagnosis was performed on single blastomeres. Euploid embryos were transferred or cyropreserved on day 5. To determine accuracy, 42 embryos diagnosed as chromosomally abnormal on day 3 were re-analysed by day-5 FISH using probes directed to the altered chromosomes
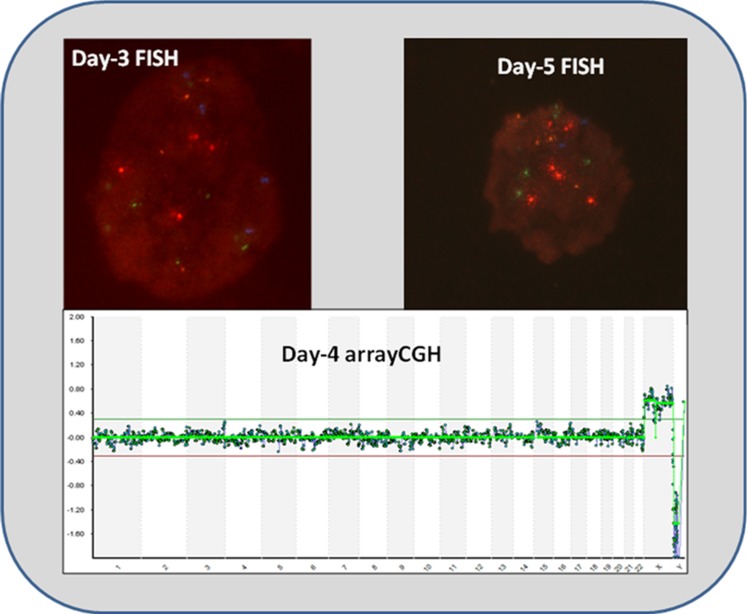
Fig. 3.
Images of one of the two discordant cases of the validation phase. Day-3 FISH show 4 signals for each chromosome: red colour for chromosome 13, Aqua for 16, Blue for 18, Green for 21 and Gold for 22 (MultiVysionTM PB panel, Vysis, Inc., Downers Grove, IL, USA), meaning it was a tetraploid cell. However, day-4 arrayCGH show a euploid pattern, and finally day-5 FISH using the same probes used on day 3 corroborate FISH diagnosis at cleavage stage. It is worth mentioned that as it was an arrested embryo on day-5, only 6 cells could be re-analysed
Results
In the pre-clinical phase, a second cell was biopsied on day 4 from 38 embryos diagnosed as abnormal by FISH on day 3. A successful single-blastomere amplification was obtained in 37/38 cells (97.4 %). Each of the 37 products of amplification produced an interpretable arrayCGH result. In 35/37 of those blastomeres (94.6 %), day-4 arrayCGH confirmed day-3 FISH diagnosis: In 62.9 % of them (22/35), arrayCGH showed only the same aneuploidies observed by FISH; while in the remaining 37.1 % (13/35), additional aneuploidies were observed by arrayCGH for chromosomes not tested by FISH. The two remaining embryos (5.4 %) exhibited discordant FISH and arrayCGH results; these embryos were re-analysed by FISH on day 5 (Table 1). One of them was diagnosed as tetraploid by FISH on day 3, but as 46, XY by arrayCGH. This embryo was arrested on day 5, and the 6 cells of the embryo were determined to be tetraploid by day-5 FISH. Thus, day-3 FISH diagnosis was confirmed in this embryo. The second discordant embryo showed monosomy 16 and trisomy XXY by day-3 FISH, but was diagnosed as 47,XY+3 by arrayCGH. The whole blastocyst was fixed on day 5, and 68 cells were analysed by FISH for chromosomes 3, 16, X, and Y. In this case, the day-4 arrayCGH result was confirmed in the whole blastocyst.
Table 1.
Comparison of discordant results obtained between day-3 FISH and day-4 arrayCGH diagnosis. In one case, day-3 FISH diagnosis was confirmed by day-5 FISH re-analysis; in the other, day-4 arrayCGH diagnosis was confirmed
| Day-3 FISH | Day-4 arrayCGH | Day-5 FISH re-analysis |
|---|---|---|
| tetraploid | 46,XY | tetraploid (6 cells)a |
| monosomy 16, trisomy XXY | 47,XY+3 | 47,XY+3 (68 cells)a |
aOnly chromosomes involved in the aneuploidies previously observed were tested for Day-5 FISH re-analysis
Regarding the type of abnormalities observed in those 37 embryos, the most common ones were chromosome loss (n = 67), followed by chromosome gain (n = 38). Of those 106 aneuploidies, 49 (46.2 %) were for chromosomes not analysed by FISH.
In the clinical phase of the study, a successful amplification was obtained in 1,736 out of 1,760 blastomeres (98.6 %). A total of 1,289 embryos were diagnosed as abnormal (74.2 %) by arrayCGH. Of those abnormal embryos, 262 (20.3 %) were diagnosed as chaotic, showing a complex pattern of aneuploidies. Excluding chaotic embryos, 31.6 % (325/1027) of the remaining abnormal embryos exhibited aneuploidies only for the 9 chromosomes analysed in our FISH program; 40.4 % (415/1027) had aneuploidies for the typically-assessed chromosomes as well as for other chromosomes; and 28 % (287/1027) of them had aneuploidies only for chromosomes different from the 9 analysed in our FISH program.
Day-5 FISH re-analysis of the abnormal embryos from the clinical phase of the study showed 97.6 % (41/42) concordance with day-3 arrayCGH diagnosis. The discordant embryo was diagnosed as 47,XX,+15 by arrayCGH on day 3, but FISH analysis of the whole embryo on day 5 detected the following abnormalities: 2n,XX in 22 cells; 4n,XXXX in 2 cells; and 48,XX,+13,+21 in one cell.
Concerning the reproductive outcome of the 230 day-3 arrayCGH cycles, chromosomally normal embryos were available for transfer in 204 cycles (63.7 %). A clinical pregnancy was observed in 123 patients (38.4 % per cycle; 60.3 % per transference), with an implantation rate of 53.5 % (161 sacs/301 transferred embryos). Thirteen miscarriages were observed (10.6 %); consequently, successful rates of ongoing pregnancy (33.1 % per cycle; 52 % per transference) and of ongoing implantation rate (47.8 %) were obtained. These results are described in Table 2 according to maternal age. Only in three of these miscarriages an hysteroembryoscopy could be performed, and all of them were chromosomally normal (one case was a gemelar gestation with 46,XY karyotype, while the other two miscarriages were 46,XX).
Table 2.
Clinical results of day-3 PGS by arrayCGH, divided into two age groups: patients up to 40 years old and patients over 40
| ≤40 years | >40 years | Total | |
|---|---|---|---|
| No. of cycles | 232 | 88 | 320 |
| Mean age (SD) | 36.3 (3.0) | 42.0 (1.1) | 37.9 (3.7) |
| % Embryo transfers | 73.3 | 38.6 | 63.7 |
| % Abnormal embryos | 68.3 | 88.1 | 73.6 |
| Pregnancy rate/transfer | 58.2 | 67.6 | 59.8 |
| Pregnancy rate/cycle | 42.7 | 26.1 | 37.5 |
| Implantation rate | 51.3 | 63.0 | 53.5 |
| Miscarriage rate | 12.1 | 3.4 | 10.6 |
Discussion
This study is focused on the evaluation of a BAC-based array CGH platform for aneuploidy detection in cleavage stage embryos and in the subsequent clinical application of this platform. In the pre-clinical phase of the study, the platform was able to detect those aneuploidies found by FISH on day 3. Moreover, arrayCGH detected additional aneuploidies for chromosomes not analysed by FISH, and the error rate for both techniques was the same (2.7 %). This error rate is in accordance with other validation studies performed using array platforms for PGS [4, 7, 24].
Two discordant results between day-3 FISH and day-4 arrayCGH were observed in the pre-clinical phase. Day-5 FISH re-analyses of the whole embryo indicated that: one case could have been a day-3 FISH misdiagnosis, because all cells on day 5 exhibited the chromosomal abnormality detected by day-4 arrayCGH instead of the one detected by day-3 FISH; while the second case could be explained as a day-4 arrayCGH misdiagnosis, likely due to the inability of the arrayCGH technique to detect full tetraploidies [7].
Later on, in the clinical application period, one discordant result was observed between day-3 arrayCGH and day-5 FISH re-analysis of the remaining cells of the embryo. Day-3 arrayCGH result was 47, XX, +15, but 22 out of the 25 cells analysed by day-5 FISH were chromosomally normal and the remaining three cells were abnormal: two tetraploid cells and one aneuploid cell (48,XX,+13,+21). This discordance might result from two main scenarios: (1) a technical artefact, or (2) a misdiagnosis due to embryo mosaicism at the cleavage stage if mosaicism was originated on day-2 and/or day-3 cell divisions and one of the abnormal cells was biopsied.
Therefore, both FISH and arrayCGH techniques have limitations. In the case of FISH, the major limitation stems from the limited number of chromosomes that can be analysed simultaneously; additionally, the technical difficulty of nuclear fixation can impede diagnosis in some cases. In arrayCGH, the ability to amplify enough DNA from one cell had been a major limitation. Improved amplification protocols, however, have led to amplification rates around 98–99 %. Nonetheless, arrayCGH remains unable to detect all tetraploid embryos. Some tetraploidies will be detected (e.g., if they carry an imbalance of sex chromosomes), and software devices are continuously being implemented to improve the accuracy of tetraploidy detection. In our pre-clinical study, the error rate due to this limitation was 2.7 %, but the number of samples analysed was very low. Further, some types of polyploid embryos are more prone to arrest during preimplantation development [21], and therefore would not have been selected for transfer. In fact, the tetraploid embryo diagnosed as euploid by day-4 arrayCGH in our study was arrested. Taking into account all these aspects, the estimated misdiagnosis rate due to non-detection of a polyploidy is below 0.2 % [7].
Day-3 arrayCGH results of the clinical application phase showed aneuploidies involving all chromosomes. Importantly, 28.0 % of the embryos analysed would have been diagnosed as normal by FISH using our 9-chromosome panel (chromosomes 13, 15, 16, 17, 18, 21, 22, X, and Y). The number of abnormal embryos detected by arrayCGH was 74.3 %, and 63.7 % of the cycles had euploid embryos for transference. Moreover the higher pregnancy rate per transfer was achieved in patients over 40 years old, probably reflecting that in older patients the main problem to obtain a healthy baby at home is their higher risk of producing aneuploid embryos.
In conclusion, this study shows a BAC-based arrayCGH platform that offers a rapid and standardised method for the screening of all chromosomes in PGS. Although the false negative rate could not be detected with this study design, the false positive rate was 2.4 %. Moreover, good clinical results were obtained, confirming the usefulness of this platform for single-cell analysis in routine clinical diagnosis. Despite the limitations of single-cell FISH technology, it has been shown in this work that the re-analysis of day-5 embryos by FISH can be used to corroborate if the anomalies observed at cleavage stage were still present on day-5 embryos.
Electronic supplementary material
(DOC 53kb)
Acknowledgments
The authors would like to thank all the embryologists in charge of the embryo biopsy, the biologists working in PGS analysis (both for FISH and arrayCGH diagnosis) and also to all clinicians involved in our PGS program.
Footnotes
Capsule
False positive rate of an arrayCGH platform was calculated for single-blastomere analysis, resulting in high efficiency and accuracy of the platform. Furthermore, an optimal reproductive outcome was obtained when applied to a clinical program.
References
- 1.Capalbo A, Wright G, Themaat L, Elliott T, Rienzi L, Nagy ZP. FISH reanalysis of inner cell mass and trophectoderm samples of previously array-CGH screened blastocysts reveals high accuracy of diagnosis and no sign of mosaicism or preferential allocation. Fertil Steril. 2012;96(No. 3). Supplement, September 2011.
- 2.Cohen J, Grifo JA. Multicentre trial of preimplantation genetic screening reported in the New England Journal of Medicine: an in-depth look at the findings. Reprod BioMed Online. 2007;15(4):365–366. doi: 10.1016/S1472-6483(10)60358-6. [DOI] [PubMed] [Google Scholar]
- 3.Colls P, Escudero T, Cekleniak N, Sadowy S, Cohen J, Munne S. Increased efficiency of preimplantation genetic diagnosis for infertility using “no result rescue”. Fertil Steril. 2007;88(1):53–61. doi: 10.1016/j.fertnstert.2006.11.099. [DOI] [PubMed] [Google Scholar]
- 4.Colls P, Escudero T, Fischer J, Cekleniak NA, Ben-Ozer S, Meyer B, et al. Validation of array comparative genome hybridization for diagnosis of translocations in preimplantation human embryos. Reprod BioMed Online. 2012;24(6):621–629. doi: 10.1016/j.rbmo.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Colls P, Sandalinas M, Pagidas K, Munne S. PGD analysis for aneuploidy in a patient heterozygous for a polymorphism of chromosome 16 (16qh-) Prenat Diagn. 2004;24(9):741–744. doi: 10.1002/pd.887. [DOI] [PubMed] [Google Scholar]
- 6.Frumkin T, Malcov M, Yaron Y, Ben-Yosef D. Elucidating the origin of chromosomal aberrations in IVF embryos by preimplantation genetic analysis. Mol Cell Endocrinol. 2008;282(1–2):112–119. doi: 10.1016/j.mce.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Gutierrez-Mateo C, Colls P, Sanchez-Garcia J, Escudero T, Prates R, Ketterson K, et al. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril. 2011;95(3):953–958. doi: 10.1016/j.fertnstert.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Handyside AH, Harton GL, Mariani B, Thornhill AR, Affara N, Shaw MA, et al. Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J Med Genet. 2010;47(10):651–658. doi: 10.1136/jmg.2009.069971. [DOI] [PubMed] [Google Scholar]
- 9.Harper JC, Coonen E, Ramaekers FC, Delhanty JD, Handyside AH, Winston RM, et al. Identification of the sex of human preimplantation embryos in two hours using an improved spreading method and fluorescent in-situ hybridization (FISH) using directly labelled probes. Hum Reprod (Oxf Engl) 1994;9(no. 4):721–724. doi: 10.1093/oxfordjournals.humrep.a138577. [DOI] [PubMed] [Google Scholar]
- 10.Magli MC, Sandalinas M, Escudero T, Morrison L, Ferraretti AP, Gianaroli L, et al. Double locus analysis of chromosome 21 for preimplantation genetic diagnosis of aneuploidy. Prenat Diagn. 2001;21(12):1080–1085. doi: 10.1002/pd.248. [DOI] [PubMed] [Google Scholar]
- 11.Mastenbroek S, Twisk M, van der Veen F, Repping S. Preimplantation genetic screening: a systematic review and meta-analysis of RCTs. Hum Reprod Updat. 2011;17(4):454–466. doi: 10.1093/humupd/dmr003. [DOI] [PubMed] [Google Scholar]
- 12.Mercader A, Garcia-Velasco JA, Escudero E, Remohi J, Pellicer A, Simon C. Clinical experience and perinatal outcome of blastocyst transfer after coculture of human embryos with human endometrial epithelial cells: a 5-year follow-up study. Fertil Steril. 2003;80(5):1162–1168. doi: 10.1016/S0015-0282(03)01178-6. [DOI] [PubMed] [Google Scholar]
- 13.Milán M, Cobo AC, Rodrigo L, Mateu E, Mercader A, Buendía P, et al. Redefining advanced maternal age as an indication for preimplantation genetic screening. Reprod BioMed Online. 2010;21(5):649–657. doi: 10.1016/j.rbmo.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Mir P, Martín J, Rubio C. Preimplantation diagnosis for aneuploidy screening: a new era beyond FISH. In: Weingarten CN, Jefferson SE, editors. Sex chromosomes: Genetics, abnormalities, and disorders. New York, NY: Nova Science Publishers, Inc., 2009. pp. 95–122.
- 15.Mir P, Rodrigo L, Mateu E, Peinado V, Milan M, Mercader A, et al. Improving FISH diagnosis for preimplantation genetic aneuploidy screening. Hum Reprod (Oxf Engl) 2010;25(no. 7):1812–1817. doi: 10.1093/humrep/deq122. [DOI] [PubMed] [Google Scholar]
- 16.Munne S, Cohen J, Simpson JL. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357(no. 17):1769–1770. doi: 10.1056/NEJMc076314. [DOI] [PubMed] [Google Scholar]
- 17.Munne S, Dailey T, Finkelstein M, Weier HU. Reduction in signal overlap results in increased FISH efficiency: implications for preimplantation genetic diagnosis. J Assist Reprod Genet. 1996;13(2):149–156. doi: 10.1007/BF02072537. [DOI] [PubMed] [Google Scholar]
- 18.Northrop LE, Treff NR, Levy B, Scott RT., Jr SNP microarray-based 24 chromosome aneuploidy screening demonstrates that cleavage-stage FISH poorly predicts aneuploidy in embryos that develop to morphologically normal blastocysts. Mol Hum Reprod. 2010;16(8):590–600. doi: 10.1093/molehr/gaq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigo L, Rubio C, Mateu E, Simon C, Remohi J, Pellicer A, et al. Analysis of chromosomal abnormalities in testicular and epididymal spermatozoa from azoospermic ICSI patients by fluorescence in-situ hybridization. Hum Reprod (Oxf Engl) 2004;19(no. 1):123. doi: 10.1093/oxfordjournals.humrep.a002246. [DOI] [PubMed] [Google Scholar]
- 20.Rubio C, Gimenez C, Fernandez E, Vendrell X, Velilla E, Parriego M, et al. The importance of good practice in preimplantation genetic screening: critical viewpoints. Hum Reprod (Oxf Engl) 2009;24(no. 8):2045–2047. doi: 10.1093/humrep/dep188. [DOI] [PubMed] [Google Scholar]
- 21.Rubio C, Rodrigo L, Mercader A, Mateu E, Buendia P, Pehlivan T, et al. Impact of chromosomal abnormalities on preimplantation embryo development. Prenat Diagn. 2007;27(no. 8):748–756. doi: 10.1002/pd.1773. [DOI] [PubMed] [Google Scholar]
- 22.Simpson JL. Randomized clinical trial in assessing PGS: necessary but not sufficient. Hum Reprod (Oxf Engl) 2008;23:2179–2181. doi: 10.1093/humrep/den250. [DOI] [PubMed] [Google Scholar]
- 23.Treff NR, Levy B, Su J, Northrop LE, Tao X, Scott RT., Jr SNP microarray-based 24 chromosome aneuploidy screening is significantly more consistent than FISH. Mol Hum Reprod. 2010;16(8):583–589. doi: 10.1093/molehr/gaq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Treff NR, Su J, Tao X, Levy B, Scott RT., Jr Accurate single cell 24 chromosome aneuploidy screening using whole genome amplification and single nucleotide polymorphism microarrays. Fertil Steril. 2010;94(6):2017–2021. doi: 10.1016/j.fertnstert.2010.01.052. [DOI] [PubMed] [Google Scholar]
- 25.Uher P, Baborova P, Kralickova M, Zech MH, Verlinsky Y, Zech NH. Non-informative results and monosomies in PGD: the importance of a third round of re-hybridization. Reprod BioMed Online. 2009;19(4):539–546. doi: 10.1016/j.rbmo.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Velilla E, Escudero T, Munne S. Blastomere fixation techniques and risk of misdiagnosis for preimplantation genetic diagnosis of aneuploidy. Reprod BioMed Online. 2002;4(3):210–217. doi: 10.1016/S1472-6483(10)61808-1. [DOI] [PubMed] [Google Scholar]
- 27.Verlinsky Y, Ginsberg N, Lifchez A, Valle J, Moise J, Strom CM. Analysis of the first polar body: preconception genetic diagnosis. Hum Reprod (Oxf Engl) 1990;5(no. 7):826–829. doi: 10.1093/oxfordjournals.humrep.a137192. [DOI] [PubMed] [Google Scholar]
- 28.Wells D, Delhanty JD. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod. 2000;6(11):1055–1062. doi: 10.1093/molehr/6.11.1055. [DOI] [PubMed] [Google Scholar]
- 29.Wilton L, Thornhill A, Traeger-Synodinos J, Sermon KD, Harper JC. The causes of misdiagnosis and adverse outcomes in PGD. Hum Reprod (Oxf Engl) 2009;24(no. 5):1221–1228. doi: 10.1093/humrep/den488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 53kb)