Abstract
The repair of DNA double-strand breaks (DSBs) is crucial for maintaining genome stability. The Saccharomyces cerevisiae protein Tbf1, which is characterized by a Myb domain and is related to mammalian TRF1 and TRF2, has been proposed to act as a transcriptional activator. Here, we show that Tbf1 and its interacting protein Vid22 are new players in the response to DSBs. Inactivation of either TBF1 or VID22 causes hypersensitivity to DSB-inducing agents and shows strong negative interactions with mutations affecting homologous recombination. Furthermore, Tbf1 and Vid22 are recruited to an HO-induced DSB, where they promote both resection of DNA ends and repair by non-homologous end joining. Finally, inactivation of either Tbf1 or Vid22 impairs nucleosome eviction around the DSB, suggesting that these proteins promote efficient repair of the break by influencing chromatin identity in its surroundings.
Keywords: chromatin, DSB, NHEJ, resection, Tbf1, Vid22
Introduction
Saccharomyces cerevisiae Tbf1 is a DNA-binding protein that contains a C-terminal ‘telobox’ DNA-binding domain (a variant of the Myb family motif) that is also found in the mammalian telomeric proteins TRF1 and TRF2, as well as in the fission yeast telomeric protein Taz1 (Brigati et al, 1993; Bilaud et al, 1996; Pitt et al, 2008).
Tbf1 is essential for cell viability, but its essential function remains enigmatic. Several studies have proposed a regulatory role for Tbf1 at telomeres, where it associates with the subtelomeric T2AG3-repeat sequences that are frequently located between the so-called ‘core X’ sequence and the telomeric TG1–3 repeats (Liu and Tye, 1991; Brigati et al, 1993; Koering et al, 2000). Binding sites for Tbf1 have also been identified in the STARs (subtelomeric antisilencing regions) of the subtelomeric X and Y′ elements (Fourel et al, 1999). Tbf1 regulates telomere length (Brevet et al, 2003; Berthiau et al, 2006; Arnerić and Lingner, 2007) and it blocks checkpoint activation at a DNA double-strand break (DSB) flanked by T2AG3-repeat arrays, suggesting that it plays a backup role in protecting telomeres that have lost the terminal TG1–3 repeats (Ribaud et al, 2011; Fukunaga et al, 2012). In any case, the Tbf1 essential function is unrelated to telomere maintenance, as Tbf1 appears to influence only mildly telomere homeostasis in both budding and fission yeasts (Berthiau et al, 2006; Hediger et al, 2006; Cockell et al, 2009).
Notably, Tbf1 has been proposed to act as a transcriptional activator. It associates to the upstream regions of most small nucleolar (sno) RNA genes and to ∼100 additional promoters, including those of genes involved in ribosome biogenesis, where it binds a RCCCT consensus sequence (Preti et al, 2010). Tbf1 localizes at these non-snoRNA promoters together with its interacting proteins Vid22 and Env11 (Preti et al, 2010), which are both dispensable for cell viability. Interestingly, Tbf1 was shown to promote formation of nucleosome-free regions at least at non-snoRNA promoters (Badis et al, 2008; Preti et al, 2010), suggesting a role for Tbf1 in modulating chromatin organization. Consistent with such a role, Tbf1-binding sites have been shown to act as insulator elements capable of regulating chromatin accessibility and delimiting independent chromatin domains (Fourel et al, 1999, 2001; Vogelmann et al, 2011).
Here, we provide evidence that Tbf1 and its interactor Vid22 are involved in the response to DSBs. In fact, they are recruited to an HO-induced DSB, where they are needed both to generate 3′-ended single-stranded DNA (ssDNA) ends and to religate the DSB ends by non-homologous end joining (NHEJ). The DSB repair defects caused by Tbf1 and Vid22 dysfunction are likely due to an altered chromatin structure at damage sites, as Tbf1 and Vid22 participate in nucleosome displacement around the DSB.
Results
Tbf1 and Vid22 are involved in DSB repair
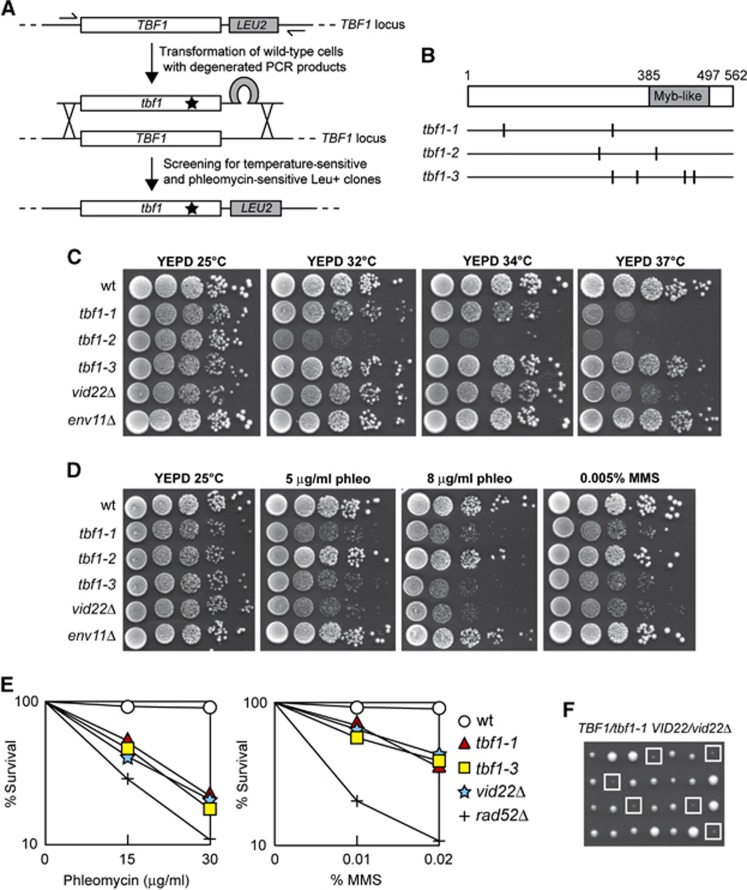
To develop suitable reagents for Tbf1 characterization, we used low-fidelity PCR to random mutagenize the TBF1 gene and searched for temperature-sensitive tbf1 alleles. Because several proteins regulating chromatin dynamics are involved in the DNA damage response (DDR), we also searched for mutant tbf1 alleles causing decreased viability in the presence of the radiomimetic drug phleomycin. Linear TBF1 degenerated PCR products were transformed into the cells in order to replace the corresponding wild-type sequence with the mutagenized DNA fragments (Figure 1A). Transformant clones showing decreased viability at 37°C and/or in the presence of phleomycin compared with the untransformed strain were chosen for further analysis.
Figure 1.
tbf1 and vid22 mutants are defective in the response to DSBs. (A) Strategy used to generate tbf1 mutants. Genomic DNA from a strain carrying the LEU2 gene located downstream of the TBF1 stop codon was used as the template to amplify by mutagenic PCR the LEU2 gene flanked by TBF1 sequences spanning from −165 to +1805, bp from the translation start codon (including 116 bp of 3′ non-coding sequence) on one side, and from +1806 to +1990, bp (3′ non-coding sequence) on the other side. The PCR products were transformed into wild-type cells in order to replace the corresponding TBF1 wild-type sequence. Leu+ transformant clones were selected and then assayed for the ability to grow at 37°C or at 25°C in the presence of 10 μg/ml phleomycin. (B) Schematic representation of the Tbf1 protein. The vertical lines indicate the position of the amino-acid substitutions caused by the indicated tbf1 mutant alleles. (C, D) Exponentially growing cultures of strains with the indicated genotypes were serially diluted (1:10) and each dilution was spotted out onto YEPD plates that were incubated at the indicated temperatures (C). Cells were also spotted out onto YEPD plates with or without phleomycin and MMS at the indicated concentrations and were incubated at 25°C (D). (E) Exponentially growing cultures of strains with the indicated genotypes were incubated for 2 h at 25°C in YEPD containing the indicated amounts of phleomycin or MMS and then appropriate dilutions were plated on YEPD plates to determine colony-forming units. (F) Meiotic tetrads from diploid cells with the indicated genotype were dissected on YEPD plates that were incubated at 25°C for 4 days, followed by spore genotyping. Clones from double mutant spores are highlighted by squares.
This procedure allowed us to isolate three independent mutants that we called tbf1-1, tbf1-2 and tbf1-3. DNA sequencing of the corresponding tbf1 alleles revealed that multiple amino-acid substitutions were carried by the mutant variants: F82S and R299H in Tbf1-1, C285Y and N398S in Tbf1-2, K297E, D357V, Q453H and K480R in Tbf1-3 (Figure 1B). The tbf1-1 and tbf1-2 mutants showed reproducible growth defects at high temperatures, although to different extents, whereas tbf1-3 cells did not display growth defects at any tested temperature (Figure 1C). When cells were spotted on plates containing phleomycin or the alkylating agent methylmethane sulphonate (MMS), both tbf1-1 and tbf1-3 mutant cells formed colonies less efficiently than wild-type cells (Figure 1D). Furthermore, both tbf1-1 and tbf1-3 cells lost viability when they were exposed to different amount of phleomycin or MMS for 2 h and then plated on YEPD, although their hypersensitivity was less severe than that observed in rad52Δ cells (Figure 1E). By contrast, phleomycin or MMS treatment did not impair viability of tbf1-2 mutant cells (Figure 1D). Both the DNA damage and the temperature sensitivity caused by each allele were recessive, being fully complemented by one wild-type copy of TBF1 (data not shown). The finding that the tbf1-2 mutant is unable to grow at 37°C, but it does not display sensitivity to DNA damaging agents, whereas the tbf1-3 mutant is hypersensitive to genotoxic treatments without showing detectable growth defects at 37°C suggests that the essential and DDR functions of Tbf1 are genetically separable.
Tbf1 has been shown to form a stable complex with the uncharacterized non-essential proteins Vid22 and Env11 (Krogan et al, 2006), which colocalize with Tbf1 at non-snoRNA promoters (Preti et al, 2010). When we analysed the sensitivity to DNA damaging agents of vid22Δ and env11Δ mutant cells, vid22Δ cells displayed a pattern of sensitivity to MMS and phleomycin similar to that of tbf1-1 and tbf1-3 cells (Figure 1D and E). By contrast, env11Δ cells did not show detectable growth defects in the presence of the same genotoxic agents (Figure 1D). Thus, both Tbf1 and Vid22 appear to participate in the DDR, whereas Env11 does not.
While Tbf1 is essential for cell viability, the lack of Vid22 slightly impaired colony formation only at 37°C (Figure 1C), indicating that Tbf1 plays a major role in supporting cell viability compared with Vid22. To investigate further the functional connections between Tbf1 and Vid22, we analysed the consequences of deleting VID22 in tbf1-1 cells. When meiotic tetrads from a diploid strain heterozygous for the tbf1-1 and vid22Δ alleles were analysed for growth at 25°C, all the tbf1-1 vid22Δ double mutant spores exhibited severe growth defects, whereas tbf1-1 and vid22Δ single mutant spores formed colonies of almost wild-type size (Figure 1F). The finding that Vid22 contributes to cell viability when Tbf1 activity is compromised further highlights a functional connection between Tbf1 and Vid22.
TBF1 and VID22 show strong functional interactions with homologous recombination genes
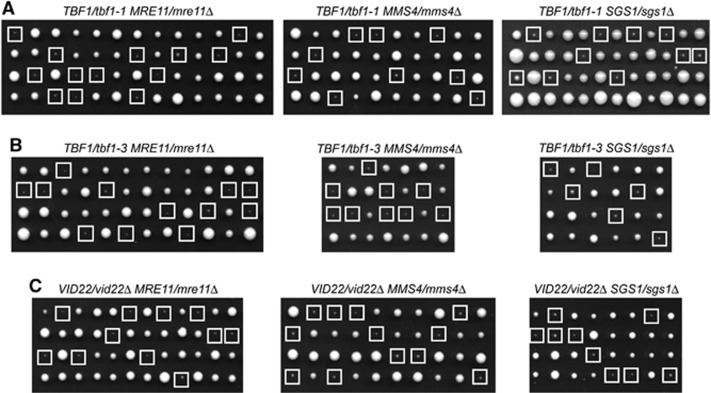
We further investigated the role of Tbf1 and Vid22 in the DDR by combining the tbf1-1, tbf1-3 and vid22Δ mutant alleles with mutations affecting homologous recombination (HR). To this end, we constructed diploid strains heterozygous for either tbf1 or vid22 mutation and the mre11Δ allele, causing the lack of the Mre11 subunit of the MRX complex that is necessary to initiate HR (Pâques and Haber, 1999). Furthermore, tbf1-1, tbf1-3 and vid22Δ alleles were combined with the sgs1Δ or mms4Δ alleles, impairing the Sgs1–Top3–Rmi1 and the Mus81–Mms4 complexes, respectively, which are both required for processing HR repair intermediates. When meiotic tetrads from these diploids were analysed for spore viability on YEPD plates at 25°C, all tbf1-1 mre11Δ, tbf1-1 mms4Δ and tbf1-1 sgs1Δ double mutant spores formed smaller colonies than each single mutant spores (Figure 2A). Similar results were obtained also when tbf1-3 (Figure 2B) or vid22Δ (Figure 2C) was combined with mre11Δ, mms4Δ or sgs1Δ mutations. Thus, Tbf1 and Vid22 support cell viability even in the absence of exogenous DNA damage when HR is defective.
Figure 2.
Functional interactions of the tbf1 and vid22Δ alleles with mutations impairing DSB processing and repair. (A–C) Meiotic tetrads from diploids with the indicated genotypes were dissected on YEPD plates that were incubated at 25°C for 4 days, followed by spore genotyping. Clones from double mutant spores are highlighted by squares.
Tbf1 and Vid22 are required for DSB repair by SSA
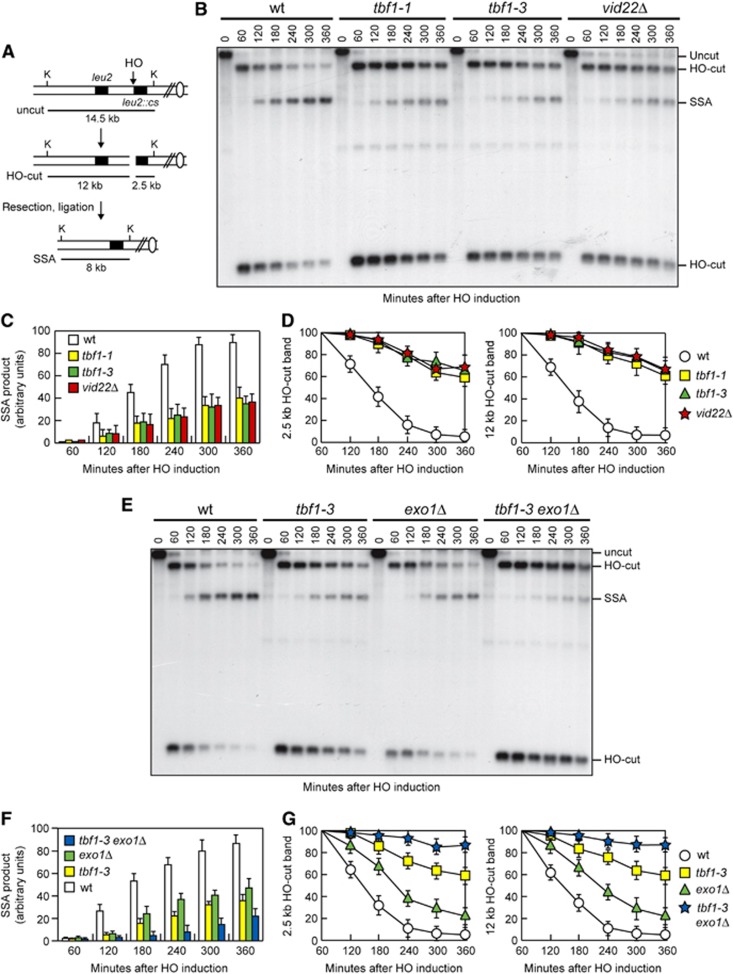
In order to investigate the possible role of Tbf1 and Vid22 in HR, we took advantage of the fact that repair of a DSB made between tandem DNA repeats occurs primarily by the HR single-strand annealing (SSA) pathway. Notably, SSA requires resection of the DSB DNA ends followed by Rad52-dependent annealing of the resulting complementary ssDNA ends (Fishman-Lobell et al, 1992). We deleted VID22 or introduced the tbf1-1 or tbf1-3 allele in a strain carrying tandem repeats of the LEU2 gene, which were located 4.6 kb apart, with a recognition site for the HO endonuclease adjacent to the centromere-proximal repeat (Figure 3A) (Vaze et al, 2002). The strain also carries a galactose-inducible GAL-HO construct that provides regulated HO expression. HO was induced by galactose addition to G2-arrested cells that were kept arrested in G2 with nocodazole for the subsequent 6 h to minimize the effect of cell-cycle progression on the repair capacity. Galactose was maintained in the medium in order to continuously produce HO and thus recleave the HO sites eventually reconstituted by NHEJ. The HO-induced break is repaired mainly by SSA, because homology is restricted to only one DSB end (Figure 3A). When kinetics of DSB repair was monitored by Southern blot analysis of KpnI-digested DNA with a LEU2 probe, accumulation of the 8-kb SSA repair product was defective in tbf1-1, tbf1-3 and vid22Δ cells compared with wild-type cells (Figure 3B and C), indicating that Tbf1 and Vid22 promote DSB repair by SSA.
Figure 3.
Tbf1 and Vid22 are required for DSB repair by SSA. (A) Schematic representation of the YMV45 chromosome III region, where a unique HO-cut site is adjacent to the leu2::cs sequence, which is 4.6 kb apart from the homologous leu2 sequence. HO-induced DSB formation results in generation of 12 and 2.5 kb DNA fragments (HO-cut) that can be detected by Southern blot analysis with a LEU2 probe of KpnI-digested genomic DNA. DSB repair by SSA generates a product of 8 kb (SSA). K, KpnI. (B–D) Exponentially growing YEPR cell cultures were arrested in G2 (time zero) with nocodazole and transferred to YEPRG in the presence of nocodazole. (B) Southern blot analysis of KpnI-digested genomic DNA. (C, D) Densitometric analysis of the SSA (C) and the HO-cut band signals (D). Plotted values are the mean values±s.d. from three independent experiments as in (B). (E–G) Exponentially growing YEPR cell cultures were arrested in G2 (time zero) with nocodazole and transferred to YEPRG in the presence of nocodazole. (E) Southern blot analysis of KpnI-digested genomic DNA. (F, G) Densitometric analysis of the SSA (F) and the HO-cut band signals (G). Plotted values are the mean values±s.d. from three independent experiments as in (E). The intensity of each band was normalized with respect to a loading control (not shown).
Hybridization with the LEU2 probe also allows to monitor 5′–3′ nucleolytic processing at each side of the break by following the disappearance of the HO-cut 2.5 and 12 kb DNA fragments. This process appeared to be defective in tbf1-1, tbf1-3 and vid22Δ cells, because the HO-cut band signals decreased less efficiently in these mutants than in wild-type cells (Figure 3B and D). Thus, the inability of tbf1 and vid22 mutant cells to repair the DSB by SSA may be due to a failure of 5′–3′ resection to reach the homologous distal leu2 sequence.
The MRX complex initiates resection of the 5′ strand possibly through an endonucleolytic cleavage, and the resulting partially resected 5′ DNA end can be further processed by the action of either Exo1 or Sgs1 (Mimitou and Symington, 2008; Zhu et al, 2008). We therefore asked whether Tbf1 and Vid22 might work in either one or both Exo1- and Sgs1-dependent pathways. Unfortunately, we were unable to analyse SSA in tbf1 sgs1Δ and vid22Δ sgs1Δ double mutants because the lack of Sgs1 severely impaired growth of tbf1-1, tbf1-3 and vid22Δ mutants even in the absence of exogenous DNA damage (Figure 2). On the other hand, the pathway by which Tbf1 and Vid22 promote SSA appears to be different from that involving Exo1. In fact, deletion of EXO1 exacerbated the SSA defects of both tbf1-3 (Figure 3E and F) and vid22Δ cells (data not shown). Furthermore, the HO-cut band signals persisted longer in tbf1-3 exo1Δ double mutants than in the corresponding single mutants (Figure 3E and G).
To investigate other possible functions of Tbf1 and Vid22 in HR, we analysed formation of crossover and non-crossover products by using a haploid strain that bears a MATa sequence on chromosome V and an uncleavable MATa-inc sequence on chromosome III (Supplementary Figure S1A) (Saponaro et al, 2010). Upon galactose addition, the HO-induced DSB can be repaired using the MATa-inc sequence as a donor, resulting in crossover and non-crossover products that can be distinguished on the basis of restriction fragment size (Supplementary Figure S1A). Neither Tbf1 nor Vid22 appeared to be required for crossover/non-crossover generation, as both the overall DSB repair efficiency and the proportion of crossover/non-crossovers of tbf1-1 and vid22Δ cells were similar to those observed in wild-type cells (Supplementary Figure S1B and C). As tbf1 and vid22 mutants were defective in DSB resection, this finding also supports previous observations, indicating that the ectopic recombination detected by this system does not require extensive resection of the DSB ends (Trovesi et al, 2011; Chen et al, 2012; Costelloe et al, 2012).
Tbf1 and Vid22 promote resection of DSB ends
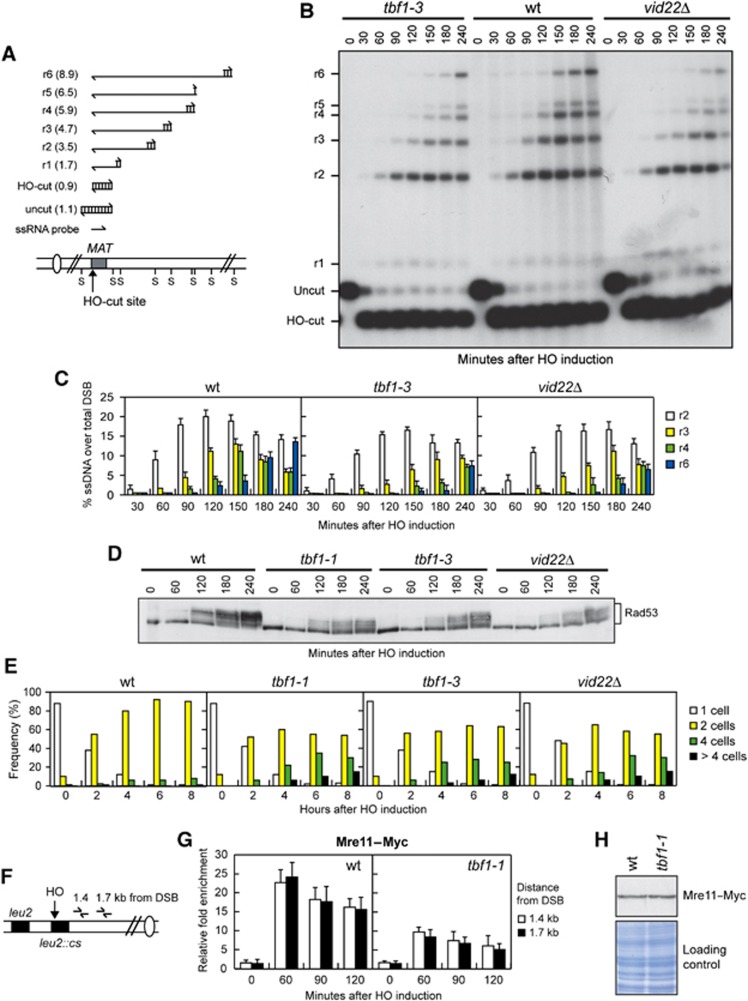
To confirm the involvement of Tbf1 and Vid22 in DSB resection, we directly monitored the generation of ssDNA at the DSB ends. To this purpose, we deleted VID22 or introduced the tbf1 alleles in a haploid strain where a DSB can be induced by HO at the MAT locus (Lee et al, 1998; Figure 4A). To prevent repair by HR, and therefore minimize the effect of DSB repair on ssDNA formation, the HML and HMR homologous donor sequences have been deleted. Because ssDNA is resistant to cleavage by restriction enzymes, a Southern blot analysis under alkaline conditions using a ssRNA probe annealing on one side of the break allows to follow the loss of SspI restriction fragments as a measure of 5′ strand resection (Figure 4A). The appearance of the resection ssDNA intermediates (r1–r6 in Figure 4A and B) after the HO-cut was delayed in nocodazole-arrested galactose-induced tbf1-3 and vid22Δ mutant cells compared with wild-type cells (Figure 4B and C). Similar results were obtained also when ssDNA formation was analysed in the tbf1-1 mutant (data not shown).
Figure 4.
tbf1 and vid22 mutants are defective in DSB end resection and Mre11 recruitment to DSB ends. (A) System used to detect DSB resection. Gel blots of SspI-digested genomic DNA separated on alkaline agarose gel were hybridized with a single-stranded MAT probe specific for the unresected strand. 5′–3′ resection progressively eliminates SspI sites (S), producing larger SspI fragments (r1 through r6, kb size in parenthesis) detected by the probe. (B–D) Exponentially growing YEPR cell cultures of wild-type JKM139 and its derivative tbf1-1, tbf1-3 and vid22Δ strains were arrested in G2 with nocodazole (time zero) and transferred to YEPRG in the presence of nocodazole. (B) Analysis of ssDNA formation as described in (A). (C) Densitometric analysis of the resection products. Plotted values are the mean values±s.d. from three independent experiments as in (B). (D) Western blot analysis with anti-Rad53 antibodies. (E) YEPR G1-arrested cell cultures of wild-type JKM139 and its derivative mutant strains were plated on galactose-containing plates (time zero). At the indicated time points, 200 cells for each strain were analysed to determine the frequency of single cells and of cells forming microcolonies of 2, 4 or >4 cells. (F) Schematic representation of the qPCR primer sets located at the indicated distance from the HO cleavage site at the LEU2 locus. (G) Exponentially growing YEPR cell cultures carrying the HO system described in (F) and expressing a fully functional Mre11–Myc fusion protein were arrested in G2 (time zero) with nocodazole and transferred to YEPRG in the presence of nocodazole. Relative fold enrichment of Mre11–Myc at the indicated distance from the HO cleavage site was evaluated after ChIP with an anti-Myc antibody. Plotted values are the mean values±s.d. from three independent experiments. (H) Western blot analysis with anti-Myc antibody of protein extracts from exponentially growing wild-type and tbf1-1 cells. The same amount of protein extracts was subjected to SDS–PAGE and stained with Coomassie blue (loading control).
It is well known that DSBs trigger the DNA damage checkpoint, whose activation requires phosphorylation of the effector kinase Rad53 that is detectable as a decrease of its electrophoretic mobility. Checkpoint activation after a single DSB depends on Mec1, which recognizes 3′-ended ssDNA tails arising from DSB processing and activates Rad53 by phosphorylation events (Zou and Elledge, 2003). We then asked whether tbf1-1, tbf1-3 and vid22Δ mutant cells were defective in Rad53 phosphorylation and checkpoint activation after induction of the HO-induced DSB at the MAT locus. Indeed, the amount of HO-induced Rad53 phosphorylation in tbf1-1, tbf1-3 and vid22Δ cells was lower than in wild-type cells (Figure 4D). Furthermore, when G1-arrested cell cultures were spotted on galactose containing plates, most wild-type cells were arrested at the two-cell dumbbell stage by the checkpoint within 2–4 h after HO induction and were still arrested after 8 h (Figure 4E). By contrast, tbf1-1, tbf1-3 and vid22Δ cells started to form colonies with four or more cells 4 h after HO induction (Figure 4E), indicating a defective DNA damage checkpoint response. Collectively, these data indicate that Tbf1 and Vid22 contribute to resect the DSB ends and to activate the checkpoint in response to a single DSB.
The MRX complex binds rapidly to newly formed DSBs, where it plays a key role in end resection (Longhese et al, 2010; Symington and Gautier, 2011). As Tbf1 and Vid22 promote DSB resection, we monitored Mre11 recruitment near an HO-induced DSB in wild-type and tbf1-1 cells expressing fully functional Myc-tagged Mre11. HO expression was induced by galactose addition to G2-arrested cells that were kept arrested in G2 with nocodazole throughout the experiment. After chromatin immunoprecipitation (ChIP) with an anti-Myc antibody, quantitative real-time PCR (qPCR) was designed to monitor Mre11 association near the repairable HO-induced DSB at LEU2 locus (Figure 4F). As resection of the DSB ends leads to loss of input DNA (Chen et al, 2008), the ChIP signals were normalized to the corresponding input for each time point. Mre11 association at the HO-induced DSB was decreased, but not abolished, in tbf1-1 G2 cells compared with wild-type cells (Figure 4G). This decreased Mre11 recruitment was not due to lower Mre11 levels in tbf1-1 cells compared with wild-type, as similar Mre11 amounts could be detected in protein extracts prepared from both cell types (Figure 4H). Thus, we can conclude that Tbf1 facilitates Mre11 binding to DSBs. As the MRX complex is required to initiate DSB resection, this decreased binding might account for the resection defects displayed by tbf1 and, possibly, vid22Δ mutant cells.
Functional interactions of TBF1 and VID22 with HR and NHEJ genes
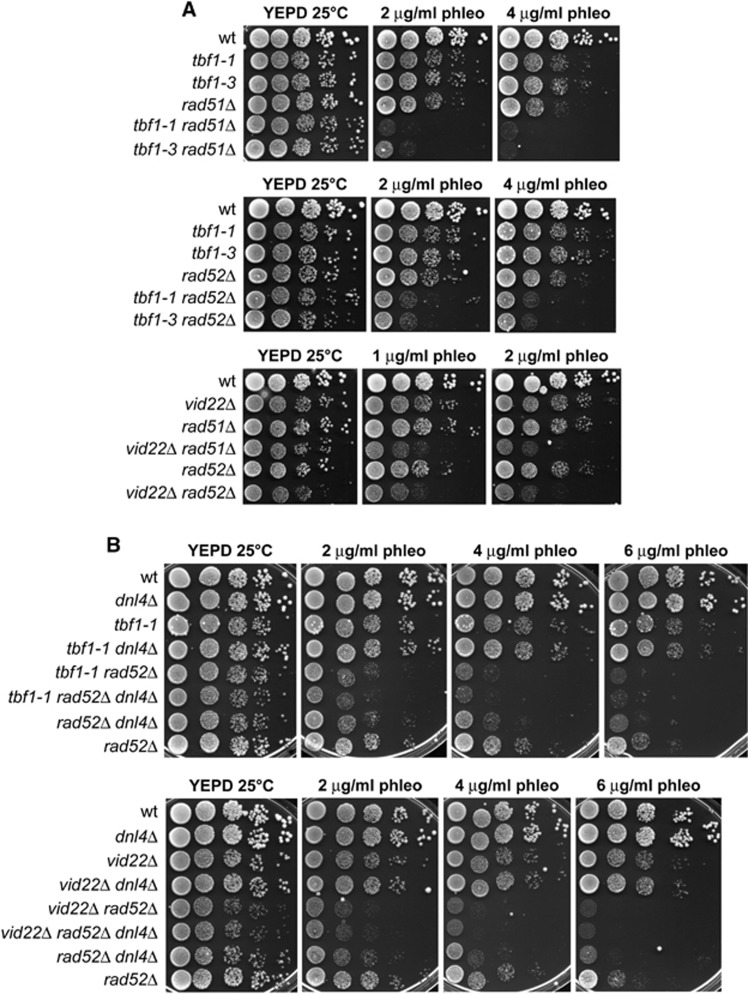
To investigate whether the Tbf1 and Vid22 role in DSB repair was limited to HR, we analysed the genetic interactions between their loss-of-function mutations and deletions of the major HR genes RAD51 and RAD52. As shown in Figure 5A, the tbf1-1, tbf1-3 and vid22Δ alleles exacerbated the sensitivity to phleomycin of rad51Δ and rad52Δ cells, suggesting that Tbf1 and Vid22 have other functions in DSB repair besides promoting HR.
Figure 5.
Functional interactions of the tbf1 and vid22Δ alleles with mutations impairing HR or NHEJ. (A, B) Exponentially growing cultures of strains with the indicated genotypes were serially diluted (1:10) and each dilution was spotted out onto YEPD plates with or without phleomycin at the indicated concentrations. Plates were then incubated at 25°C for 3 days.
Concomitant abolition of the two main DSB repair pathways, NHEJ and HR, leads to synergistic cell sensitivity to DNA damaging agents. For example, cells harbouring deletions of both DNL4 and RAD52 genes, which are essential for NHEJ and HR, respectively, displayed higher sensitivity to phleomycin than each single mutant (Figure 5B). Thus, we deleted DNL4 in tbf1-1 and vid22Δ cells and tested the sensitivity of the resulting double mutants to phleomycin. Deletion of DNL4 did not exacerbate the sensitivity to phleomycin of either tbf1-1 or vid22Δ cells (Figure 5B), which is consistent with the possibility that Dnl4, Tbf1 and Vid22 act in the same genetic pathway. Yet, tbf1-1 rad52Δ dnl4Δ and vid22Δ rad52Δ dnl4Δ triple mutants were as sensitive to phleomycin as tbf1-1 rad52Δ and vid22Δ rad52Δ double mutants, respectively. Therefore, the enhanced sensitivity of tbf1-1 rad52Δ and vid22Δ rad52Δ double mutants compared with rad52Δ single mutant can be explained by the inability of the double mutants to perform NHEJ. Interestingly, the sensitivity to phleomycin of tbf1-1 rad52Δ dnl4Δ and vid22Δ rad52Δ dnl4Δ triple mutants was even higher than that of dnl4Δ rad52Δ cells (Figure 5B), suggesting that the role of Tbf1 and Vid22 in the maintenance of genome integrity is not limited to facilitating resection and NHEJ.
Tbf1 and Vid22 are required for DSB repair by NHEJ
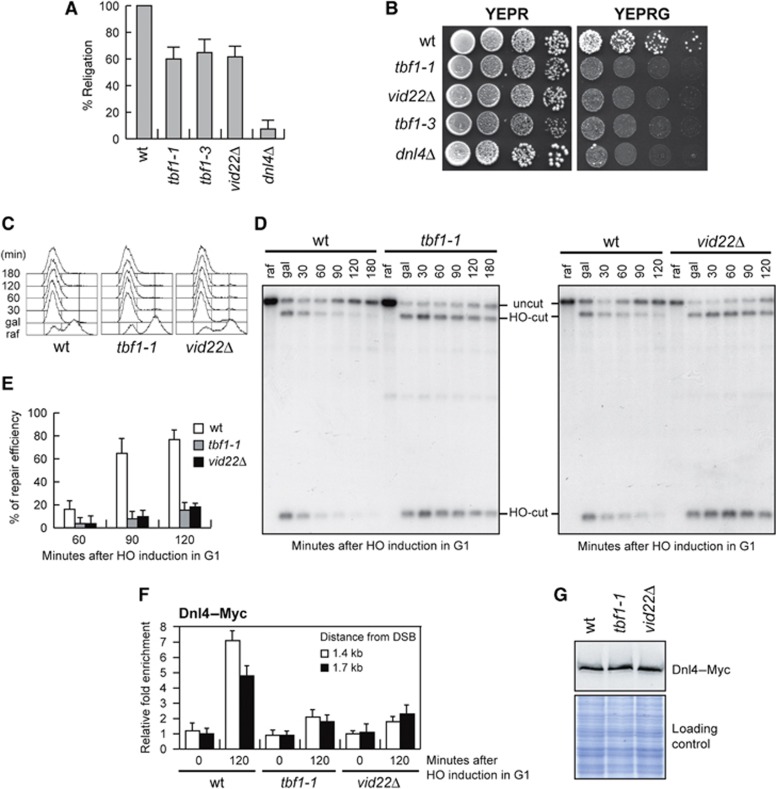
We further investigated whether tbf1-1 and vid22Δ mutant cells were defective in NHEJ-mediated DSB repair by analysing the ability of cells to religate a plasmid that was linearized before being transformed into the cells (Lee et al, 1999). As shown in Figure 6A, tbf1-1, tbf1-3 and vid22Δ mutant cells religated the plasmid less efficiently than wild-type cells, although more efficiently than dnl4Δ cells, which lack the NHEJ enzyme responsible for ligation of the DSB ends.
Figure 6.
Efficient DSB repair by NHEJ requires Tbf1 and Vid22. (A) Plasmid religation assay. The same amount of BamH1-linearized or uncut pRS316 plasmid DNA was transformed into wild-type, tbf1-1, tbf1-3, vid22Δ and dnl4Δ cells, followed by transformant selection for the pRS316 nutritional marker. Data are expressed as percentage of religation relative to wild-type after normalization to the corresponding transformation efficiency of the uncut plasmid. (B) Saturated cultures of strains with the indicated genotypes, all carrying at the MAT locus an HO site, whose galactose-induced cleavage is lethal on galactose unless it is repaired by error-prone NHEJ (see text for details), were serially diluted (1:10) and each dilution was spotted out onto YEPR and YEPRG plates. Plates were then incubated at 25°C for 3 days. (C–E) Exponentially growing YEPR cultures of cells carrying the HO system in Figure 3A were arrested in G1 with α-factor (raf) and transferred to YEPRG in the presence of α-factor. After 30 min (gal), cells were shifted to YEPD medium in the presence of α-factor to allow NHEJ. (C) FACS analysis of DNA content. (D) Southern blot analysis of KpnI-digested genomic DNA using a LEU2 probe as in Figure 3A and B. (E) Densitometric analysis of the uncut band signals (see Materials and methods for details). Plotted values are the mean values±s.d. from three independent experiments as in (D). (F) Exponentially growing YEPR wild-type, tbf1-1 and vid22Δ cell cultures carrying the HO system described in Figure 3A and expressing a fully functional Dnl4–Myc fusion protein were arrested in G1 (time zero) with α-factor and transferred to YEPRG in the presence of α-factor. Relative fold enrichment of Dnl4–Myc at the indicated distance from the HO cleavage site at the LEU2 locus was calculated after ChIP with an anti-Myc antibody. Plotted values are the mean values±s.d. from three independent experiments. (G) Western blot analysis with anti-Myc antibody of protein extracts from exponentially growing wild-type, tbf1-1 and vid22Δ cells. The same amount of protein extracts was subjected to SDS–PAGE and stained with Coomassie blue (loading control).
In addition, we used the GAL-HO strain described in Figure 4A, where HO induction by galactose addition generates at the MAT locus a DSB that cannot be repaired by HR. In fact, this strain lacks the homologous donor sequences HML and HMR (Lee et al, 1998) and therefore it can repair the HO-induced DSB only by NHEJ. Survival of these cells on galactose, which continuously induces HO cleavage, requires an error-prone NHEJ repair that mutates the HO site, thus preventing subsequent rounds of cleavage. As shown in Figure 6B, the viability of tbf1-1, tbf1-3 and vid22Δ cells on galactose-containing plates was lower than that of wild-type cells, further supporting the hypothesis that tbf1-1, tbf1-3 and vid22Δ mutant cells are defective in NHEJ-mediated DSB repair.
To confirm the NHEJ defect in these mutants, we monitored DSB repair by NHEJ at the molecular levels by using the strains previously used to monitor DSB repair by SSA (Figure 3A). HO expression was induced for 30 min by galactose addition and was then rapidly shut off by the addition of glucose, to allow both error-prone and error-free NHEJ-mediated repair of the DSB. To ensure that repair of the HO-induced DSB occurred mainly by NHEJ, HO was induced in G1-arrested cells that were kept arrested in G1 with α-factor throughout the experiment (Figure 6C). In fact, the low Cdk1 activity in G1 cells prevents resection of the HO-induced DSB and therefore its repair by SSA (Aylon et al, 2004; Ira et al, 2004). Strikingly, the 14.5-kb uncut band resulting from NHEJ-mediated ligation reaccumulated less efficiently in G1-arrested tbf1-1 and vid22Δ than in similarly treated wild-type cells (Figure 6D and E), which also showed the expected decrease of both the 2.5- and 12-kb HO-cut band signals due to NHEJ repair events. Thus, Tbf1 and Vid22 are required for NHEJ-mediated DSB repair.
To investigate the NHEJ defect of tbf1 and vid22 mutants at the molecular level, we monitored recruitment at the HO-induced DSB of the NHEJ DNA ligase Dnl4. HO expression was induced by galactose addition to G1-arrested cells that were kept arrested in G1 with α-factor throughout the experiment and expressed fully functional Dnl4–Myc. After ChIP with an anti-Myc antibody, qPCR was designed to monitor Dnl4 association near the repairable HO-induced DSB at LEU2 locus. Both tbf1-1 and vid22Δ G1 cells showed decreased Dnl4 association at the HO-induced DSB compared with similarly treated wild-type cells (Figure 6F). This decreased binding was not due to lower Dnl4 levels in the mutant cells compared with wild-type, as similar Dnl4 amounts could be detected in protein extracts from wild-type, tbf1-1 and vid22 cells (Figure 6G). Thus, we can conclude that Tbf1 and Vid22 facilitate the binding of Dnl4 to DSBs, suggesting that defective Dnl4 recruitment at DSBs might account for the NHEJ defects displayed by tbf1 and vid22Δ mutant cells.
Tbf1 and Vid22 proteins are recruited to DSBs
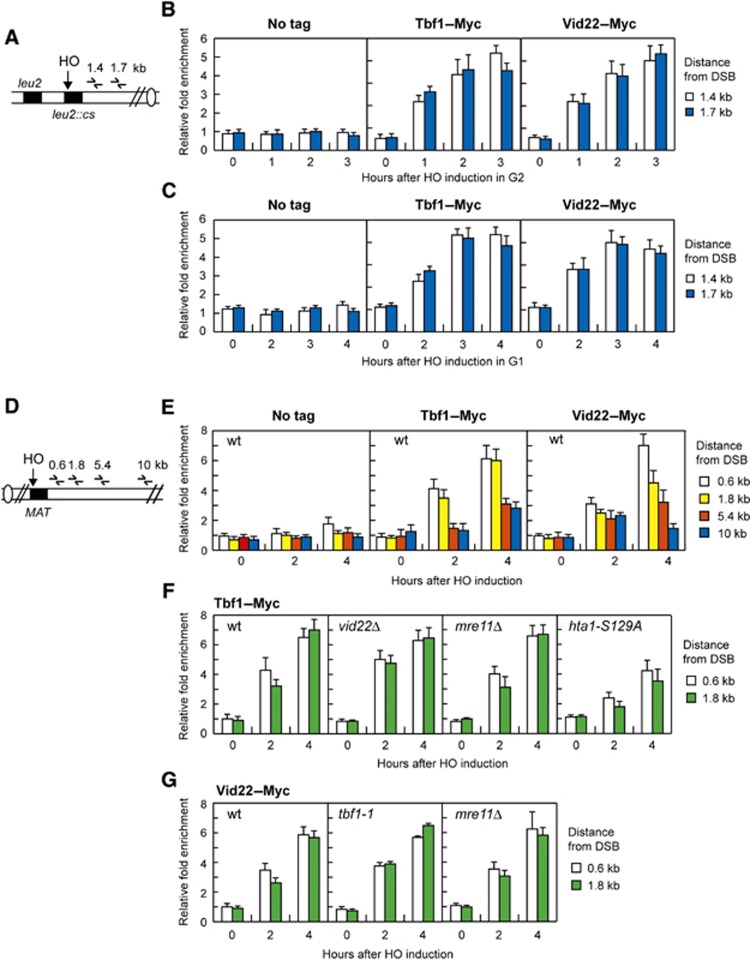
To investigate whether Tbf1 and Vid22 were directly involved in DSB repair, we asked if they were recruited near an HO-induced DSB in strains expressing fully functional Myc-tagged variants of either Tbf1 or Vid22. qPCR was performed to monitor Tbf1 and Vid22 association near the repairable DSB at the LEU2 locus (Figure 7A). HO expression was induced by galactose addition to G2-arrested cells that were kept arrested in G2 with nocodazole throughout the experiment. Following HO induction by galactose addition, Tbf1 and Vid22 were efficiently recruited close to the cut site as early as 1 h after HO induction in G2 (Figure 7B). Association of Tbf1 and Vid22 at DSBs seemed to occur independently of DSB resection, as they were efficiently recruited near the DSB site even in G1-arrested cells (Figure 7C), where DSB resection is inhibited due to the low Cdk1 activity (Aylon et al, 2004; Ira et al, 2004).
Figure 7.
Tbf1 and Vid22 recruitment at chromosomal DSBs. (A) Schematic representation of the qPCR primer sets located at the indicated distance from the HO cleavage site at the LEU2 locus. (B) Exponentially growing YEPR wild-type cells carrying the HO system in (A) and expressing fully functional Tbf1–Myc or Vid22–Myc fusion proteins or not expressing any Myc-tagged protein (no tag) were arrested in G2 (time zero) with nocodazole and transferred to YEPRG in the presence of nocodazole. Relative fold enrichment of Tbf1–Myc or Vid22–Myc at the indicated distance from the HO cleavage site was calculated after ChIP with an anti-Myc antibody. (C) As in (B) but showing relative fold enrichment of Tbf1–Myc or Vid22–Myc fusion proteins in G1-arrested wild-type cells. (D) Schematic representation of the qPCR primer sets located at the indicated distance from the HO cleavage site at the MAT locus. Donor HML and HMR loci are deleted. (E) Exponentially growing YEPR wild-type cell cultures carrying the HO system in (D) and expressing fully functional Tbf1–Myc or Vid22–Myc fusion proteins or not expressing any Myc-tagged protein (no tag) were transferred to YEPRG. Relative fold enrichment of Tbf1–Myc and Vid22–Myc at the indicated distance from the HO cleavage site at the MAT locus was calculated after ChIP with an anti-Myc antibody. (F) As in (E) but showing relative fold enrichment of Tbf1–Myc fusion protein in wild-type, vid22Δ, mre11Δ and hta1-S129A hta2Δ cells. (G) As in (E) but showing relative fold enrichment of Vid22–Myc fusion protein in wild-type, tbf1-1 and mre11Δ cells. In all graphs, data are expressed as relative fold enrichment of Myc-tagged proteins found at the HO-cut site over that found at a non-cleavable locus after normalization of each ChIP signals to the corresponding input for each time point. Plotted values are the mean values±s.d. from three independent experiments.
To analyse the kinetics and the extent of Tbf1 and Vid22 association independently of DSB repair, we monitored their recruitment also at the irreparable HO-induced DSB at the MAT locus (Figure 7D). Both Tbf1 and Vid22 were recruited near this HO-induced DSB and their binding increases over 4 h, spreading to 4–5 kb from the HO cleavage site (Figure 7E). Interestingly, Tbf1 and Vid22 associated to DSBs independently of each other, as recruitment of Tbf1 in vid22Δ cells (Figure 7F) or Vid22 in tbf1-1 cells (Figure 7G) was as efficient as in wild-type cells. Consistent with our finding that Tbf1 and Vid22 association at the DSB does not require generation of ssDNA, deletion of MRE11, which impairs DSB resection (Lee et al, 1998; Tsubouchi and Ogawa, 1998), did not affect the binding of either Tbf1 or Vid22 at the HO-induced DSB (Figure 7F and G).
One of the earliest events in the response to DNA damage is the phosphorylation of histone H2A (γH2A) by either Tel1 or Mec1 (Downs et al, 2000; Shroff et al, 2004). As one of the γH2A functions in DSB repair is to promote the recruitment of chromatin remodelling complexes at DSB ends, we introduced the tbf1-1 allele in an hta1-S129A mutant strain, where the H2A Ser129 was replaced with a non-phosphorylatable alanine residue. Because both the HTA1 and the HTA2 genes encode H2A, the hta1-S129A strain also carried the deletion of HTA2. Interestingly, recruitment of Tbf1 at the DSB site was significantly reduced in hta1-S129A hta2Δ cells compared with wild-type (Figure 7F), indicating that γH2A helps the enrichment of Tbf1 at sites near a DSB.
Tbf1 and Vid22 functionally interact with chromatin remodelers and histone-modifying enzymes
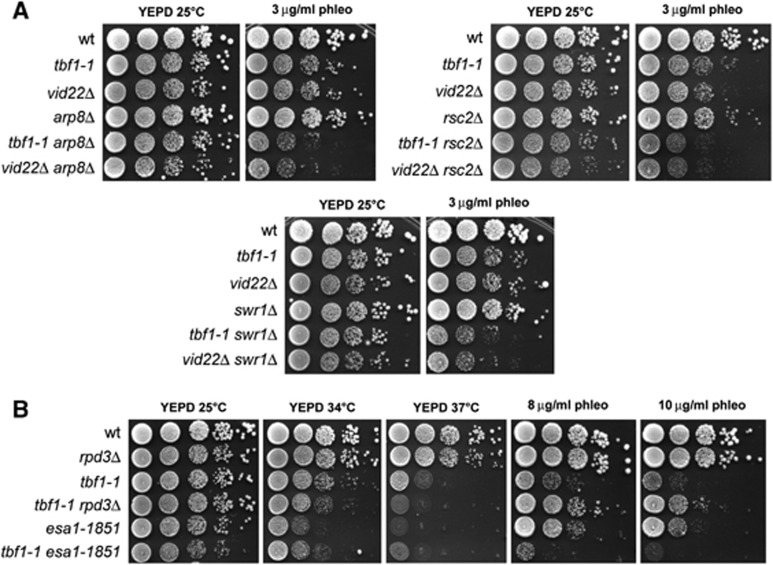
DSBs are accompanied by changes in chromatin organization in the surroundings of the break site that hamper the access to DNA for repair/recombination proteins (Sinha and Peterson, 2009; Soria et al, 2012). Mobilization of nucleosomes flanking the break is regulated by the ATP-dependent remodelling complexes RSC, INO80 and SWR1, as well as by histone-modifying enzymes. As Tbf1 was proposed to counteract nucleosome formation at some promoters (Badis et al, 2008; Preti et al, 2010), we investigated whether Tbf1 and Vid22 might participate in DSB resection and NHEJ by promoting chromatin remodelling at the DSB site. To explore this possibility, we first analysed the possible interactions between their loss of function mutations and deletions of the ARP8, RSC2 and SWR1 genes, which encode for components of the chromatin remodelling complexes INO80, RSC and SWR1, respectively. As shown in Figure 8A, each of these deletions exacerbated the sensitivity to phleomycin of both tbf1-1 and vid22Δ mutant cells, indicating that Tbf1 and Vid22 support cell viability in face of DNA lesions when chromatin remodelling complexes are not fully functional.
Figure 8.
Interactions of the tbf1-1 and vid22Δ alleles with mutations impairing chromatin remodelers and histone-modifiers. (A, B) Exponentially growing cultures of strains with the indicated genotypes were serially diluted (1:10) and each dilution was spotted out onto YEPD plates with or without phleomycin at the indicated concentrations. Unless otherwise indicated, plates were then incubated at 25°C for 3 days.
Among the histone-modifying enzymes, histone acetylation promotes the formation of relaxed chromatin structures by neutralizing the negative charge on lysines, and therefore by decreasing histone–DNA interactions within the nucleosome (Xu and Price, 2011). The acetyltransferase NuA4 complex is important to support DSB repair by both HR and NHEJ (Bird et al, 2002; Tamburini and Tyler, 2005), and its acetylation activity is counteracted by the histone deacetylase Rpd3 (Yang and Seto, 2008). Thus, we investigated the consequences of disabling either the NuA4 catalytic subunit Esa1 or Rpd3 in tbf1-1 mutant cells. Because Esa1 is essential for cell viability, we used the temperature-sensitive esa1-1851 mutation, which is defective in DSB repair (Bird et al, 2002). We found that the esa1-1851 allele exacerbated the sensitivity to phleomycin of tbf1-1 cells, whereas RPD3 deletion suppressed it (Figure 8B). Notably, the lack of RPD3 did not suppress the temperature sensitivity of tbf1-1 cells (Figure 8B). Given that histone acetylation and deacetylation are thought to exert opposite effects in the regulation of chromatin accessibility, these genetic interactions suggest that the DSB repair defects in tbf1 and vid22 mutants may be due to an altered chromatin structure at the damaged sites.
Tbf1 and Vid22 are involved in chromatin remodelling near a DSB
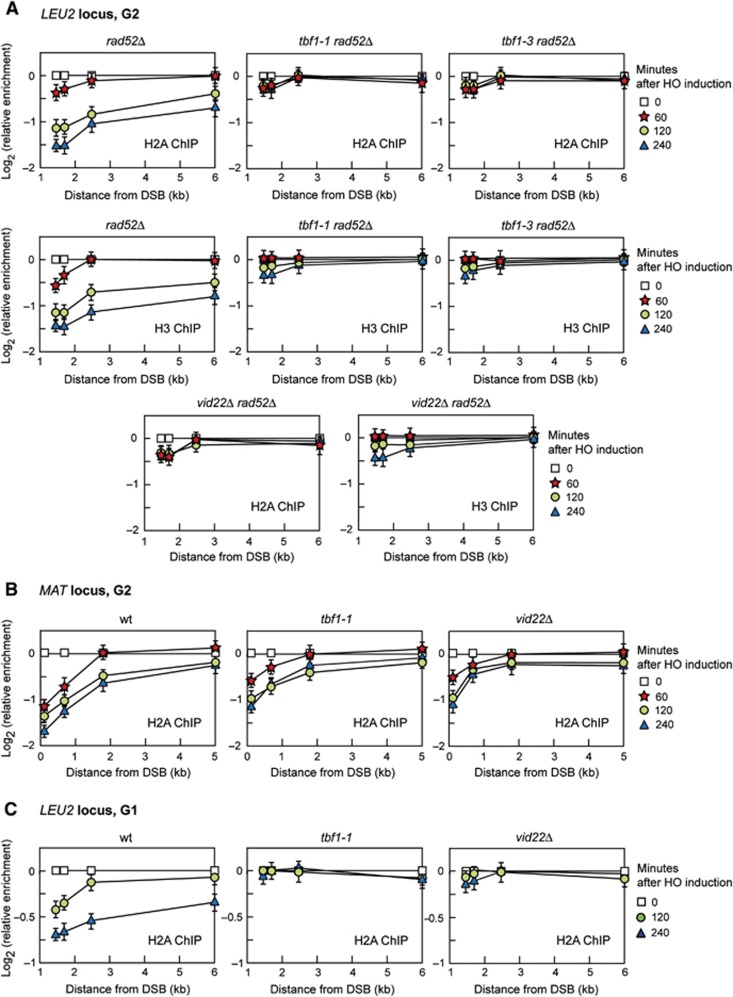
We asked whether the DSB repair defects displayed by tbf1 and vid22Δ cells might be related to the retention of nucleosomes around a DSB by using ChIP analysis and qPCR to evaluate H2A and H3 occupancy centromere-proximal to the HO cleavage site at the LEU2 locus. To exclude that possible differences in histone occupancy were due to different repair kinetics, DSB repair by SSA was abolished in tbf1 and vid22Δ mutant cells by deleting RAD52. HO expression was induced by galactose addition to G2-arrested cells that were kept arrested in G2 with nocodazole for the subsequent 4 h. As resection of the DSB ends leads to loss of input DNA (Chen et al, 2008), the ChIP signals were normalized to the corresponding input for each time point. As shown in Figure 9A, the amount of H2A and H3 bound near the DSB was significantly higher in tbf1-1 rad52Δ, tbf1-3 rad52Δ and vid22Δ rad52Δ than in rad52Δ cells, suggesting that histone removal around a DSB is defective in cells lacking functional Tbf1 or Vid22. Similarly to what was observed for certain chromatin remodelling enzymes (van Attikum et al, 2007), the severity of this defect was locus-dependent, because tbf1-1 and vid22Δ G2 cells still removed H2A centromere-distal to the HO-induced DSB at the MAT locus, although less efficiently than wild-type cells (Figure 9B).
Figure 9.
Tbf1 and Vid22 are involved in histone removal from the surroundings of a DSB. (A) HO expression was induced at time zero by galactose addition to G2-arrested cells carrying the HO system at the LEU2 locus described in Figures 3A and 7A. Cells were kept arrested in G2 by nocodazole throughout the experiment. ChIP was performed with anti-H2A or anti-H3 antibody and DNA was analysed by qPCR using primer pairs located at different distances from the HO cleavage site. In all graphs, data are expressed as relative fold enrichment of H2A or H3 found at the HO-cut site over that found at a non-cleavable locus after normalization of each ChIP signals to the corresponding input for each time point. (B) As in (A) but the HO-induced DSB is at the MAT locus as depicted in Figure 7D. (C) As in (A) but the HO-cut at the LEU2 locus is induced in G1-arrested cells. In all graphs, plotted values are the mean values±s.d. from three independent experiments. All the cell-cycle arrests were verified by FACS analysis (not shown).
DSB resection is inhibited in the G1 phase of the cell cycle, when Cdk1 activity is low, because Cdk1 activity is necessary to resect the break (Aylon et al, 2004; Ira et al, 2004). To assess whether the defects in nucleosome removal displayed by tbf1-1 and vid22Δ cells was not a consequence of their resection defect, we monitored H2A occupancy at the LEU2 DSB in G1-arrested wild-type, tbf1-1 and vid22Δ cells that were kept arrested in G1 with α-factor throughout the experiment. Although the overall H2A loss from the DSB was lower in G1 than in G2, the amount of DSB-bound H2A was higher in G1-arrested tbf1-1 and vid22Δ cells than in similarly treated wild-type cells (Figure 9C). Altogether, these data suggest that Tbf1 and Vid22 participate in nucleosome displacement from a DSB.
Discussion
We provide evidence that the Myb domain protein Tbf1 and its interacting protein Vid22 are new players in the response to DNA DSBs. In fact, loss of function tbf1 and vid22 mutations cause sensitivity to DSB-inducing agents and show strong negative interactions with mutations affecting HR. Furthermore, Tbf1 and Vid22 facilitate the conversion of a DSB end to a 3′ ssDNA overhang, which is necessary to initiate HR. Tbf1 and Vid22 might regulate resection directly, because we observed the recruitment at DSBs of Tbf1 and Vid22, which spread along the DSB end coincident with resection. Interestingly, binding of these proteins near the DSB ends seems to occur independently of ssDNA generation, supporting the idea that they directly interact with nucleosomes to facilitate ssDNA generation at the DSB ends. We also found that Tbf1 and Vid22 are required for efficient NHEJ-mediated religation of the broken ends. While the resection defect caused by Tbf1 and Vid22 dysfunction could be due to a decreased recruitment of Mre11 at the DSBs, a less efficient binding of the NHEJ ligase Dnl4 at the broken ends may account for the NHEJ defects displayed by tbf1 and vid22 mutant cells. Altogether, these observations highlight a broad role for Tbf1 and Vid22 in both HR and NHEJ pathways for DSB repair.
Tbf1 is also essential for cell viability and its essential function is commonly assumed to involve its role as transcriptional regulator (Koering et al, 2000; Preti et al, 2010). On the other hand, no promoters of genes directly implicated in DNA repair/recombination/checkpoint are so far known to be bound by Tbf1 or Vid22 (Preti et al, 2010). Furthermore, tbf1 and vid22Δ cells do not show altered levels of the DSB repair proteins Mre11 and Dnl4, suggesting that the DNA repair defects of tbf1 mutants are not likely due to transcriptional defects. Indeed, not only Tbf1 and Vid22 are recruited to DSBs, but the essential and DSB repair functions of Tbf1 are genetically separable. In fact the tbf1-2 mutant is unable to grow at 37°C, but it does not display sensitivity to DNA damaging agents, whereas the tbf1-3 mutant is hypersensitive to genotoxic treatments without showing detectable growth defects at 37°C. Moreover, the lack of the histone deacetylase Rpd3 suppresses the sensitivity to DSB-inducing agents of tbf1-1, but not the temperature sensitivity of the same mutant. Thus, although we cannot exclude that Tbf1/Vid22-mediated changes in the levels of repair gene transcription might also partially contribute to DSB repair efficiency, our data argue strongly that Tbf1 and Vid22 participate directly in the response to DSBs.
Tbf1 is known to bind T2AG3-repeat sequences located in the subtelomeric regions of most chromosomes (Liu and Tye, 1991; Brigati et al, 1993; Koering et al, 2000). How can a sequence-specific DNA-binding protein be recruited to randomly occurring DSBs? A recent work has shown that the consensus site for Tbf1 binding appears quite flexible in terms of sequence and orientation. In fact, Tbf1 was found to bind a RCCCT consensus sequence at promoters of snoRNAs and of additional 136 genes (Preti et al, 2010). As this sequence can be found at ∼23 000 sites scattered into the S. cerevisiae genome, random DSBs can easily occur in proximity of these sites. In any case, Tbf1–DNA association appears to be regulated, as Tbf1 was found to bind only 197 of these RCCCT sites in the absence of DNA damaging agents (Preti et al, 2010) and Tbf1 recruitment at DSBs is enhanced by γH2A.
Interestingly, it has been recently shown that long arrays of T2AG3 repeats block 5′-end resection and Mre11 localization in a Tbf1-dependent manner (Ribaud et al, 2011), while our data indicate that tbf1 mutants are defective in both Mre11 binding at DSBs and DSB end resection. Furthermore, the insertion of subtelomeric sequences with binding sites for Tbf1 adjacent to a TG81-end inhibits Mre11 association to this end (Fukunaga et al, 2012), suggesting that Tbf1 is involved in telomere protection. However, this capping function of Tbf1 appears to be specific for telomeric ends. In fact, the introduction of the same subtelomeric sequences did not affect Mre11 binding at non-telomeric DNA ends (Fukunaga et al, 2012). Moreover, the artificial tethering of Tbf1 close to an HO cutting site is not sufficient to inhibit Mre11 recruitment to the HO-induced DSB, which requires the co-tethering of the telomeric protein Rap1 (Fukunaga et al, 2012). Finally, Vid22, which acts together with Tbf1 in the DSB response, is not required for the Tbf1-mediated protection of long T2AG3-flanked ends (Ribaud et al, 2011).
Tbf1 has been shown to decrease nucleosome occupancy at some promoters (Badis et al, 2008; Preti et al, 2010). Notably, the sensitivity of tbf1 mutant cells to DSB-inducing agents is exacerbated by the lack of the NuA4 acetyltransferase complex, whereas is suppressed by deletion of the deacetylase Rpd3, which is known to counteract NuA4 activity. As histone acetylation/deacetylation regulates chromatin compaction at DSBs, the above data suggest that alterations in chromatin structure can account for the DSB repair defects caused by Tbf1 or Vid22 dysfunction. Tbf1 and Vid22 might facilitate resection by modulating histone occupancy or by increasing the access to DNA of repair proteins within DSB-associated chromatin. The first possibility is more likely, because we show by quantitative ChIP analysis that H2A and H3 histone removal around the DSB is defective in both tbf1 and vid22 mutant cells, although we cannot exclude that the slower DSB resection in tbf1 and vid22 mutants can contribute to the observed histone occupancy changes.
We also show that Mre11 and Dnl4 association at DSBs is defective in tbf1 mutants, suggesting that these reduced associations might account for the defects in resection and NHEJ displayed by tbf1 and vid22 cells. The finding that Mre11 and Dnl4 association is not completely abolished in tbf1 and vid22 mutants suggests that Tbf1 and Vid22 do not act upstream of MRX and Dnl4. Instead, as Tbf1 has been proposed to serve as boundary factor at telomeres, preventing the propagation of silent chromatin (Fourel et al, 1999, 2001), we propose that Tbf1 and Vid22 might facilitate the persistence of MRX, Dnl4 and other DSB processing enzymes at the site of damage by counteracting the reposition of nucleosomes near a DSB. The maintenance of such a nucleosome-free region around the DSB could promote both the NHEJ repair and the conversion of the DSB to a 3′ ssDNA overhang.
In summary, we have identified Tbf1 and Vid22 as novel regulators of DSB processing in the context of chromatin remodelling and of DDR activation. Moreover, this work highlights the functional importance of two so far poorly characterized proteins in the very early stages of DSB repair, defining Tbf1 and Vid22 as DDR proteins that function to promote genomic stability.
Materials and methods
Search for tbf1 mutants
Genomic DNA from strain YLL2920, carrying the LEU2 gene located 116 bp downstream of the TBF1 stop codon, was used as the template to amplify by PCR under mutagenic conditions a TBF1 region spanning from position −165 to +1990, bp from the TBF1 translation initiation codon. Thirty independent PCR reaction mixtures were prepared, each containing 5 U of EuroTaq DNA polymerase (Euroclone), 10 ng of template genomic DNA, 500 ng each primer, 0.5 mM each dNTP (dATP, dTTP, dCTP), 0.1 mM dGTP, 0.5 mM MnCl2, 10 mM βmercaptoethanol, 10 mM Tris–HCl (pH 9), 50 mM KCl and 1.5 mM MgCl2. The resulting PCR amplification product contained the LEU2 gene flanked by TBF1 sequences spanning from −165 to +1805, bp (including 116 bp of 3′ non-coding sequence) on one side, and from +1806 to +1990, bp (3′ non-coding sequence) on the other side. A wild-type strain was transformed with the PCR products in order to replace the corresponding TBF1 wild-type sequences with the mutagenized PCR products. 3000 transformants were selected on leucine-lacking synthetic complete medium, and then assayed by drop tests for the ability to grow at 37°C on YEPD plates or at 25°C on YEPD plates containing 10 μg/ml phleomycin.
Yeast strains and media
Strain genotypes are listed in Supplementary Table S1. Strains JKM139 and YMV45, used to detect DSB resection and SSA, respectively, were kindly provided by J Haber (Brandeis University, Waltham, USA). Strain tGI354, used to detect crossovers and non-crossovers, was kindly provided by G Liberi (IFOM, Milano, Italy) and J Haber. PCR one-step tagging was used to obtain strains carrying fully functional Myc-tagged MRE11, TBF1, VID22 and DNL4 alleles. To induce a persistent G1 arrest with α-factor, some strains used in this study carried the deletion of the BAR1 gene, which encodes a protease that degrades the α-factor. Gene disruptions were generated by one-step PCR disruption method. Unless otherwise indicated, cells were grown in YEP medium (1% yeast extract, 2% bactopeptone) supplemented with 2% glucose (YEPD), 2% raffinose (YEPR) or 2% raffinose and 3% galactose (YEPRG). All the synchronization experiments have been performed at 25°C.
Plasmid religation assay
The plasmid religation assay was performed as described in Lee et al (1999). The centromeric plasmid pRS316 was digested with the BamHI restriction enzyme before being transformed into the cells. Parallel transformation with undigested pRS316 DNA was used to determine the transformation efficiency. Efficiency of religation was determined by the number of colonies that were able to grow on medium selective for the plasmid marker and was normalized by the transformation efficiency for each sample. The religation efficiency in mutant cells was compared with that of wild-type cells that was set up to 100%.
DSB resection and repair
DSB formation and repair in the YMV45 strain were detected as described in Trovesi et al (2011). DSB end resection at the MAT locus in JKM139 derivative strains was analysed on alkaline agarose gels as described in Clerici et al (2006), by using a single-stranded RNA probe complementary to the unresected DSB strand. Quantitative analysis of DSB resection was performed by calculating the ratio of band intensities for ssDNA and total amount of DSB products. The NHEJ repair efficiency was normalized with respect to the efficiency of DSB formation by subtracting the value of the uncut band at 30 min after HO induction (maximum efficiency of DSB formation) from the values of the same band calculated at the subsequent time points after glucose addition. For each time point, the uncut band has been normalized relative to a loading control.
ChIP analysis
ChIP analysis was performed as described in Viscardi et al (2007). H2A and H3 histones were immunoprecipitated by using anti-H2A and H3 antibodies from Active Motif. Input and immunoprecipitated DNA were purified and analysed by qPCR using a Biorad MiniOpticon. Data are expressed as fold enrichment at the HO-induced DSB over that at the non-cleavable ARO1 locus, after normalization of each ChIP signals to the corresponding input for each time point. Fold enrichment was then normalized to the efficiency of DSB induction. For histone loss, the fold enrichment from each sample after HO induction was divided by the fold enrichment from uninduced cells, and log2 of the resulting values was calculated. Primer sequences are available upon request.
Supplementary Material
Acknowledgments
We thank J Haber, G Liberi for providing yeast strains and J Diffley for anti-Rad53 antibodies. We are grateful to E Sala for preliminary data, and G Dieci for useful discussions. This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC) (Grant IG11407) to MPL, Cofinanziamento 2009 MIUR/Università di Milano-Bicocca to MC and Cofinanziamento 2009 MIUR/Università di Milano-Bicocca to GL. SA was supported by the EU contract PITN-GA-2008-215148 Image DDR.
Author contributions: DB, MC and MPL designed the experiments. DB, SA and MC performed the experiments. DB, GL, MC and MPL analysed the data. GL, MC and MPL wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arnerić M, Lingner J (2007) Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep 8: 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23: 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, Gebbia M, Talukder S, Yang A, Mnaimneh S, Terterov D, Coburn D, Li Yeo A, Yeo ZX, Clarke ND, Lieb JD et al. (2008) A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthiau AS, Yankulov K, Bah A, Revardel E, Luciano P, Wellinger RJ, Géli V, Gilson E (2006) Subtelomeric proteins negatively regulate telomere elongation in budding yeast. EMBO J 25: 846–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E (1996) The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res 24: 1294–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF (2002) Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415 [DOI] [PubMed] [Google Scholar]
- Brevet V, Berthiau AS, Civitelli L, Donini P, Schramke V, Géli V, Ascenzioni F, Gilson E (2003) The number of vertebrate repeats can be regulated at yeast telomeres by Rap1-independent mechanisms. EMBO J 22: 1697–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigati C, Kurtz S, Balderes D, Vidali G, Shore D (1993) An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol Cell Biol 13: 1306–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK (2008) Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, Ira G (2012) The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489: 576–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP (2006) The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep 7: 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell MM, Lo Presti L, Cerutti L, Cano Del Rosario E, Hauser PM, Simanis V (2009) Functional differentiation of tbf1 orthologues in fission and budding yeasts. Eukaryot Cell 8: 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, van Attikum H, Llorente B (2012) The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489: 581–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Lowndes NF, Jackson SP (2000) A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J, Rudin N, Haber JE (1992) Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol 12: 1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Boscheron C, Revardel E, Lebrun E, Hu YF, Simmen KC, Müller K, Li R, Mermod N, Gilson E (2001) An activation-independent role of transcription factors in insulator function. EMBO Rep 2: 124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J 18: 2522–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga K, Hirano Y, Sugimoto K (2012) Subtelomere-binding protein Tbf1 and telomere-binding protein Rap1 collaborate to inhibit localization of the Mre11 complex to DNA ends in budding yeast. Mol Biol Cell 23: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F, Berthiau AS, van Houwe G, Gilson E, Gasser SM (2006) Subtelomeric factors antagonize telomere anchoring and Tel1-independent telomere length regulation. EMBO J 25: 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koering CE, Fourel G, Binet-Brasselet E, Laroche T, Klein F, Gilson E (2000) Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res 28: 2519–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrín-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409 [DOI] [PubMed] [Google Scholar]
- Lee SE, Pâques F, Sylvan J, Haber JE (1999) Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol 9: 767–770 [DOI] [PubMed] [Google Scholar]
- Liu ZP, Tye BK (1991) A yeast protein that binds to vertebrate telomeres and conserved yeast telomeric junctions. Genes Dev 5: 49–59 [DOI] [PubMed] [Google Scholar]
- Longhese MP, Bonetti D, Manfrini N, Clerici M (2010) Mechanisms and regulation of DNA end resection. EMBO J 29: 2864–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt CW, Valente LP, Rhodes D, Simonsson T (2008) Identification and characterization of an essential telomeric repeat binding factor in fission yeast. J Biol Chem 283: 2693–2701 [DOI] [PubMed] [Google Scholar]
- Preti M, Ribeyre C, Pascali C, Bosio MC, Cortelazzi B, Rougemont J, Guarnera E, Naef F, Shore D, Dieci G (2010) The telomere-binding protein Tbf1 demarcates snoRNA gene promoters in Saccharomyces cerevisiae. Mol Cell 38: 614–620 [DOI] [PubMed] [Google Scholar]
- Ribaud V, Ribeyre C, Damay P, Shore D (2011) DNA-end capping by the budding yeast transcription factor and subtelomeric binding protein Tbf1. EMBO J 31: 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saponaro M, Callahan D, Zheng X, Krejci L, Haber JE, Klein HL, Liberi G (2010) Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet 6: e1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M (2004) Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol 14: 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Peterson CL (2009) Chromatin dynamics during repair of chromosomal DNA double-strand breaks. Epigenomics 1: 371–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G (2012) Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell 46: 722–734 [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271 [DOI] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK (2005) Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol 25: 4903–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trovesi C, Falcettoni M, Lucchini G, Clerici M, Longhese MP (2011) Distinct Cdk1 requirements during single-strand annealing, noncrossover, and crossover recombination. PLoS Genet 7: e1002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H (1998) A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol 18: 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM (2007) Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J 26: 4113–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE (2002) Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell 10: 373–385 [DOI] [PubMed] [Google Scholar]
- Viscardi V, Bonetti D, Cartagena-Lirola H, Lucchini G, Longhese MP (2007) MRX-dependent DNA damage response to short telomeres. Mol Biol Cell 18: 3047–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann J, Valeri A, Guillou E, Cuvier O, Nollmann M (2011) Roles of chromatin insulator proteins in higher-order chromatin organization and transcription regulation. Nucleus 2: 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Price BD (2011) Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle 10: 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E (2008) The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 9: 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.