Abstract
The journey of many viruses to infect cells begins when the virus first binds to cell surface heparan sulfate (HS). The initial step of cell attachment or binding during herpes simplex virus type-1 (HSV-1) entry is mediated by envelope glycoprotein B (gB) and C (gC). The binding is followed by fusion between virus envelope and cell membrane during which HSV-1 glycoprotein D (gD) interacts with a modified form of HS know as 3-O-sulfated heparan sulfate (3-OS HS). The rare modification of 3-O-sulfation on HS chain is governed by enzymes known as 3-O-sulfotransferase (3-OST). Currently, there are seven isoforms of human 3-OSTs that have been identified, and with the exception of 3-OST-1, all other 3-OST isoforms allow HSV-1 entry and spread. Recently, the product of the zebrafish (ZF)-encoded 3-OST-3 was also recognized as a gD receptor, which mediates HSV-1 entry and cell-cell fusion similar to human 3-OST-3. Interestingly, the ZF system expresses multiple isoforms of 3-OST which could be very useful for studying the involvement of HS and 3-OS HS in virus tropism and virus-induced inflammation. In addition, therapeutic targeting of 3-OST generated HS is likely to bring about novel interventions against HSV-1. In this review we have taken a closer look at the potential of both human and ZF encoded 3-OSTs as valuable tools in HSV entry and inflammation studies.
Keywords: HSV, viral entry, heparan sulfate, modified form of heparan sulfate.
BIOSYNTHESIS OF HEPARAN SULFATE: A TEAM EFFORT
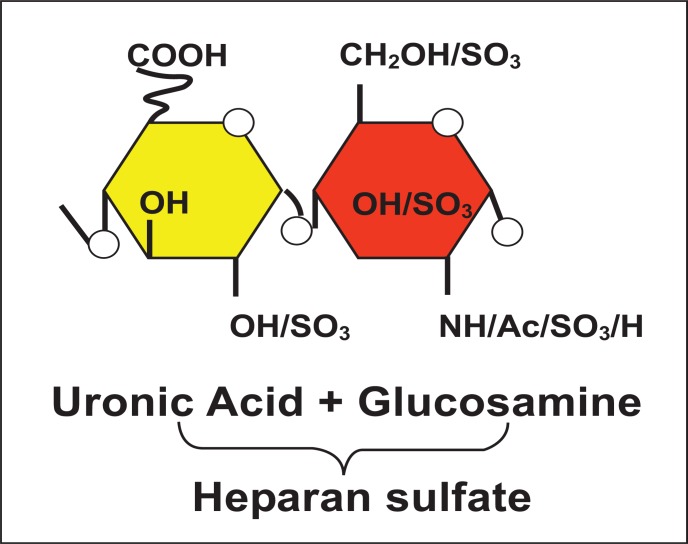
The synthesis of heparan sulfate (HS) is a multistep process which begins with the addition of tetrasaccharide linker region (GlcA-Gal-Gal-Xyl) on a serine residue of the protein core [1, 2]. Once the glucosamine (GlcNAc) residue is added to the HS chain, polymerization begins with the alternating GlcA and GlcNAc residues (Fig. 1). The last step during HS synthesis is the specific and rare modifications [1]. The key players in this process are specific enzymes such as glycosyltransferases, an epimerase, and sulfotrans-ferases. First, glycosyltransferases bring N-deacetylation and N-sulfation of GlcNAc which results in the conversion to N-sulfoglucosamine (GlcNS). This reaction is followed by C5 epimerization of GlcA to IdoA. Finally, O-sulfations are catalyzed by 2 -O- sulfotransferases (2-OST), 6-O-sulfotransferases (6-OST), and 3-O-sulfotransferases (3-OST). Therefore, 2-O-sulfation of IdoA and GlcA is carried by 2-OST, then this modification 6-O-sulfation of GlcNAc and GlcNS units is followed by 6-OST. The last step of HS modification is 3-O- sulfation of GlcN residues [1]. The above modifications results in unique protein binding sites in modified HS chains which gives specificity to both structures and functions including herpes simplex virus (HSV) membrane fusion and penetration [3, 4] (Fig. 2). It has become clear that extensive molecular changes within the HS chain provide functional diversity. The final product of modified HS is a highly sulfated polysaccharide with alternating repeating disaccharide units of uronic acid and D-glucosamine (Fig. 2). On the cell surface HS is expressed as a polysaccharide chain, which are covalently attached to a protein core with the help of a tetrasaccharide linker. Therefore, the fine tuning of HS expands the potential of HS functions in many biological processes such as cell adhesions, cell signaling, embryonic development, wound healing, and angiogenesis [5-9]. Chains of HS have been shown to be a receptor for multiple viruses and pathogens of medical importance [10-13].
HS AND HSV-1: A GROWING RELATIONSHIP
Ubiquitous expression of HS in a majority of the cell-types provides universal docking sites for viruses. The presence of a negative charge on the highly sulfated HS allows binding of specific viral proteins carrying positive charged amino acids [14]. Evidence further suggests that specificities within the saccharide sequences generate potentially high affinity sites for viral binding and penetration [3, 4]. In addition, it is becoming increasingly clear that in order to have efficient viral entry and spread, the presence of fine-tuned HS structure is critical. HS is not only a receptor for HSV-1 but many other medically important viruses including human papillopma virus (HPV), human immunodeficiency virus (HIV), Dengue virus and hepatitis C [15-18] viruses.
HSV-1 is a member of alphaherpesvirus subfamily of herpesviruses [19]. After invading the host cells, the virus establishes latent infection in the peripheral nervous system, which is a hallmark of HSV infection and also a requirement to establish neurotropism. The common symptoms include mucocutaneous lesions that are most apparent around the lip area. Occasionally, the virus can cause ocular infections leading to significant vision loss. More severe life-threatening symptoms of HSV-1 include encephalitis and meningitis [19]. It is possible that primary infections of HSV-1 may remain asymptomatic; however the continued presence of the virus in the neurons results in recurrent symptomatic infections and associated complications. In any case, interaction of HSV-1 with HS represents a first crucial step in pathogenesis and it may be targeted for controlling the complications associated with the infection.
During cell entry, HSV-1 exhibits several key features that are common among many enveloped viruses. The most common among them is the fact that viral envelope glycoprotein(s) trigger fusion with the host cell membrane. HSV-1 also uses its envelope glycoproteins to induce fusion. Another common mechanism is endocytosis. It has been suggested that many enveloped viruses can enter via endocytosis, which is also observed with HSV-1. The virus can also enter via a phagocytic-like uptake in which receptor-mediated endocytosis mediates viral trafficking [13]. In either situation, the virus-cell interactions via HS are critical for successful infection.
HSV-1 was the first virus demonstrated to exploit cell surface HS during attachment [20]. Since then many pieces of evidence have supported its important role in attachment and entry. For instance, soluble heparin competitively binds to HSV-1 glycoproteins, and therefore inhibits viral binding to host cells [21]. In addition, heparinase treatment to the cells significantly reduces viral binding and HSV-1 infection [20]. Finally, the cells that are defective in HS biosynthesis but not in chondrotin sulfate (CS) biosynthesis show reduced level of infectivity [22, 23]. It has also been demonstrated that spinoculation, a low speed centrifugation enhances HSV-1 binding and infection in a mutant cell line defective in HS biosynthesis [24].
The initial interaction of virus with a HS chain on the host cell surface is mediated by HSV-1 envelope glycoprotein’s B (gB) and /or C (gC) [25], This interaction results in HSV-1 glycoprotein D (gD) interacting with one of its receptors to trigger fusion between host and viral envelope. The fusion requires participation of additional envelope glycoprotein’s (gB, gH and gL).
It has been suggested that HSV-1 glycoprotein C (gC) may further enhance viral binding through HS, but it is not essentially required for cell entry [26]. The mutant viral lacking gC is able to bind and infect a cell. However, the mutant HSV lacking both gB and gC binding is significantly reduced compared to gC deficient virus, and the virus is not able to mediate infection [27, 28]. Further experiments have demonstrated that soluble forms of both gB, and gC or gB and gC proteins isolated from virions can bind heparin and HS [28-30]. In addition, soluble gC is able to inhibit HSV-1 binding to cells [28, 31]. The specific amino acid residues in gC involved in HS binding are Arg-143, Arg-145, Arg-147, Thr-150 and Gly-247 [32]. It has been suggested that these arginine residues may interact with 6-O and 2-O-sulfate regions in HS chain [33]. A study further demonstrates that basic and hydrophobic residues localized in gC (Cyst-127-Cys144) is a critical domain for HS binding [34]. Similarly, in gB a short lysine-rich region has been shown critical for HSV-1 attachment [35], but is not critical for fusion during cell entry. Beside gB and gC, a third glycoprotein D (gD) interacts with three receptors of unrelated families to trigger fusion between host and cell membrane [36]. The gD receptors include nectin-1 and nectin-2, which belong to the immunoglobulin (Ig) superfamily [37], while herpesvirus entry mediator (HVEM) is a TNFR family member [38]. The third gD receptor is a modified form of HS which is also known as 3-O-sulfated HS (3-OS HS) [3].
SIGNATURE OF 3-O-SULFATION: MULTIPLE FACES OF HS
The specificity of any given HS-protein interaction is determined largely by placement of sulfate groups along the chain’s length, which allows HS chains to interact with a wide range of functionally diverse proteins, such as growth factors, cytokines, chemokines, proteases, lipases and cell-adhesion molecules. The 3-O-sulfation of HS accounts for 0.5% of the total sulfation in an HS chain and is the final and relatively rare modification of HS. It is also the one that generates unique HSV-1 gD binding activity within HS [3]. Thus, the order and ring position of sulfate substituents creates distinct oligosaccharide sequences and defines the corresponding biological activities. Members of the family of 3-OSTs are responsible for the addition of sulfate groups at the 3-OH position of glucosamine in HS. These highly homologous enzymes are present as several isoforms expressed from different genes at different levels in various tissues and cells. The 3-OSTs modify HS late during biosynthesis, and each isoform recognizes as a substrate glucosamine residues in regions of the HS chains having specific, but different prior modifications, including epimerization and sulfations at other positions. Thus, different 3-OSTs generate different potential protein-binding sites in HS. For example, the 3-OST-1 isoform, but not other isoforms (3-OST-2, 3-OST-3A, and 3-OST-3B), generates antithrombin (AT) binding sites in HS, similarly 3-OST-3 isoforms and not 3-OST-1 isoform can generate HSV-1 gD binding site [39]. It is interesting to mention that a recently discovered isoform, 3-OST-5, can generate both gD receptor and AT binding sites. The modification of 3-O-sulfation in HS is mediated by family members of 3-O-sulfotransferases (3-OSTs) [40]. To date there are seven members of the 3-OSTs: 3-OST-1,-2,-3A, 3B,-4,-5, and 6 [40-43]. The 3-OST-3A and 3-OST-3B are identical in terms of amino acid sequences in sulfotransferase domain and therefore generate similar 3-O-sulfated disaccharides [44, 45]. Interestingly, sulfotransferase domain is conserved among all the 3-OST isoforms except N-terminal and a C-terminal [45]. It is the sulfotransferase domain that generates a unique sequence in each 3-OST isoforms [45, 46]. The 3-OST isoforms select specific saccharide sequences around the modification site [45]. This gives an opportunity to 3-OST to create their own distinct 3-O-sulfated motifs. The unique expression pattern of 3-OSTs in different cells and tissues together with specific functions for each 3-OST isoform make this enzyme a key regulator for diverse HS function
EXPANDING THE POTENTIAL OF 3-OS HS IN HSV-1 INFECTION
To date all the characterized 3-OST isoforms have been shown to generate products that interact with gD and therefore allow HSV-1 entry except 3-OST-1 isoform [40-43]. Our group first provided visual evidence for colocalization of 3-OS HS and HSV-1 gD during virus fusion process using primary culture of human corneal fibroblasts [47]. In addition, the role of 3-OS HS to trigger fusion process, not to anchor HSV-1 to a specific location on the cell surface was confirmed by soluble 3-OS HS [42, 48]. A similar result was presented using soluble forms of nectin-1 and nectin-2 to trigger the entry into HSV resistant cells [49]. The fractions of soluble 3-OS HS generated by 3-OST-3 were shown to trigger HSV-1 entry into resistant cells similar to soluble nectins [50]. Further evidence for the role of 3-OS HS in fusion rather than attachment was demonstrated by using cells that are deficient in glycosaminoglycan (GAG) synthesis. In the experiment GAG cells allowed HSV-1 entry when spinoculated in the presence of 3-OS HS [50]. Therefore soluble 3-OS HS can trigger both cell-cell fusion and entry [48]. HSV-1 spreads in the host via cell-to-cell fusion process. During spread virally infected cells express envelope glycoprotein’s required for the fusion. As a result the infected cells bind to an uninfected neighboring cell which leads to the formation of large multinucleated giant cells or syncytia. 3-OS HS generated by 3-OST-1 cannot trigger entry or cell-to-cell fusion but soluble 3-OS HS generated by 3-OST-3 results in cell-to-cell fusion. The example of 3-OST-3 mediated cell-to-cell fusion is observed during co-culture of infected human corneal fibroblasts with uninfected corneal fibroblasts or during HSV-1 glycoprotein induced cell-to-cell fusion with primary cultures of human corneal fibroblasts [47]. It has been shown that expression of 3-OS HS is required for syncytia formation in human corneal fibroblasts, furthermore recent studies demonstrate that unmodified HS is not required for cell-to-cell fusion to occur [51]. In addition, studies have been done to identify the regions involved in 3-OS HS-gD binding [52]. It has been postulated that there are two possible 3-OS HS binding sites on gD after evaluating the crystal structure of the HSV-1 gD-HVEM [52]. It has been suggested that one region with a positively charged deep surface pocket and one region with the flat surface with numerous amino acid residues are involved in binding steps. The interaction between HSV-1 gD and 3-OS HS is an electrostatic interaction involving positively charged amino acid residues from gD and the negatively charged sulfate groups from 3-OS HS. A demonstration of an octasaacharide developed from HS modified via 3-OST-3 was shown to bind HSV-1 gD [53]. The mutations in amino acid N-terminal region of the gD that had a negative impact on its ability to interact with 3-OS HS are consistent with the proposed 3-OS HS gD binding sites [54].
ZEBRAFISH: AN EMERGING MODEL TO STUDY HOST-PATHOGEN INTERACTION AND INFLAMMATION
In recent years, the Zebrafish (ZF) has become a favorite model organism for biologists studying developmental processes in vertebrates. Its rapid embryonic development, the transparency of its embryos, the large number of offspring together with several other advantages make it ideal for discovering and understanding the genes that regulate embryonic development as well as the physiology of the adult organism. These characteristics are now exploited by researchers to understand host-pathogen interaction at the level of inflammation and innate immune response to infectious disease and, accordingly, there is a growing literature on the use of ZF to model viral disease including HSV [55-60]. While the study of disease processes in the ZF is a powerful tool in its own right, the development of zebrafish models of viral entry and spread is also a first step for their use in screening of inhibitors against HSV. ZF’s competitive advantage over other model systems is optical clarity in a vertebrate embryo amenable to large-scale screening, receptor-specific and small viral entry inhibitor molecules. Therefore, the transparency of ZF embryos and early larvae permits observations to be made in vivo on intact animals, whereas similar procedures in rodents would require surgery or other invasive monitoring techniques. A further advantage of using ZF assays over traditional mammalian models is the short duration of such assays. For example, screens for compounds that are effective in blocking viral entry/replication/egress can be performed in intact ZF in one week [61]. By comparison, in rodents, the assays for viral entry/replication will take a period between 3-5 weeks and the fate of the inhibitor could not be examined in the real-time.
MULTIPLE ISOFORMS OF 3-OS HS IN ZEBRAFISH: A NEW TOOL TO UNDERSTAND VIRUS-CELL INTERACTION
Recently, the expression pattern of multiple isoforms of 3-OSTs and their significance was reported in ZF [62-66]. Cadwallader and Yost (2006) reported in vivo characterization of eight 3-OST family members in ZF with seven genes showing homology to known 3-OST genes in mouse and humans [62]. Interestingly, two ZF genes, 3-OST-3X and 3-OST-3Z, were found highly similar to human 3-OST-3A and 3-OST-3B respectively. They are likely the true homologs since their catalytic domains are near 100% identical. Such a high degree of conservation points to highly conserved functions as well. In addition, it was noted that members of ZF 3-OST family share at least 63% similarity within the catalytic domain to the corresponding human isoform, with the exception of ZF 3-OST-5, which showed only 53% similarity to human 3-OST-5. In terms of ZF 3-OST expression, most family members showed extensive brain expression which was restricted to very specific brain subdivisions. For instance, ZF 3-OST-2 was expressed in developing brain, otic vesicle, and olfactory areas during early ZF development, while 3-OST-3X was observed in neural tube and lateral plate mesoderm [67]. Similarly, ZF 3-OST-6 was expressed at high level in hindbrain with no expression in spinal cord region [68]. Interestingly, structural and cell adhesion properties of ZF HS bearing syndecan proteins were reported to be shared with higher vertebrates [64]. The diversity in the expression of 3-OST family members in a ZF system provided us an opportunity to examine the role of ZF 3-OST-3 in terms of HSV-1 entry. Using CHO-K1 cells that lack endogenous 3-O-sulfation, we demonstrated the role of ZF 3-OST-3 in HSV-1 entry and spread. More direct and visual evidence for HSV-1 entry was demonstrated by using green fluorescent protein (GFP)-tagged HSV-1 (K26GFP) virions infecting ZF 3-OST-3 expressing CHO-K1 cells [69].
Further studies with the characterization of all other members of ZF 3-OST for HSV infection is needed to rule out their role in HSV tropism and gD binding. For example, those ZF 3-OST members, 3-OST-3Z, 3-OST-4 and 3-OST-6, that are highly expressed in eye could contribute during HSV-1 infection in the eye as reported for human 3-OST-3 isoform during ocular HSV infection [70]. Because ZF 3-OSTs are widely expressed in brain and HSV is a neurotropic virus, they are likely to be very important for neuro-pathologies associated with HSV infection. Therefore, use of ZF embryo model to understand in-vivo significance of 3-OS HS and its interaction with gD during HSV-1 entry/spread is innovative and further enhances our ability to understand the critical regions of HS and modified HS involved in HSV pathogenesis. The current murine and rabbit models used to study HSV infection suffer from the limitation of tracking viral trafficking (entry and spread) in real-time. The “optical clarity” in ZF embryos along with the expression of HS and 3-OS HS moieties in the ZF provides an excellent opportunity to study real time events during HSV entry and spread in relation to receptor usage. Furthermore, the advantage of using a ZF model is to test 3-OST isoform receptor-specific inhibitors in short duration of time, which again is not possible with present murine and rabbit models against HSV infection. For instance, HSV-1 entry inhibitors generated against HS and 3-OST modified HS by our group [73, 74] may also turn useful to study HSV-1 induced pathological damages especially during ocular corneal infection or neuronal damages along with recurrent infections in ZF model because HS and 3-OS HS have been widely implicated for their role in assisting HSV-1 entry and spread in both ocular and neuronal cells [70, 73].
CONCLUSION
In conclusion, significant new information generated in the past decade strongly implicates a wider role for HS and modified form of HS (3-O sulfated HS; 3-OS HS) during HSV-1 entry into cells (Table 1). Using both in vitro cell culture and in vivo model system, our previous studies have highlighted the role of 3-OST-3 modified HS during HSV-1 entry and spread [70, 73]. Although 3-OST modified forms of HS are widely expressed in human cells and tissues (Table 2), however some forms of 3-OSTs such as 3-OST-2 and 3-OST-4 are exclusively expressed in human (Table 2) and ZF brain [62] which opens an important questions regarding the role of 3-OST receptors in HSV-1 neurotropism.
Table 1.
The Heparan Sulfate D-Glucosaminyl 3-O-Sulfotransferases (3-OSTs): Tissue Distribution and Functions
| Isoforms | Sulfated Residue | Upstream Uronic acid | Tissue Distribution | Known Function(s) |
|---|---|---|---|---|
| 3-OST-1 | GlcNS | GlcUA | Heart, brain, lung, kidney | AT binding |
| 3-OST-2 | GlcNS | IdoUA 2S/GlcA 2S | Brain | HSV-1 entry |
| 3-OST-3A | GlcNS | IdoUA 2S | Heart, placenta, lung, liver, kidney | HSV-1 entry |
| 3-OST-3B | GlcNS | IdoUA 2S | Heart, brain, placenta, liver, kidney, pancreas | HSV-1 entry |
| 3-OST-4 | Unknown | Unknown | Brain | HSV-1 entry |
| 3-OST-5 | GlcNS | IdoUA 2S/GlcUA | Skeletal muscle, fetal brain | HSV-1 entry, AT binding |
| 3-OST-6 | GlcNS | IdoUA 2S | Kidney, liver | HSV-1 entry |
Abbreviations: GlcNS: N-acetlyglucosamine; GlcUA: Glucornic acid; IdoUA: Iduronic acid; AT: antithrombin.
Table 2.
Development on the Role of Heparan Sulfate (HS) and Modified HS in HSV-1 Entry
| Year | Chronological Development on the Role of HSPGs (Heparan Sulfate Proteoglycans) in HSV-1 Entry and Spread |
|---|---|
| 1992 | HSV-1 uses cell surface HS for cell binding/attachment [23] |
| 1999 | 3-OST-3 modifications of HS generates HSV-1 gD receptor [3] |
| 2002 | 3-OST-5 modified HS allows HSV-1 entry and spread [40] |
| 2004 | 3-OST-6 modified HS is a receptor for HSV-1 entry and spread [43] |
| 2005 | 3-OST-4 modified HS is a receptor for HSV-1 entry and spread [42] |
| 2005 | 3-OST-2 modified HS is a receptor for HSV-1 entry and spread [42] |
| 2006 | Clinical significance of 3-OST-3 in HSV-1 infection to human corneal fibroblasts [70] |
| 2006 | Role of HS in HSV-1 surfing on filopodia [71] |
| 2010 | Role of syndecans in HSV-1 infection [72] |
| 2011 | Zebrafish encoded 3-OST-3 also function as HSV-1 entry receptor [69] |
| 2011 | In vivo significance of 3-OST-3 receptor during corneal HSV-1 infection [73] |
| 2012 | Role of anti-3-OS HS peptides targeting HSV-2 entry [74] |
Our ability to understand the structural features of HS required for viral infection is hampered due to complex nature of HS structure. However, recent expanding roles of HS and 3-O sulfated HS and its association with viral fusion mechanism in primary cultures of human corneal fibroblasts during entry and spread, viral surfing on filopodia during entry and in vivo significance during corneal HSV-1 infection has laid a strong new possibility that specificity within HS chains may be crucial for its function as a pathogen receptor [71, 72-74]. More structure-function analysis of 3-OST isoforms will be needed to properly understand the true significance of HS and 3-OS HS in the entry process. Since HS and 3-OS HS are associated with many aspects of HSV-1 infection [4, 13], molecules that interfere with HS and 3-OS HS have considerable potential as novel anti-HSV agents. In this regard our group has generated phage display derived anti-HS and anti-3-OS HS peptides that showed promising results both in vitro and in vivo model of HSV infections [73-74]. In parallel, ZF is already emerging as an important tool to study inflammation and innate immune response against infectious diseases [60]. One recent report highlights the usage of ZF as a potential model organism for drug testing against hepatitis C virus [75]. Therefore, significant future work will be needed to establish the critical structure-function understanding of HS generated by ZF and human forms of 3-OSTs that not only allows HSV-1 entry and spread but also plays potentially significant roles in cells and tissue damage via inflammation.
Fig. (1).
Back bone of cell surface heparan sulfate (HS): Shown is the representative of disaccharide unit consisting of a glucuronic acid (GlcA) and N-acetylated glucosamine (GlcNAc) residue.
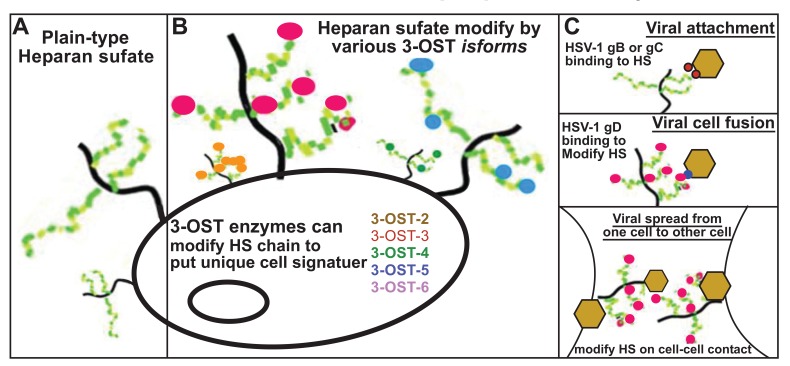
Fig. (2).
Panels A-B showing types of heparan sulfate (HS) on cell surface: (Panel A) plain vanilla-type of unmodified HS and (Panel B) modified form of HS. The modified form of HS is represented by 3-O-sulfation by 3-OST-2, 3, 4, 5 and 6 isoforms. (Panel C): Multiple roles of HS during HSV-1 pathogenesis. This includes initial binding of HSV-1 glycoprotein B (gB) and C (gC) to plain-vanilla-type HS (upper panel), viral cell fusion in which modified from of HS, 3-OS HS, interacts with HSV-1 gD to mediate this process (middle panel) and finally both HS and modified HS are involved in viral spread from cell-to-cell in which four HSV-1 glycoprotein’s: gB, gD, gH and gL are involved (lower panel).
ACKNOWLEDGEMENTS
The work presented in this review article was supported from NIH grants to DS (AI057860, AI081869) and VT (NIH 1R15 AI 088429-01A1). JB was supported by Midwestern University (Downers Grove, IL) sponsored Kenneth A. Suarez Summer Research Fellowship (10-2014-8172).
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1. Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001; 108 (2):169–73. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lindahl U, Kusche-Gullberg M, Kjellén L. Regulated diversity of heparan sulfate. J Biol Chem. 1998; 273 (39 ):24979–82. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 3. Shukla D, Liu J, Blaiklock P, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999; 99 (1 ):13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 4. Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001; 108(4 ):503–10. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iozzo RV, San Antonio JD. Heparan sulfate proteoglycans: heavy hitters in the angiogenesis arena. J Clin Invest. 2001; 108(3 ):349–55. doi: 10.1172/JCI13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002; 41(3 ):391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 7. Stringer SE. The role of heparan sulphate proteoglycans in angiogenesis. Biochem Soc Trans. 2006; 34(Pt 3 ):451–3. doi: 10.1042/BST0340451. [DOI] [PubMed] [Google Scholar]
- 8. Parish CR. The role of heparan sulphate in inflammation. Nature Rev Immunol. 2006; 6:633–43. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- 9. Gorsi B, Stringer SE. Tinkering with heparan sulfate sulfation to steer development. Trends Cell Biol. 2007; 17(4 ):173–7. doi: 10.1016/j.tcb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 10. Liu J, Thorp SC. Cell surface heparan sulfate and its roles in assisting viral infections. Med Res Rev. 2002; 22(1 ):1–25. doi: 10.1002/med.1026. [DOI] [PubMed] [Google Scholar]
- 11. O'Donnell CD, Shukla D. The Importance of Heparan Sulfate in Herpesvirus Infection. Virol Sin. 2008; 23(6 ):383–93. doi: 10.1007/s12250-008-2992-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aquino RS, Lee ES, Park PW. Diverse functions of glycosaminoglycans in infectious diseases. Prog Mol Biol Transl Sci. 2010; 93:373–94. doi: 10.1016/S1877-1173(10)93016-0. [DOI] [PubMed] [Google Scholar]
- 13. Tiwari V, Maus E, Sigar IM, Ramsey KH, Shukla D. Role of heparan sulfate in sexually transmitted infections. Glycobiology . 2012 doi: 10.1093/glycob/cws106. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trybala E, Liljeqvist JA, Svennerholm B, et al. Herpes simplex virus types 1 and 2 differ in their interaction with heparan sulfate. J Virol. 2000; 74 (19 ):9106–14. doi: 10.1128/jvi.74.19.9106-9114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barth H, Schafer C, Adah MI, et al. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003; 278 (42 ):41003–12. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 16. Chen Y, Maguire T, Hileman RE, et al. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997; 3 (8 ):866–71. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 17. Giroglou T, Florin L, Schafer F, et al. Human papillomavirus infection requires cell surface heparan sulfate. J Virol. 2001; 75 (3 ):1565–70. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tyagi M, Rusnati M, Presta M, et al. Internalization of HIV-1 Tat requires cell surface heparan sulfate proteoglycans. J Biol Chem . 2001; 276 (5 ):3254–61. doi: 10.1074/jbc.M006701200. [DOI] [PubMed] [Google Scholar]
- 19. Corey L, Spear PG. Infections with herpes simplex viruses. N Engl J Med. 1986; 314 (11 ):686–91. doi: 10.1056/NEJM198603133141105. [DOI] [PubMed] [Google Scholar]
- 20. WuDunn D, Spear PG. Initial interaction of herpes simplex virus with cells is binding to heparan sulphate. J Virol. 1989; 63(1 ):52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nahmias AJ, Kibrick S. Inhibitory effect of heparin on herpes simplex virus. J Bacteriol. 1964; 87(5 ):1060–66. doi: 10.1128/jb.87.5.1060-1066.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gruenheid S, Gatzke L, Meadows H, et al. Herpes simplex virus infection and propagation in a mouse L cell mutant lacking heparan sulfate proteoglycans. J Virol. 1993; 67(1 ):93–100. doi: 10.1128/jvi.67.1.93-100.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shieh MT, WuDunn D, Montgomery RI, et al. Cell surface receptors for herpes simplex virus are heparan sulphate proteoglycans. J Cell Biol. 1992; 116(5 ):1273–81. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Scanlan PM, Tiwari V, Bommireddy S, et al. Spinoculation of heparan sulfate deficient cells enhances HSV-1 entry, but does not abolish the need for essential glycoproteins in viral fusion. J Virol Methods. 2005; 128(1-2 ):104–12. doi: 10.1016/j.jviromet.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 25. Spear PG, Shieh MT, Herold BC, et al. Heparan sulfate glycosaminoglycans as primary cell surface receptors for herpes simplex virus. Adv Exp Med Biol. 1992; 313:341–53. doi: 10.1007/978-1-4899-2444-5_33. [DOI] [PubMed] [Google Scholar]
- 26. Spear PG, Longnecker R. Herpesvirus entry: an update. J Virol . 2003; 77 (19 ):10179–85. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herold BC, Visalli RJ, Susmarski N, et al. Glycoprotein Cindependent binding of herpes simplex virus to cells requires cell surface heparan sulphate and glycoprotein B. J Gen Virol. 1994; 75(Pt 6 ):1211–22. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 28. Herold BC, WuDunn D, Soltys N, et al. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991; 65(3 ):1090–98. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tal-Singer R, Peng C, Ponce De Leon M, et al. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J Virol. 1995; 69(7 ):4471–83. doi: 10.1128/jvi.69.7.4471-4483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trybala E, Liljeqvist JA, Svennerholm B, et al. Herpes simplex virus types 1 and 2 differ in their interaction with heparan sulfate. J Virol. 2000; 74(19 ):9106–14. doi: 10.1128/jvi.74.19.9106-9114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Svennerholm B, Jeansson S, Vahlne A. Involvement of glycoprotein C (gC) in adsorption of herpes simplex virus type 1 (HSV-1) to the cell. Arch Virol. 1991; 120(3-4 ):273–9. doi: 10.1007/BF01310482. [DOI] [PubMed] [Google Scholar]
- 32. Trybala E, Bergstrom T, Svennerholm B, et al. Localization of a functional site on herpes simplex virus type 1 glycoprotein C involved in binding to cell surface heparan sulphate. J Gen Virol . 1994; 75(Pt 4 ):743–52. doi: 10.1099/0022-1317-75-4-743. [DOI] [PubMed] [Google Scholar]
- 33. Feyzi E, Trybala E, Bergstrom T, et al. Structural requirement of heparan sulphate for interaction with herpes simplex virus type 1 virions and isolated glycoprotein C. J Biol Chem. 1997; 272(40 ):24850–7. doi: 10.1074/jbc.272.40.24850. [DOI] [PubMed] [Google Scholar]
- 34. Mardberg K, Trybala E, Glorioso JC, et al. Mutational analysis of the major heparan sulfate-binding domain of herpes simplex virus type 1 glycoprotein C. J Gen Virol. 2001; 82(Pt 8 ):1941–50. doi: 10.1099/0022-1317-82-8-1941. [DOI] [PubMed] [Google Scholar]
- 35. Laquerre S, Argnani R, Anderson DB, et al. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J Virol. 1998; 72(7 ):6119–30. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Campadelli-Fiume G, Cocchi F, Menotti L, et al. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev Med Virol. 2000; 10(5 ):305–19. doi: 10.1002/1099-1654(200009/10)10:5<305::aid-rmv286>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 37. Geraghty RJ, Krummenacher C, Cohen GH, et al. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998; 280(5369 ):1618–20. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 38. Montgomery RI, Warner MS, Lum BJ, et al. Herpes simplex virus- 1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996; 87(3 ):427–36. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 39. O'Donnell CD, Shukla D. A novel function of heparan sulfate in the regulation of cell-cell fusion. J Biol Chem. 2009; 284(43 ):29654–65. doi: 10.1074/jbc.M109.037960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xia G, Chen J, Tiwari V, et al. Heparan sulfate 3-Osulfotransferase isoform 5 generates both an anti-thrombin-binding site and an entry receptor for herpes simplex virus, type 1. J Biol Chem. 2002; 277(40 ):37912–19. doi: 10.1074/jbc.M204209200. [DOI] [PubMed] [Google Scholar]
- 41. O’Donnell CD, Tiwari V, Oh MJ, et al. A role for heparan sulfate 3-O sulfotransferase isoform 2 in herpes simplex virus type 1 entry and spread. Virology. 2006; 346(2 ):452–9. doi: 10.1016/j.virol.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 42. Tiwari V, O’Donnell CD, Oh MJ, et al. A role for 3-Osulfotransferase isoform-4 in assisting HSV-1 entry and spread. Biochem Biophys Res Commun. 2005; 338(2 ):930–7. doi: 10.1016/j.bbrc.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 43. Xu D, Tiwari V, Xia G, et al. Characterization of heparan sulphate 3-O sulphotransferase isoform 6 and its role in assisting the entry of herpes simplex virus type 1. Biochem J. 2005; 385(Pt 2 ):451–9. doi: 10.1042/BJ20040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu J, Shworak NW, Sinay P, et al. Expression of heparan sulphate D glucosaminyl 3-O sulphotransferase isoforms reveals novel substrate specificities. J Biol Chem. 1999; 274(8 ):5185–92. doi: 10.1074/jbc.274.8.5185. [DOI] [PubMed] [Google Scholar]
- 45. Shworak NW, Liu J, Petros LM, et al. Multiple isoforms of heparan sulfate D glucosaminyl 3-O-sulfotransferase. Isolation, characterization, and expression of human cDNAs and identification of distinct genomic loci. J Biol Chem. 1999; 274(8 ):5170–84. doi: 10.1074/jbc.274.8.5170. [DOI] [PubMed] [Google Scholar]
- 46. Yabe T, Shukla D, Spear PG, et al. Portable sulphotransferase domain determines sequence specificity of heparan sulphate 3-Osulphotransferases. Biochem J. 2001; 359(Pt 1 ):235–41. doi: 10.1042/0264-6021:3590235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tiwari V, ten Dam GB, Yue BY, et al. Role of 3-O-sulfated heparan sulfate in virus-induced polykaryocyte formation. FEBS Lett. 2007; 581(2-3 ):4468–72. doi: 10.1016/j.febslet.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tiwari V, Clement C, Duncan MB, et al. A role for 3-O-sulphated heparan sulphate in cell fusion induced by herpes simplex virus type 1. J Gen Virol. 2004; 85(Pt 4 ):805–9. doi: 10.1099/vir.0.19641-0. [DOI] [PubMed] [Google Scholar]
- 49. Kwon H, Bai Q, Baek HJ, et al. Soluble V domain of Nectin- 1/HveC enables entry of herpes simplex virus type 1 (HSV-1) into HSV-resistant cells by binding to viral glycoprotein D. J Virol . 2006; 80(1 ):138–48. doi: 10.1128/JVI.80.1.138-148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tiwari V, O’Donnell C, Copeland RJ, et al. Soluble 3-O-sulfated heparan sulfate can trigger herpes simplex virus type 1 entry into resistant Chinese hamster ovary (CHO-K1) cells. J Gen Virol. 2007; 88:1075–9. doi: 10.1099/vir.0.82476-0. [DOI] [PubMed] [Google Scholar]
- 51. Pertel PE, Fridberg A, Parish ML, et al. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology. 2001; 279(1 ):313–24. doi: 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- 52. Carfi A, Willis SH, Whitbeck JC, et al. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell. 2001; 8(1 ):169–79. doi: 10.1016/s1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 53. Liu J, Shriver Z, Pope RM, et al. Characterization of a heparan sulphate octasaccharide that binds to herpes simplex viral type 1 glycoprotein D. J Biol Chem. 2002; 277(36 ):33456–67. doi: 10.1074/jbc.M202034200. [DOI] [PubMed] [Google Scholar]
- 54. Yoon M, Zago A, Shukla D, et al. Mutations in the N-termini of herpes simplex virus type 1 and 2 gDs alter functional interactions with the entry/fusion receptors HVEM, Nectin-2, and 3-Osulphated heparan sulphate but not with Nectin-1. J Virol. 2003; 77(17 ):9221–31. doi: 10.1128/JVI.77.17.9221-9231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Burgos JS, Ripoll-Gomez, et al. Zebrafish as a new model for herpes simplex virus type 1 infection. Zebrafish. 2008; 5:323–33. doi: 10.1089/zeb.2008.0552. [DOI] [PubMed] [Google Scholar]
- 56. Liu X, Collodi P. Novel form of fibronectin from zebrafish mediates infectious hematopoietic necrosis virus infection. J Virol . 2002; 76:492–98. doi: 10.1128/JVI.76.2.492-498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Phelan PE, et al. Pressley Characterization of snakehead rhadbovirus infection in zebrafish (Danio rerio) J Virol. 2005; 79:1842–52. doi: 10.1128/JVI.79.3.1842-1852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu X, et al. Zhang A zebrafish (Danio rerio) model of infectious spleen and kidney necrosis virus (ISKNV) infection. Virology . 2008; 376:1–12. doi: 10.1016/j.virol.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 59. Biacchesi S, LeBerre M, Lamoureux A, et al. MAVS plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J Virol. 2009; 83(16 ):7815–27. doi: 10.1128/JVI.00404-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sullivan C, Kim CH. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 2008; 25:341–50. doi: 10.1016/j.fsi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 61. Ding CB, Zhang JP, Zhao Y, Peng ZG, Song DQ, Jiang JD. Zebrafish as a potential model organism for drug test against hepatitis C virus. PLoS One. 2011; 6(8 ):e22921. doi: 10.1371/journal.pone.0022921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cadwallander AB, Yost HJ. Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: I. The 3-Osulfotransferase family. Dev Dyn. 2006; 235(12 ):3423–31. doi: 10.1002/dvdy.20991. [DOI] [PubMed] [Google Scholar]
- 63. Samson SC, Ferrer T, Tristani-Firouzi M. 3-O-sulfotransferase is required for cardiac development and phyisology in zebrafish. Dev Biol. 2008; 319:599–610. [Google Scholar]
- 64. Whiteford JR, Ko S, Lee W, Couchman JR. Structural and cell adhesion properties of zebrafish syndecan-4 are shared with higher vertebrates. J Biol Chem. 2008; 283:29322–30. doi: 10.1074/jbc.M803505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bink RJ, Habuchi H, Lele Z, et al. Heparan sulfate 6-Osulfotransferae is essential for muscle dvelopment in zebrafish. J Biol Chem. 2003; 278:3118–27. doi: 10.1074/jbc.M213124200. [DOI] [PubMed] [Google Scholar]
- 66. Liu I-H, Zhang C, Kim MJ, Cole GJ. Retina development in zebrafish requires the heparan sulfate proteoglycan agrain. Dev Neuro. 2008:877–98. doi: 10.1002/dneu.20625. [DOI] [PubMed] [Google Scholar]
- 67. Cadwallader AB, Yost HJ. Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: III. 2-O-sulfotransferase and C5-epimerases. Dev Dyn. 2007; 236(2 ):581–6. doi: 10.1002/dvdy.21051. [DOI] [PubMed] [Google Scholar]
- 68. Cadwallader AB, Yost HJ. Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: II. The 6-Osulfotransferase family. Dev Dyn. 2006; 235(12 ):3432–7. doi: 10.1002/dvdy.20990. [DOI] [PubMed] [Google Scholar]
- 69. Hubbard S, Darmani NA, Thrush GR, et al. Zebrafish-encoded 3- O-sulfotransferase-3 isoform mediates herpes simplex virus type 1 entry and spread. Zebrafish. 2010; 7(2 ):181–7. doi: 10.1089/zeb.2009.0621. [DOI] [PubMed] [Google Scholar]
- 70. Tiwari V, Clement C, Xu D, et al. Role for 3-O-sulfated heparan sulfate as the receptor for herpes simplex virus type 1 entry into primary human corneal fibroblasts. J Virol. 2006; 80(18 ):8970–80. doi: 10.1128/JVI.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Oh MJ, Akhtar J, Desai P, Shukla D. A role for heparan sulfate in viral surfing. Biochem Biophys Res Commun. 2010; 391(1 ):176–81. doi: 10.1016/j.bbrc.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bacsa S, Karasneh G, Dosa S, Liu J, Valyi-Nagy T, Shukla D. Syndecan-1 and syndecan-2 play key roles in herpes simplex virus type-1 infection. J Gen Virol. 2011; 92(Pt 4 ):733–43. doi: 10.1099/vir.0.027052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tiwari V, Liu J, Valyi-Nagy T, Shukla D. Anti-heparan sulfate peptides that block herpes simplex virus infection in vivo. J Biol Chem. 2011; 286(28 ):25406–15. doi: 10.1074/jbc.M110.201103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ali MM, Karasneh GA, Jarding MJ, Tiwari V, Shukla D. A 3-Osulfated heparan sulfate binding peptide preferentially targets herpes simplex virus 2-infected cells. J Virol. 2012; 86(12 ):6434–43. doi: 10.1128/JVI.00433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ding CB, Zhang J-P, Zhao Y, Peng Z-G, Song D-Q, Jiang J-D. Zebrafish as a potential model organism for drug test against hepatitis C virus. Plos One. 2011; 6(8 ):e22921. doi: 10.1371/journal.pone.0022921. [DOI] [PMC free article] [PubMed] [Google Scholar]