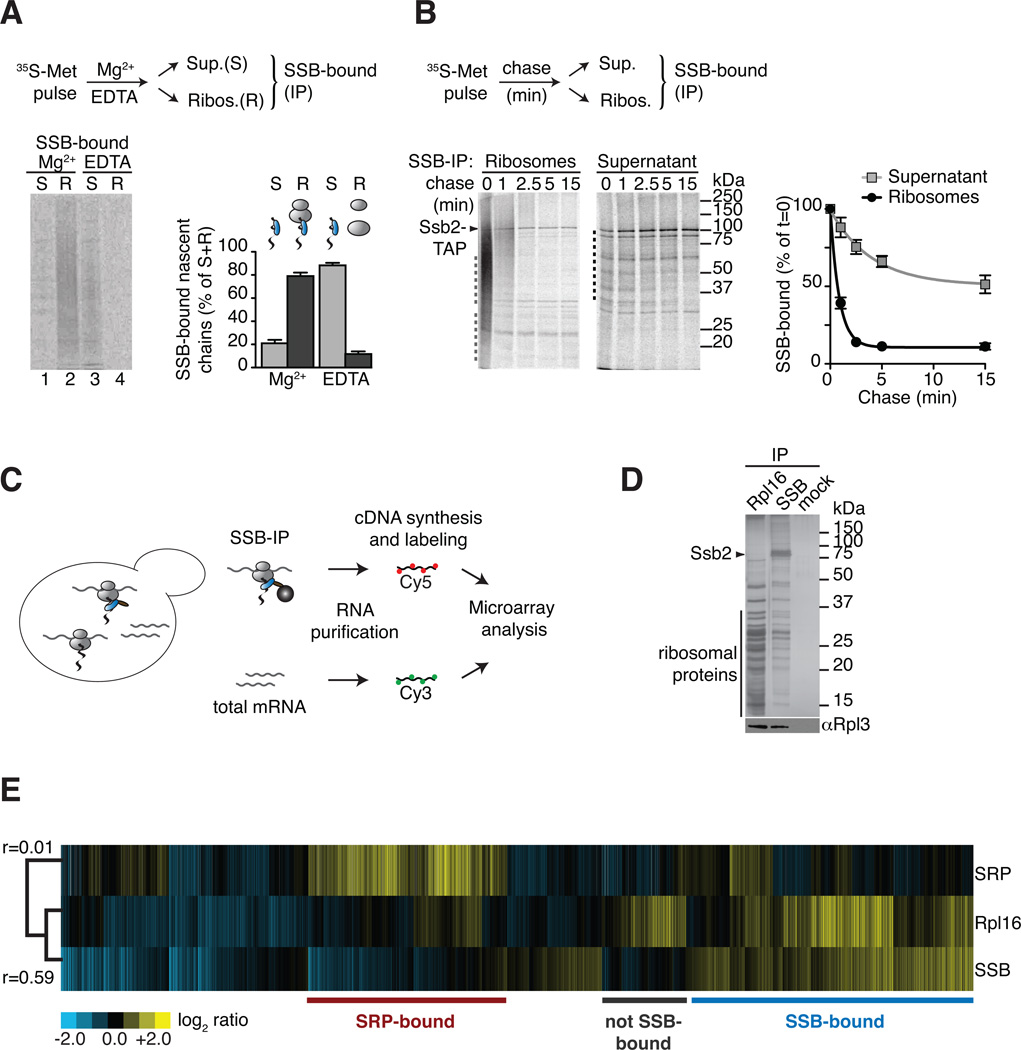
Figure 1. Global identification of physiological SSB-associated nascent polypeptides.
(A) SSB directly binds nascent polypeptides. Nascent polypeptides were pulse labeled with 35S-methionine (35S-Met), ribosome-nascent chain complexes (RNC) were either stabilized (Mg2+) or dissociated using 25 mM EDTA (EDTA) and fractionated by centrifugation in ribosome-bound (R) and soluble (S). SSB-nascent chain interactions in each fraction were determined after Ssb2-TAP immunopurification (IP), SDS-PAGE and autoradiography, followed by quantification of SSB-bound labeled nascent chains (mean ± SEM, n=3). (B) Kinetics of newly translated polypeptide flux through SSB. Nascent polypeptides were 35S-labeled and chased with cold methionine, samples at indicated time points were processed as in (A). Quantification of SSB-bound polypeptides in ribosome and soluble fractions reflects their co-/post-translational flux through SSB (mean ± SEM, n=4). For totals of (A) and (B) see Figure S1. (C) Scheme: Global identification of cotranslational SSB substrates. SSB-bound RNCs and total mRNAs were isolated, reverse transcribed, and labeled for subsequent microarray analysis. (D) Top: SDS-PAGE and silver staining of IPs of TAP-tagged Rpl16, Ssb2 and untagged cells (mock). Bottom: Immunoblot of ribosomal Rpl3. Ssb2-TAP IP depleted all Ssb2 from the lysate (see Figure S1D). (E) Translation profile of SRP-bound and SSB-bound mRNAs compared to the total Translatome identified by ribosome isolation (Rpl16). Hierarchically clustered heat map showis the average values of three individual experiments in rows; columns represent genes. mRNAs enriched over total RNAs in yellow, mRNAs depleted in blue. Pearson correlation coefficients are shown next to the tree. Comparison with the Translatome shows that some RNCs are preferentially enriched for SSB binding (blue) others are enriched in SRP binding (red). See also Figure S1.