SUMMARY
The host response to a virus is determined by intracellular signaling pathways that are modified during infection. These pathways converge as networks and produce interdependent phenotypes, making it difficult to link virus-induced signals and responses at a systems level. Coxsackievirus B3 (CVB3) infection induces death of cardiomyocytes, causing tissue damage and virus dissemination, through incompletely characterized host cell signaling networks. We built a statistical model that quantitatively predicts cardiomyocyte responses from time-dependent measurements of phosphorylation events modified by CVB3. Model analysis revealed that CVB3-stimulated cytotoxicity involves tight coupling between the host ERK and p38 MAPK pathways, which are generally thought to control distinct cellular responses. The kinase ERK5 requires p38 kinase activity and inhibits apoptosis caused by CVB3 infection. By contrast, p38 indirectly promotes apoptosis via ERK1/2 inhibition but directly causes CVB3-induced necrosis. Thus, the cellular events governing pathogenesis are revealed when virus-host programs are monitored systematically and deconvolved mathematically.
INTRODUCTION
Coxsackievirus B3 (CVB3) is among the most common causes of viral myocarditis-associated heart failure in infants and young children (Esfandiarei and McManus, 2008). A major component of CVB3 pathogenesis is cell death of infected cardiomyocytes, which leads to immediate tissue damage and the subsequent release of virulent CVB3 progeny that furthers disease progression (Kawai, 1999). Intervening at the early stages of CVB3 cytotoxicity could potentially reduce the severity of the disease and the need for heart transplantation in patients with viral myocarditis.
Throughout infection, CVB3 modulates various cell-signaling pathways that enable virus propagation (Esfandiarei and McManus, 2008; Garmaroudi et al., 2010). Inhibiting these pathways may provide a therapeutic opportunity to restrict CVB3 pathogenesis. But, an important hurdle is our fragmentary understanding of how the CVB3 infection cycle intersects with the host network. Viruses such as CVB3 have evolved to modulate cell-signaling networks in ways that allow them simultaneously to evade host defenses, promote cell entry, and undergo replication in a changing environment (Esfandiarei and McManus, 2008; Ribet and Cossart, 2010). Blocking individual signaling pathways in host cells often reduces CVB3 infectivity but does not prevent infection entirely (Garmaroudi et al., 2010). It remains unclear whether such “partly required” pathways converge upon a common set of host effectors or instead make independent contributions to pathogenesis (Miller-Jensen et al., 2007). The challenge is that CVB3 adaptively perturbs a collection of host pathways, which must be examined concurrently with time to understand how they interact and give rise to viral functions.
Here, we took a multi-pathway systems approach to connect signaling and host-cell responses in an in vitro model of CVB3 infection (Tan et al., 2007). The dynamics of eight signaling phospho-proteins were monitored together with six CVB3-induced host-cell readouts at five different dosings of CVB3. We then linked CVB3-induced signaling to host-cell readouts by building a predictive data-driven model using two time-dependent combinations of measured phospho-proteins. The results of this analysis revealed unexpected connections between the ERK1/2, ERK5, and p38 MAPK pathways related to their control of apoptotic caspases and overall cell death induced by CVB3. Combined perturbations of these pathways validated the predictions of the model and deconstructed the CVB3 response as a mixture of apoptosis (involving ERK5) and necrosis (involving p38). Our results illustrate how viruses such as CVB3 hijack multiple host signaling pathways simultaneously but use relatively straightforward strategies to manipulate host responses.
RESULTS
CVB3-Induced Phospho-Protein Dynamics Quantitatively Predict Host-Cell Outcomes
To determine whether known CVB3-induced signaling events were sufficient to predict viral propagation and host-cell toxicity, we sought to build a predictive mathematical model based entirely on quantitative experiments. Data-driven modeling identifies higher-order statistical covariations that can be used for prediction and analysis (Janes and Yaffe, 2006). Unlike other modeling formalisms (Aldridge et al., 2006), data-driven approaches can accommodate variegated datasets and make predictions without detailed knowledge of the underlying biochemical mechanisms.
To build the model, we systematically assembled a host-cell signaling and response dataset in virus-infected cardiomyocytes. At five different CVB3 multiplicities of infection (M.O.I.), we profiled eight signaling phospho-proteins by ELISA at six time points over 24 hr together with six CVB3-induced host-cell readouts at three time points over 24 hr (Figures 1A and 1B). Each phospho-protein and host-cell readout was selected based on previous studies suggesting that they were critical for CVB3 pathogenesis (Table S1). Analyzing the information contained in this mechanism-rich signature would then allow us to examine how host-cell pathways were coordinately perturbed during CVB3 infection.
Figure 1. A Predictive Data-driven Model of CVB3-induced Host-cell Responses.
(A) Dynamic phospho-proteins signatures measured by phospho (p)-ELISA that were used as predictor variables in the data-driven model.
(B) Host-cell outcomes that were to be predicted in the data-driven model. Caspase (C)−3, −8 and −9 activities were measured by activity assays with fluorogenic substrates, VP1 capsid protein expression was measured by immunoblotting, released viral progeny (RVP) titer was measured by plaque assay, and cell death was measured by MTS assay at the indicated time points. All outcome data were non-dimensionalized as z scores for comparison of relative changes across conditions.
(C and D) Accurate predictions of host-cell responses with a partial-least-squares model using two principal components. (C) Percentage of information captured with one or two principal components. Information was measured by the percentage of variance in host-cell outcomes that was captured by the model. Note the small-but-detectable increase in information capture after inclusion of the second principal component (dashed arrow). (D) Correlation between crossvalidated predictions of biological responses by partial least squares regression (x-axis) and observed biological responses (y-axis). Marker color corresponds to the post-infection (p.i.) time point at 8 h (white), 16 h (gray), and 24 h (black).
HL1 cells were infected with CVB3 at one of five multiplicities of infection and then assessed for the indicated phospho-proteins and biological responses at six and three time points, respectively, over 24 h. For (A) and (B), data are shown as the z-score standardized mean of three independent experiments as described in the Experimental Procedures. For (C), data are shown as median information captured (red) after fivefold leave-one-out cross-validation (black). For (D), data are shown as the median ± range of 3–4 biological replicates (vertical) or four model cross-validation runs (horizontal). See also Figure S1 and Table S1.
We found that CVB3-induced host-cell responses showed time and dose dependencies that were expected for end-stage readouts (Figure 1B). Activation of the initiator caspases, caspase-8 and caspase-9, was accelerated with increasing M.O.I., corresponding to more-complete activation of the effector caspase for apoptosis, caspase-3 (Riedl and Shi, 2004). Interestingly, readouts of CVB3 propagation, such as expression of the VP1 capsid protein and the titers of released viral progeny (RVP), did not accelerate appreciably as they increased with CVB3 M.O.I.. This finding suggests intrinsic limits to the timing of the CVB3 replication cycle downstream of the M.O.I.-dependent rate processes of viral docking and internalization. The pattern of overall CVB3 cytotoxicity fell in between that of caspase and viral readouts, showing some acceleration as host-cell viability dropped with increasing CVB3 titers. Thus, CVB3 infection of cardiomyocytes elicits a collection of host-cell and viral phenotypes that are monotonic in time but differ in their kinetics and dose-dependent behaviors.
By comparison, we found that the dynamic patterns of protein phosphorylation stimulated by CVB3 were substantially more complex than the associated phenotypic readouts (Figures 1A and 1B). As before, we observed accelerated phosphorylation of some CVB3-induced pathways with increasing M.O.I., such as p38 and Hsp27, but not others, such as ERK (Figure 1A). In addition, biphasic activation patterns were common, and many individual activation peaks appeared or disappeared above a critical threshold of CVB3 M.O.I. (e.g., ATF2, CREB, and IκBα). Sham infection with 0 M.O.I. did not lead to any meaningful changes when compared to 0.5 M.O.I. (Figure S1), confirming that the measured signaling events were due to CVB3 infection. The internal consistency of our phospho (p)-ELISA measurements was also verified by the strong correlations between p-Akt and p-GSK3β (R = 0.6), a direct substrate of Akt (Cross et al., 1995), and between p-p38 and p-Hsp27 (R = 0.8), a direct substrate of the MK2 kinase that is a substrate of p38 (Rouse et al., 1994). The p-ELISA signatures thus provided a reliable starting point for connecting CVB-induced signaling to host-cell outcomes.
One way that the observed CVB3-induced pattern of readouts could be coordinated is if each phospho-protein contributed incrementally to the pattern based on its extent of phosphorylation. Host cells would then “integrate” the intracellular state established by the level of CVB3 infection and gauge their responses accordingly. To test the feasibility of this network mechanism, we used partial-least-squares modeling to link linear combinations of measured phospho-proteins to observed CVB3-induced readouts (Janes et al., 2005; Janes and Yaffe, 2006). In a partial-least-squares model, linear combinations take the form of principal components, which are latent dimensions in the underlying dataset that are derived to be optimally efficient at predicting response outcomes (Jensen and Janes, 2012).
To build the model, we first subdivided the phospho-protein time courses into early (0–8 hr) and late (8–24 hr) phases and then time-integrated each early and late phospho-protein measurement for every CVB3 M.O.I.. This subdivision allowed us to separate biphasic activation profiles into early and late peaks. Using the phospho-protein data as a set of predictor variables, we next sought a partial-least-squares model that could predict all of the CVB3-induced readouts accurately and simultaneously. We found that a model with the two leading principal components could capture all of the measured readouts to within 97% accuracy (Figure 1C). Importantly, this model also accurately predicted readouts for individual M.O.I. conditions that were left out of the model training during crossvalidation (Figure 1D). The model thus supported a network mechanism in which multiple intracellular pathways work together by independently contributing to CVB3-induced readouts.
Intracellular Crosstalk between the ERK and p38 Pathways
Principal components can be further analyzed by plotting the weighted linear combinations of the original measurements that provided the basis for accurate model predictions (Figure 2A) (Janes et al., 2005; Janes and Yaffe, 2006; Jensen and Janes, 2012). In this mapping, early and late phospho-proteins are depicted together with CVB3-induced readouts. Clusters of phospho-proteins and readouts indicate measurements with close association in principal-component space and highlight correlations in the data that are most worthy of follow-up experiments (Janes et al., 2006; Jensen and Janes, 2012).
Figure 2. Model Principal Components Identify Crosstalk between ERK and p38 Pathways.
(A) Model projections of early phospho-proteins (0–8 h p.i., circles), late phospho-proteins (8–24 h p.i., clubs), and host-cell responses (contours) onto the principal components derived in Figure 1C.
(B) Dynamics of phospho (p)-ERK1/2 and p-ERK5 over a 24-hr time course of CVB3 infection. HL1 cells were infected with CVB3 at M.O.I. = 9 and then assessed for p-ERKs at the indicated times p.i. by immunoblotting with tubulin used as a loading control.
(C) ERK p-ELISA measurements are a convolution of p-ERK1/2 and p-ERK5. HL1 cells were pretreated with DMSO, PD to inhibit p-ERK1/2, or U0 to inhibit p-ERK1/2 and p-ERK5 and then infected with sham or CVB3 at M.O.I. = 9. The p-ERK signals were assessed by p-ELISA at 0.17 hr p.i.
(D and E) Early- and late-phase activation of p-p38, p-ERK1/2, and effector kinases upon pretreatment with SB and infection with CVB3.
(F) Densitometry of late-phase p-ERK1/2 in response to SB.
(G) Prolonged SB inhibition leads to ERK1/2 phosphorylation independently of CVB3 infection. (H and I) Early- and late-phase phosphorylation of ERK5 is blocked upon pretreatment with SB and infection with CVB3.
(J) Densitometry of late-phase p-ERK5 in response to SB.
For (D–J), HL1 cells were pretreated with SB203580 (SB, 20 μM) for one hour, infected with sham or CVB3 at M.O.I. = 9, and then assessed for phospho-proteins at the indicated times post-infection by immunoblotting with either tubulin or β-actin used as a loading control. For (C), (F), and (J), data were normalized to CVB3-infected cells without inhibitor and are shown as the mean ± s.e.m. of three (C) or four (F,J) biological replicates. Asterisk indicates p < 0.05 by Welch's one-sided t test. See also Figure S2.
Inspection of this principal-component mapping revealed that all CVB3-induced readouts were densely clustered in one region (Figure 2A, contours), suggesting that they were tightly coupled. Within the cluster lay the transcription factor ATF2, which is critical for CVB3 pathogenesis in vivo (Reimold et al., 2001), and p38, a MAPK that we recently showed is the dominant ATF2 kinase during CVB3 infection (Garmaroudi et al., 2010). We also found p-Hsp27 in the cluster, which was expected because of its strong concordance with p-p38 (see above). Conversely, we were surprised to find p-ERK located in the cluster together with p-p38, because the ERK1/2 and p38 pathways are generally thought to be activated by distinct stimuli and often serve antagonistic functions (Xia et al., 1995). Nevertheless, their tight association in the model suggested that ERKs and p38 might be functionally interlinked during CVB3 infection.
An important consideration for this prediction was the high-throughput data upon which the model was founded (Janes and Yaffe, 2006). The commercial p-ELISA used to assemble the p-ERK dataset is marketed as specific for ERK1/2. However, ERK1 and ERK2 share ~50% identity with ERK5, a third MAPK whose regulation is distinct (Nishimoto and Nishida, 2006). All three ERKs have a Thr–Glu–Tyr motif that is bis-phosphorylated upon activation, and the sequence surrounding this motif is so similar that many p-ERK1/2 antibodies will cross-react with ERK5 (K.J.J. and K.A.J., unpublished observations). p-ERK5 cross-reactivity is readily distinguished from p-ERK1/2 during immunoblotting (ERK5 ~ 80–100 kDa vs. ERK1 ~ 44 kDa, ERK2 ~ 42 kDa), but the ELISA format cannot resolve proteins by molecular weight. Because ERK5 signaling is important for cardiovascular tissues (Regan et al., 2002), we decided to investigate the individual contributions of ERK1/2 and ERK5 by independent methods.
We first monitored the kinetics of ERK1/2 and ERK5 phosphorylation by blotting with antibodies that were specific for each pathway (Figure 2B). Both ERK1/2 and ERK5 were strongly phosphorylated shortly after CVB3 infection at 0.17 h p.i. and also after host cytotoxicity was evident at 24 h p.i. However, ERK5 showed a more-sustained phosphorylation up to 1 h p.i. and p-ERK1/2 exhibited a second peak at 8 h p.i., illustrating differences in their regulatory kinetics. The multiphase activation of ERK1/2 was further confirmed by measuring phosphorylation of RSK, a specific ERK1/2 substrate (Figure S2A) (Sturgill et al., 1988). Next, we used a pair of MEK inhibitors (PD184352 [PD] and U0126 [U0]) to separate the ERK1/2 and ERK5 contributions to the ERK p-ELISA. PD at low concentrations selectively blocks MEK1/2 and ERK1/2 phosphorylation, whereas U0 inhibits MEK1/2-ERK1/2 and MEK5-ERK5 equally (Davies et al., 2000) (Figures S2B and S2C). Thus, the contribution of ERK5 can be inferred from the difference between PD (ERK1/2 inhibition) and U0 (ERK1/2 + ERK5 inhibition). When cells were preincubated with U0 and treated with CVB3 for 10 min, we found that the measured p-ERK ELISA signal was reduced to background levels (Figure 2C). By contrast, pretreatment with PD reduced the ELISA signal by only ~30%, despite that ERK1/2 phosphorylation was completely inhibited (Figures 2C and S2C). This indicated that the p-ERK ELISA data was a convolution of ERK1/2 and ERK5 pathway activities and further implied that the predicted ERK–p38 associations (Figure 2A) could be between ERK1/2 and p38 or ERK5 and p38, or both.
We tested for crosstalk between p38 and ERKs by using SB203580 (SB), an ATP-competitive small-molecule inhibitor of p38 (Lee et al., 1994). We monitored p-ERK1/2, p-ERK5, and p-p38, as well as the major ERK1/2 and p38 effector kinases, RSK and MK2 (Rouse et al., 1994; Sturgill et al., 1988). We found that SB potently inhibited p38 activity in cardiomyocytes, as expected, blocking phosphorylation of MK2 at early and late times after CVB3 infection (Figures 2D and 2E). Acute SB treatment was also specific, because we did not observe any effect on early CVB3-induced ERK1/2 phosphorylation or activity (Figure 2D). Upon prolonged SB treatment, however, we observed a modest-but-reproducible increase in ERK1/2 phosphorylation (Figures 2E and 2F). We attribute this to secondary inhibition of PP1 and PP2A phosphatases, which are normally activated by p38 signaling and serve to dephosphorylate MEK1/2 upstream of ERK1/2 (Westermarck et al., 2001). Subsequent control experiments showed that SB-induced upregulation of ERK1/2 was independent of CVB3 treatment (Figure 2G). Thus, p38 signaling antagonizes late ERK1/2 signaling, prompting a reevaluation of earlier p38-inhibition experiments involving CVB3 (see below) (Si et al., 2005).
A second finding from these experiments was that SB treatment potently blocked both early- and late-phase phosphorylation of ERK5 (Figures 2H–J). The p38–ERK5 coupling was consistent with predictions of the model (Figure 2A), but such crosstalk had not previously been reported. To exclude the possibility that ERK5 inhibition was caused by off-target effects of SB, we repeated the experiments with BIRB796 (BIRB), an allosteric inhibitor of p38 (Pargellis et al., 2002) (Figure S2D). SB and BIRB have completely different mechanisms of inhibition and off-target signatures; therefore, a common phenotype with SB and BIRB strongly implicates p38 signaling (Bain et al., 2007). When cardiomyocytes were pretreated with BIRB, we observed the same blockade of CVB3-induced ERK5 phosphorylation as with SB (Figures S2E and S2F). Last, to examine the generality of the p38–ERK5 connection, we treated human embryonic kidney cells with sorbitol as an osmotic stress that activates both p38 and ERK5 (Abe et al., 1996; Raingeaud et al., 1995). SB and BIRB each blocked hyperosmolarity-induced ERK5 phosphorylation (Figure S2G), suggesting that p38 is generally required for proper activation of the MEK5–ERK5 pathway. Taken together, the molecular consequences of SB and BIRB indicate that p38 is functionally interconnected with both ERK1/2 and ERK5, as predicted by the model of CVB3 pathogenesis (Figure 2A).
Deconvolution of the ERK- and p38-Dependent Apoptotic Response
An important category of host-cell responses in the starting dataset was the activity of apoptotic caspases (Figure 1B). ERKs and p38 mapped closely to these readouts in the model and could conceivably control CVB3-induced apoptosis directly (Figure 2A). Both ERK1/2 and p38 have been reported to be important for proper caspase activation (Luo et al., 2002; Si et al., 2005). However, these earlier studies used a dual MEK1/2–MEK5 inhibitor (U0) and were not aware of the antagonism between p38 and ERK1/2 (Figures 2E–G and S2C). We thus pursued follow-up studies using gain- and loss-of-function approaches for ERK–p38 together with direct measurements of caspase processing.
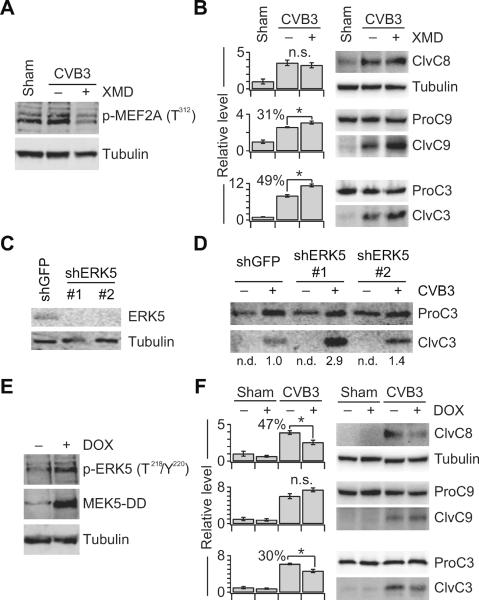
We began with ERK5, as it inhibits cardiac apoptosis in other contexts (Kimura et al., 2010; Yan et al., 2007) but had not been previously implicated in CVB3 infection. To block ERK5 signaling, we used the specific ATP-competitive ERK5 inhibitor, XMD8-92 (XMD) (Yang et al., 2010). XMD treatment potently reduced phosphorylation of an ERK5 substrate (MEF2A) in cardiomyocytes and significantly increased caspase-9 and caspase-3 cleavage upon CVB3 infection (p < 0.05) (Figures 3A and 3B). CVB3-induced apoptosis also increased when endogenous ERK5 was downregulated with shRNA (Figures 3C and 3D). We performed a reciprocal gain-of-function experiment by establishing stable lines expressing a doxycycline (DOX)-inducible mutant of MEK5 that was constitutively active (MEK5-DD) (Figure 3E). Upon low-level infection with CVB3 (M.O.I. = 1.5), we found that DOX treatment of MEK5-DD-expressing cells caused a significant decrease in caspase-3 cleavage (p < 0.05) (Figure 3F). Interestingly, the drop in caspase-3 cleavage was associated with changes in caspase-8 activation rather than caspase-9 activation as with XMD. We attribute this difference to the kinetics of ERK5 activation with MEK5-DD (~8 hours) versus ERK5 inhibition with XMD (< 1 hour). The MEK5-DD, XMD, and shRNA results together indicate that CVB3-induced ERK5 signaling inhibits cardiomyocyte apoptosis. This link between ERK5 and host-cell survival is unique, because virtually all other CVB3-stimulated pathways described thus far promote apoptosis rather than inhibit it (Table S1).
Figure 3. ERK5 Inhibits CVB3-induced Apoptosis.
(A) XMD inhibits ERK5 activity in cells. Samples were analyzed for phospho-MEF2A at 24 h p.i. by immunoblotting with tubulin as a loading control.
(B) CVB3-induced caspase-9 and caspase-3 processing is increased by XMD-mediated inhibition of ERK5.
(C and D) Knockdown of endogenous ERK5 increases CVB3-induced caspase-3 cleavage. HL1 cells were transduced with the indicated shRNAs and analyzed for the indicated proteins by immunoblotting with tubulin or full-length caspase-3 used as a loading control. In (D), quantitative densitometry is shown relative to shGFP control cells infected with CVB3 at M.O.I. = 9.
(E) DOX induction of MEK5-DD activates endogenous ERK5. HL1 cells stably expressing doxycycline (DOX)-inducible MEK5-DD were treated with 1 μg/ml DOX for 24 h and analyzed for the indicated proteins by immunoblotting with tubulin used as a loading control.
(F) CVB3-induced caspase-8 and caspase-3 processing is decreased upon activation of ERK5 by DOX-inducible MEK5-DD.
For (A) and (B), HL1 cells were pretreated with XMD8-92 (XMD, 5 μM) for one hour, and infected with sham or CVB3 at M.O.I. = 9. For (E) and (F), HL1 cells stably expressing doxycycline (DOX)-inducible MEK5-DD were infected with CVB3 at M.O.I. = 1.5 and treated with 1 μg/ml DOX at 0 h p.i.. Samples were analyzed for the indicated active caspase-cleavage products at 24 h p.i. by immunoblotting with tubulin or full-length caspases used as a loading control. For (B) and (F), densitometry measurements were normalized to sham-infected cells without inhibitor and data are shown as the mean ± s.e.m of four biological replicates. Asterisk indicates p < 0.05 by Welch's one-sided t test. See also Figure S3.
Next, we examined the p38 pathway by using a similar set of approaches. Consistent with an earlier report (Si et al., 2005), we found that p38 inhibition via SB profoundly reduced caspase-3 cleavage during CVB3 infection (Figure 4A). We reinforced the SB result by showing that p38 inhibition with BIRB phenocopied SB in its blockade of initiator and effector caspases (Figures S4A and S4B). The intuitive conclusion from these experiments is that p38 promotes CVB3-induced apoptosis. However, when we attempted the reciprocal gain-of-function experiment with a DOX-inducible, constitutively active mutant of MKK6 (MKK6-EE), there was no detectable change in caspase activation (Figures 4B and 4C). The apparent contradiction prompted us to re-evaluate our experiments considering the cross-communication between p38 and ERKs (Figures 2E–J).
Figure 4. p38 Inhibition Blocks CVB3-induced Apoptosis Indirectly Via ERK1/2 Hyperactivation.
(A) CVB3-induced caspase processing is blocked by SB-mediated inhibition of p38.
(B–D) p38 activation via DOX-inducible MKK6-EE does not affect CVB3-induced apoptosis but suppresses ERK1/2 phosphorylation. For (B) and (D), HL1 cells stably expressing doxycycline (DOX)-inducible MKK6-EE were treated with 1 μg/ml DOX for 8 hr and analyzed for the indicated proteins by immunoblotting with tubulin used as a loading control.
(E) SB-mediated inhibition of caspase processing is blocked by co-inhibition of ERK1/2 signaling with PD.
(F) ERK1/2 inhibition with PD does not affect CVB3-induced apoptosis.
For (A), (E), and (F), HL1 cells were pretreated with SB203580 (SB, 20 μM), PD184352 (PD, 2 μM), or SB+PD for one hour, and infected with sham or CVB3 at M.O.I. = 9. For (C), HL1 cells stably expressing DOX-inducible MKK6-EE were infected with CVB3 at M.O.I. = 1.5 and treated with 1 μg/ml DOX at 8 h p.i.. Samples were analyzed for the indicated active caspase-cleavage products at 24 h p.i. by immunoblotting with tubulin or full-length caspases used as a loading control. Densitometry measurements were normalized to sham-infected cells without inhibitor and data are shown as the mean ± s.e.m of four biological replicates. Asterisk indicates p < 0.05 by Welch's one-sided t test. See also Figure S4.
We reasoned that secondary inhibition of ERK5 would partially offset the observed SB–BIRB phenotype rather than cause it (Figures 2H–J and 3). Therefore, our attention turned to ERK1/2, which becomes hyperactivated upon prolonged p38 inhibition (Figures 2F and 2G). This negative regulation of ERK1/2 by p38 was further strengthened by the reduced p-ERK1/2 observed in DOX-treated MKK6-EE cells (Figure 4D). To determine whether the consequences of p38 inhibition were mediated through ERK1/2, we combined SB with PD to block ERK1/2 hyperactivation and found that CVB3-induced apoptosis occurred normally (Figures 4E and S4C). Remarkably, ERK1/2 inhibition by itself did not substantially affect apoptosis of CVB3-infected cells (Figures 4F and S4C), suggesting that ERK1/2 acted as a pro-survival signal only when p38 function was blocked. The p38-specific role of ERK1/2 was re-emphasized in ERK5-inhibited cells, where PD+XMD increased apoptosis as with XMD alone (Figures 3B, S3A, and S3B). Upon this re-evaluation of earlier studies using SB (Si et al., 2005), we conclude that p38 signaling does not directly control CVB3-induced apoptosis.
p38 Signaling Contributes to CVB3-induced Pathogenesis by Stimulating Necrosis
Apoptosis is but one facet of the host-cell response to CVB3 infection, raising the question of whether other aspects of pathogenesis could require p38 signaling (Carthy et al., 1998; Yuan et al., 2003). In the original dataset, overall CVB3 cytotoxicity was measured via tetrazolium reduction (see Experimental Procedures). However, this method was inadequate to read out cytotoxicity in the presence of signaling perturbations, which could also affect proliferation and metabolism. We therefore switched to a fluorescent amine-reactive dye that intensely labels cells with compromised plasma-membrane integrity irrespective of the mechanism of cell death (Perfetto et al., 2006).
We found that CVB3 infection caused a dramatic increase in the percentage of dye-labeled, non-viable cells as compared to sham infection (Figures 5A–D). The actual extent of cytotoxicity was much greater than the flow-cytometry estimate (compare Figures 5B and 5D), because many infected cells were so damaged that they were unavoidably lost during the suspension preparation. As with the earlier apoptosis experiments (Figure 4), we found that SB pretreatment strongly decreased the extent of CVB3-induced cytotoxicity, whereas PD did not have a significant impact (p > 0.05) (Figures 5B–F). Surprisingly, when SB and PD were combined, we observed a clear improvement in overall cell viability even though caspase activation was unaffected under these conditions (Figures 4E, 5B, and 5G). The pronounced result of dual p38–ERK1/2 inhibition was also reflected in significantly reduced titers of released viral progeny (p < 0.001) (Figures 6A and 6B). This raised the possibility that p38 could control alternative death pathways that were distinct from apoptosis but critically important for CVB3 pathogenesis.
Figure 5. p38 Inhibition Improves Viability of CVB3-infected Cells Independently of ERK1/2 Signaling.
(A) Representative flow-cytometry profile of CVB3-infected cardiomyocytes labeled with amine-reactive dye. Dye-positive cells (pink) are considered non-viable.
(B) SB and SB+PD inhibit CVB3-induced cytotoxicity.
(C–G) Representative images of amine-labeled adherent CVB3-infected cardiomyocytes pretreated with SB, PD, or SB+PD. Right panel of (D) shows an enlarged image highlighting vesiculated-necrotic cells (ves) and apoptotic cells (apop).
Cells were pretreated with SB203580 (SB, 20 μM), PD184352 (PD, 2 μM), or SB+PD for one hour and infected with sham or CVB3 at M.O.I. = 9. Cells were labeled with amine-reactive dye, and monitored by flow cytometry or fluorescence microscopy. For (B), data are shown as the mean ± s.e.m. of four biological replicates. Asterisk indicates p < 0.05 by Welch's one-sided t test. For (C–G), scale bar is 20 μm.
Figure 6. p38 Controls CVB3-induced Necrosis.
(A and B) SB+PD markedly inhibits released viral progeny (RVP) in CVB3-infected cells.
(C) HMGB1 is a reliable marker of CVB3-induced necrosis.
(D–F) SB blocks CVB3-induced HMGB1 release independently of ERK1/2 pathway inhibition with PD. Note that PD does not affect the inhibition of HMGB1 release caused by SB.
(G) p38 activation via DOX-inducible MKK6-EE increases CVB3-induced HMGB1 release.
For (A–F), cells were pretreated with SB203580 (SB, 20 μM), PD184352 (PD, 2 μM), SB+PD, Necrostatin-1 (Nec-1, 50 μM), or DEVD-CHO (DEVD, 0.1 μM) for one hour and infected with sham or CVB3 at M.O.I. = 9. For (A) and (B), RVP titers were determined by plaque assay. For (D–G), culture supernatants were concentrated and analyzed for HMGB1 release by immunoblotting. For (G), HL1 cells stably expressing doxycycline (DOX)-inducible MKK6-EE were infected with CVB3 at M.O.I. = 1.5 and treated with 1 μg/ml DOX at 8 h p.i.. For (B), data are shown as the median ± range of four biological replicates. Single asterisk indicates p < 0.05 and double asterisk indicates p < 0.001 by Welch's two-sided t test. See also Figure S5.
We closely examined the morphology of CVB3-infected cells by microscopy and noted a mixture of phenotypes indicative of discrete single-cell outcomes (Figure 5D). Some cells had a rounded appearance with condensed nuclei, suggesting an apoptotic fate. Others, however, remained fully spread and had aberrant lamellipodia-like projections (Figure 5D, right). These cells also had an intact nucleus along with intracellular vesicles that remained dye impermeant. Our observations suggested that a fraction of CVB3-infected cells undergo a vesiculated form of cell death with certain hallmarks of necrosis (Yuan et al., 2003).
To determine whether CVB3 infection was associated with biochemical readouts of necrosis, we examined the chromatin protein HMGB1, which is released extracellularly by necrotic cells (Scaffidi et al., 2002). We validated the marker by stimulating cardiomyocyte necrosis with hydrogen peroxide and observing pronounced HMGB1 release (Figure S5A). Importantly, we found that HMGB1 was clearly detected in supernatants from CVB3-infected cells (Figure 6C). HMGB1 release was unaffected by the apoptosis inhibitor DEVD-CHO but was slightly reduced by the necrosis inhibitor Necrostatin-1, likely as a result of CVB3-induced autocrine TNF signaling (Degterev et al., 2005; Garmaroudi et al., 2010). Thus, HMGB1 is a reliable marker of necrosis stimulated by CVB3.
Upon p38 inhibition with SB or BIRB, we observed near-complete blockade of HMGB1 release, suggesting potent inhibition of necrosis (Figures 6D and S5B). Conversely, necrosis was negligibly affected in CVB3-infected cells treated with PD to inhibit ERK1/2, consistent with the earlier labeling results (Figures 5B, 5F, and 6E). In stark contrast to the apoptotic readouts (Figure 4E), we did not observe any reversion of necrosis when CVB3-infected cells were pretreated with PD + SB (Figure 6F). Last, to test whether p38 signaling was sufficient to drive virus-induced necrosis, we returned to the inducible MKK6-EE cells and found that DOX treatment substantially augmented HMGB1 release during CVB3 infection (Figure 6G). We conclude that p38 signaling is a critical component of a necrosis pathway, which promotes CVB3 propagation independently of ERK-dependent apoptosis.
DISCUSSION
Viruses such as CVB3 activate many host-cell signaling pathways and evoke many host-cell responses. Our study here began with a holistic approach to monitor these events dynamically and as a function of CVB3 titer. By analyzing the data to make quantitative predictions of host-cell outcome, we quickly converged on ERKs and p38 as key pathways for CVB3 pathogenesis. Early-phase ERK1/2 activation stems directly from CVB3 docking to host membranes, whereas late-phase activation occurs due to cleavage of upstream signaling molecules by viral proteases (Huber et al., 1999; Luo et al., 2002). Late-phase p38 and ERK5 signaling probably lies downstream of autocrine proinflammatory cytokines, which are induced during the final stages of the viral life cycle (Figure S6) (Garmaroudi et al., 2010). Despite differences in activation, our work here shows that ERKs and p38 are strongly interconnected (Figure 7). These dependencies are important for interpreting the results of “single-pathway” perturbations that propagate through the network (Luo et al., 2002; Si et al., 2005).
Figure 7. Model for ERK–p38 signaling, apoptosis, and necrosis induced by CVB3.

Dashed line indicates context-dependent inhibition of apoptosis by ERK1/2. See also Figure S6.
Notably, we were able to uncover a role for ERK5 in CVB3 pathogenesis by modeling a dataset that did not measure ERK5 explicitly. We have shown elsewhere that quantitatively accurate signaling measurements are critical for data-driven models to reflect underlying biological mechanisms (Janes et al., 2008; Janes and Yaffe, 2006). Our results here using a pan-ERK p-ELISA indicate that measurements of specific proteins may not be as important. This is encouraging, because many modern signaling assays increase overall throughput by relaxing the specificity constraints of traditional approaches (Albeck et al., 2006).
Similarly, our work shows that agglomerated cell-outcome data may be sufficient for viral-host modeling and discovering overlooked phenotypes. The importance of CVB3-induced necrosis as a host-cell fate was revealed here without direct necrotic readouts in the model (Figure 7). This information was presumably embedded in the overall cytotoxicity measure, which depends strongly on the level of necrosis (Figures 5A–D). Interestingly, the associated RVP titers appear to be influenced by apoptosis and necrosis reciprocally. When both apoptosis and necrosis are blocked upon p38 inhibition with SB, there is a slight reduction in RVP. However, when apoptosis is restored in p38-inhibited cells by blocking ERK1/2 hyperactivation, RVP is dramatically reduced (Figure 6B). Thus, necrosis may be the preferred outcome for CVB3, which is counteracted by the host-cell drive to die by apoptosis. To isolate necrosis specifically requires targeting an upstream mediator (p38) and resetting the other secondary consequences of pathway inhibition (e.g., ERK1/2) (Figure 7). Such combinatorial anti-viral strategies would be difficult to predict without the aid of a systems model for the host-cell response to CVB3 infection.
EXPERIMENTAL PROCEDURES
Plasmids
DOX-inducible MEK5 (Boehm et al., 2007) and MKK6-EE (Raingeaud et al., 1996) were cloned by PCR into the Tet-tight entry vector pEN_TTmiRc2 (Shin et al., 2006). To generate MEK5-DD, S311 and T315 of MEK5 were both mutated to glutamate by site-directed mutagenesis (Quikchange II XL, Stratagene). All entry vectors were verified by sequencing, and lentiviral vectors were cloned by LR recombination into pSLIK neo (Shin et al., 2006). pLKO.1 puro shERK5 lentiviral vectors (TRCN0000023234 and TRCN0000023236) were obtained from Open Biosystems.
Cells and Viruses
HL1 cells were provided by Dr. William Claycomb (Louisiana State University Health Sciences Center, New Orleans, USA) (Claycomb et al., 1998). 293T cells were obtained from ATCC. CVB3 (Kandolf strain) was propagated in HeLa cells, and virus titers were determined by plaque assay. Retroviruses and lentiviruses were packaged as previously described (Wang et al., 2011). Stably transduced HL1 cells were selected with 4 μg/ml puromycin or 150 μg/ml G418 until control plates had cleared.
Viral Infection and Perturbations
HL1 cells were sham-infected with PBS or infected with CVB3 at M.O.I. = 0.5, 1.5, 4.5, 9, or 18 and cell extracts were prepared at 0, 0.17, 1, 8, 16, and 24 hr. For perturbation experiments, the following chemical inhibitors were added one hour before infection: SB203580 (20 μM, Tocris Biosciences), BIRB796 (5 μM, Selleck Chemicals), XMD8-92 (5 μM, Axon Medchem), U0126 (20 μM, Tocris Biosciences), PD184352 (2 μM, Santa Cruz Biotechnology), DEVD-CHO (0.1 μM, EMD), and Necrostatin-1 (50 μM, Calbiochem).
Plaque Assays
CVB3 titers from triplicate cell supernatants were determined on monolayers of HeLa cells by an agar overlay plaque assay as described elsewhere (Garmaroudi et al., 2010).
p-ELISA
Cell lysates were normalized to protein concentration and analyzed by p-ELISA (Biosource) for the phosphorylation levels of Akt (S473), ATF2 (T69/T71), CREB (S133), ERK1/2 (T185/Y187), GSK3β (S9), Hsp27 (S82), IκBα (S32), and p38 MAPK (T180/Y182) according to the manufacturer's instruction.
Caspase-3, -8 and -9 Activity Assays
Caspase activities were measured according to the manufacturer's instruction (R&D Systems) as described elsewhere (Si et al., 2005). Fluorescence was measured at excitation and emission wavelengths of 485 nm and 535 nm, respectively, using a Tecan GENios fluorescent reader.
Immunoblotting
Immunoblotting was performed as described previously (Garmaroudi et al., 2010) with one of the following primary antibodies: anti-p-ERK1/2 (T202/Y204, Cell Signaling, 1:1000), anti-p-ERK5 (T218/Y220, Cell Signaling, 1:1000), anti-p-p38 (T180/T182, Cell Signaling, 1:1000), anti-p-MAPKAPK2 (T334, Cell Signaling, 1:1000), anti-VP1 (Dako, 1:1000), anti-cleaved caspase-8 (Cell Signaling, 1:1000), anti-caspase-9 (Cell Signaling, 1:1000), anti-caspase-3 (Cell Signaling, 1:1000), anti-β-actin (Sigma, 1:5000), anti-p-Akt (S473, Cell Signaling, 1:1000), anti-p-GSK3β (S9, Cell Signaling, 1:1000), anti-p-ATF2 (T69/T71, Cell Signaling, 1:1000), anti-p-CREB (S133, Cell Signaling, 1:1000), anti-p-IκBα (S32, Cell Signaling, 1:1000), anti-p-Hsp27 (S82, Cell Signaling, 1:1000), anti-p-RSK (T359/S363, 1:1000), anti-MEK5 (StressGen, 1:1000), anti-HA (Roche, 1:1000), anti-HMGB-1 (Epitomics, 1:1000), anti-p-MEF2A (T312, Abcam, 1:1000), anti-cleaved caspase-8 (Cell Signaling, 1:1000), or anti-tubulin (Cell Signaling, 1:5000 or Abcam, 1:20000) for 1 hr or overnight, followed by incubation for 1 hr with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz) or infrared dye-conjugated secondary antibodies (Licor). Immunoreactive bands were visualized by enhanced chemiluminescence (Pierce, Rockford, IL) on ChemiGenius2 or ChemiDoc MP camera-based detection systems or by infrared fluorescence on an Odyssey infrared imaging system. Where indicated, band intensities were quantified by densitometry with ImageJ, and all blotting results were replicated with at least one additional set of independent biological samples.
Cell Viability Assays
HL1 cells were grown in 12-well plates and infected with CVB3 (M.O.I. = 9) for 16 and 24 hr after pretreatment with inhibitors. The MTS solutions (1:5) were added to wells for 2.5 hr and then transferred to 96-well plates. Cell viabilities of infected cells and non-infected were assessed by MTS assay (CellTiter 96; Promega, Inc., Madison, WI). Amine-reactive labeling was performed with the LIVE/DEAD fixable violet dead stain (Invitrogen) according to the manufacturer's recommendations. For flow cytometry, cells were labeled in suspension, washed with PBS + 0.1% Tween-20, and analyzed on a BD FACSCalibur equipped with 407 nm violet laser excitation. For microscopy, adherent cells were labeled, washed with PBS, permeabilized with 0.3% Triton X-100 in PBS, and counterstained with DRAQ-5 before imaging by widefield microscopy as described previously (Wang et al., 2011).
Partial Least Squares Regression
Phospho-proteins (predictor variables) and readouts (response variables) were standardized as z-scores, and the phospho-protein time course was time-integrated over early (0–8 hr) and late (8–24 hr) phases. Partial least squares regression was performed with the “plsregress” function in MATLAB by standard approaches (Janes et al., 2005; Janes and Yaffe, 2006). The stability of the model was assessed by fivefold leave-one-out cross-validation.
Statistical analysis
All hypothesis testing was performed with Welch's one- or two-sided t test at a significance level of α = 0.05.
Supplementary Material
HIGHLIGHTS
Intracellular signaling predicts the fate of coxsackievirus B3 (CVB3)-infected cells
p38 and ERK signaling pathways are tightly interconnected during CVB3 infection
CVB3-induced apoptosis of cardiomyocytes is inhibited by ERK5 and ERK1/2 pathways
CVB3 induces necrosis of cardiomyocytes via p38 activation
ACKNOWLEDGMENTS
We thank Benjamin Kuhn for technical assistance with immunoblotting and image analysis. This work was supported by the National Institutes of Health Director's New Innovator Award Program (1-DP2-OD006464 to K.A.J.), the Pew Scholars Program in the Biomedical Sciences (to K.A.J.), and the David and Lucile Packard Foundation (to K.A.J.), the Heart and Stroke Foundation of British Columbia & Yukon (to B.M.M.), and the Canadian Institutes of Health Research (CIHR) (to B.M.M.). F.S.G. is supported by a Doctoral Award from Tehran University of Medical Science-Iran. K.J.J. is partly supported by a predoctoral award from the ARCS Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abe J, Kusuhara M, Ulevitch RJ, Berk BC, Lee JD. Big mitogen-activated protein kinase 1 (BMK1) is a redox-sensitive kinase. The Journal of biological chemistry. 1996;271:16586–16590. doi: 10.1074/jbc.271.28.16586. [DOI] [PubMed] [Google Scholar]
- Albeck JG, MacBeath G, White FM, Sorger PK, Lauffenburger DA, Gaudet S. Collecting and organizing systematic sets of protein data. Nat Rev Mol Cell Biol. 2006;7:803–812. doi: 10.1038/nrm2042. [DOI] [PubMed] [Google Scholar]
- Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8:1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson AL, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Carthy CM, Granville DJ, Watson KA, Anderson DR, Wilson JE, Yang D, Hunt DW, McManus BM. Caspase activation and specific cleavage of substrates after coxsackievirus B3-induced cytopathic effect in HeLa cells. Journal of virology. 1998;72:7669–7675. doi: 10.1128/jvi.72.9.7669-7675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb WC, Lanson NA, Jr., Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- Esfandiarei M, McManus BM. Molecular biology and pathogenesis of viral myocarditis. Annu Rev Pathol. 2008;3:127–155. doi: 10.1146/annurev.pathmechdis.3.121806.151534. [DOI] [PubMed] [Google Scholar]
- Garmaroudi FS, Marchant D, Si X, Khalili A, Bashashati A, Wong BW, Tabet A, Ng RT, Murphy K, Luo H, et al. Pairwise network mechanisms in the host signaling response to coxsackievirus B3 infection. Proc Natl Acad Sci U S A. 2010;107:17053–17058. doi: 10.1073/pnas.1006478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M, Watson KA, Selinka HC, Carthy CM, Klingel K, McManus BM, Kandolf R. Cleavage of RasGAP and phosphorylation of mitogen-activated protein kinase in the course of coxsackievirus B3 replication. Journal of virology. 1999;73:3587–3594. doi: 10.1128/jvi.73.5.3587-3594.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- Janes KA, Gaudet S, Albeck JG, Nielsen UB, Lauffenburger DA, Sorger PK. The Response of Human Epithelial Cells to TNF Involves an Inducible Autocrine Cascade. Cell. 2006;124:1225–1239. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- Janes KA, Reinhardt HC, Yaffe MB. Cytokine-induced signaling networks prioritize dynamic range over signal strength. Cell. 2008;135:343–354. doi: 10.1016/j.cell.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol. 2006;7:820–828. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- Jensen KJ, Janes KA. Modeling the latent dimensions of multivariate signaling datasets. Physical biology. 2012;9:045004. doi: 10.1088/1478-3975/9/4/045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai C. From myocarditis to cardiomyopathy: mechanisms of inflammation and cell death: learning from the past for the future. Circulation. 1999;99:1091–1100. doi: 10.1161/01.cir.99.8.1091. [DOI] [PubMed] [Google Scholar]
- Kimura TE, Jin J, Zi M, Prehar S, Liu W, Oceandy D, Abe J, Neyses L, Weston AH, Cartwright EJ, et al. Targeted deletion of the extracellular signal-regulated protein kinase 5 attenuates hypertrophic response and promotes pressure overload-induced apoptosis in the heart. Circ Res. 2010;106:961–970. doi: 10.1161/CIRCRESAHA.109.209320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Laydon JT, McDonnell PC, Gallagher TF, Kumar S, Green D, McNulty D, Blumenthal MJ, Heys JR, Landvatter SW, et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- Luo H, Yanagawa B, Zhang J, Luo Z, Zhang M, Esfandiarei M, Carthy C, Wilson JE, Yang D, McManus BM. Coxsackievirus B3 replication is reduced by inhibition of the extracellular signal-regulated kinase (ERK) signaling pathway. Journal of virology. 2002;76:3365–3373. doi: 10.1128/JVI.76.7.3365-3373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–608. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- Nishimoto S, Nishida E. MAPK signalling: ERK5 versus ERK1/2. EMBO Rep. 2006;7:782–786. doi: 10.1038/sj.embor.7400755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, et al. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat Struct Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- Perfetto SP, Chattopadhyay PK, Lamoreaux L, Nguyen R, Ambrozak D, Koup RA, Roederer M. Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J Immunol Methods. 2006;313:199–208. doi: 10.1016/j.jim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. The Journal of biological chemistry. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Molecular and cellular biology. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan CP, Li W, Boucher DM, Spatz S, Su MS, Kuida K. Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc Natl Acad Sci U S A. 2002;99:9248–9253. doi: 10.1073/pnas.142293999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Kim J, Finberg R, Glimcher LH. Decreased immediate inflammatory gene induction in activating transcription factor-2 mutant mice. Int Immunol. 2001;13:241–248. doi: 10.1093/intimm/13.2.241. [DOI] [PubMed] [Google Scholar]
- Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: A re-emerging field. Cell. 2010;143:694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Rouse J, Cohen P, Trigon S, Morange M, Alonso-Llamazares A, Zamanillo D, Hunt T, Nebreda AR. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell. 1994;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci U S A. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si X, Luo H, Morgan A, Zhang J, Wong J, Yuan J, Esfandiarei M, Gao G, Cheung C, McManus BM. Stress-activated protein kinases are involved in coxsackievirus B3 viral progeny release. Journal of virology. 2005;79:13875–13881. doi: 10.1128/JVI.79.22.13875-13881.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill TW, Ray LB, Erikson E, Maller JL. Insulin-stimulated MAP-2 kinase phosphorylates and activates ribosomal protein S6 kinase II. Nature. 1988;334:715–718. doi: 10.1038/334715a0. [DOI] [PubMed] [Google Scholar]
- Tan SL, Ganji G, Paeper B, Proll S, Katze MG. Systems biology and the host response to viral infection. Nat Biotechnol. 2007;25:1383–1389. doi: 10.1038/nbt1207-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brugge JS, Janes KA. Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc Natl Acad Sci U S A. 2011;108:E803–812. doi: 10.1073/pnas.1103423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermarck J, Li SP, Kallunki T, Han J, Kahari VM. p38 mitogen-activated protein kinase-dependent activation of protein phosphatases 1 and 2A inhibits MEK1 and MEK2 activity and collagenase 1 (MMP-1) gene expression. Molecular and cellular biology. 2001;21:2373–2383. doi: 10.1128/MCB.21.7.2373-2383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- Yan C, Ding B, Shishido T, Woo CH, Itoh S, Jeon KI, Liu W, Xu H, McClain C, Molina CA, et al. Activation of extracellular signal-regulated kinase 5 reduces cardiac apoptosis and dysfunction via inhibition of a phosphodiesterase 3A/inducible cAMP early repressor feedback loop. Circ Res. 2007;100:510–519. doi: 10.1161/01.RES.0000259045.49371.9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Deng X, Lu B, Cameron M, Fearns C, Patricelli MP, Yates JR, 3rd, Gray NS, Lee JD. Pharmacological inhibition of BMK1 suppresses tumor growth through promyelocytic leukemia protein. Cancer Cell. 2010;18:258–267. doi: 10.1016/j.ccr.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JP, Zhao W, Wang HT, Wu KY, Li T, Guo XK, Tong SQ. Coxsackievirus B3-induced apoptosis and caspase-3. Cell Res. 2003;13:203–209. doi: 10.1038/sj.cr.7290165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.