Abstract
Background:
Early serum detection is of critical importance to improve the therapy for hepatocellular carcinoma (HCC), one of the most deadly cancers. Hepatitis infection is a leading cause of HCC.
Methods:
In the present study, we collected total serum samples with informed consent from 80 HCC patients with HBV (+)/cirrhosis (+), 80 patients with benign diseases (50 liver cirrhosis patients and 30 HBV-infected patients) and 60 healthy controls. Analysis was by using surface-enhanced laser desorption/ionisation-time-of-flight mass spectroscopy (SELDI-TOF-MS) to find new serum markers of HCC. SELDI peaks were isolated by SDS–PAGE, identified by LC-MS/MS and validated by immunohistochemistry (IHC) in liver tissues. Migration and invasion assay were performed to test the ability of cell migration and invasion in vitro.
Results:
SELDI-TOF-MS revealed a band at 7777 M/Z in the serum samples from HCC patients but not from healthy controls or patients with benign diseases. The protein (7777.27 M/Z) in the proteomic signature was identified as C-C motif chemokine 15 (CCL15) by peptide mass fingerprinting. A significant increase in serum CCL15 was detected in HCC patients. Functional analysis showed that HCC cell expressed CCL15, which in turn promoted HCC cell migration and invasion.
Conclusion:
CCL15 may be a specific proteomic biomarker of HCC, which has an important role in tumorigenesis and tumour invasion.
Keywords: CCL15, biomarker, HCC, migration, invasion
Primary hepatocellular carcinoma (HCC) is one of the most lethal malignancies worldwide. It is the third leading cause of cancer death in China and the sixth most common cancer in the world (Jemal et al, 2011). Prognosis of HCC remains poor, mainly due to the failure of early diagnosis of the disease in symptom-free patients(Fujiyama et al, 2002; Marrero and Lok, 2004). Early detection of HCC before the onset of clinical symptoms can lead to curative treatment, significantly improving prognosis (Witjes et al, 2010).
At present, alpha-fetoprotein (AFP) and des-γ-carboxy prothrombin (DCP) are the two most widely used tests to aid in the diagnosis of HCC. However, AFP is found to be normal in approximately one-third of patients with small (<3 cm) HCC (Jiang et al, 2011), and the specificity of AFP is only about 68.2% in detecting the patients with HCC (Johnson, 2001). Although DCP is believed to be a better marker, elevated DCP activity is only present in 44–47% of HCCs <3 cm in size (Grazi et al, 1995; Soga et al, 1998). Thus, both AFP and DCP are insufficient for early diagnosis of HCC.
There is a pressing need to find new serum biomarkers to improve the efficacy of HCC detection. Advances in proteomic analysis have offered exciting opportunities for finding novel biomarkers. Two major strategies are widely used to discover clinically useful biomarkers. One utilises a surface-enhanced laser desorption ionisation-time of flight-mass spectrometry (SELDI-TOF-MS) system (Petricoin et al, 2002; Blanc et al, 2005; Lee et al, 2006; Aivado et al, 2007), and the other is based on 2D SDS–PAGE coupled with MS (Seow et al, 2000; Ward et al, 2006). The SELDI-TOF, which generates the protein patterns by MS, has been considered as a powerful tool for the discovery of new biomarkers (Paradis et al, 2005; Ward et al, 2006). However, the protein peaks detected by SELDI are not easily identified. Thus, these findings cannot provide any information about the biological roles of the marked proteins in the pathogenesis of a disease. On the other hand, the 2D SDS–PAGE-based method provides a lot of information about proteins, including expression levels, actual p/s and molecular weights (Rocken et al, 2004). However, the 2D SDS–PAGE method is not sensitive for resolving hydrophobic, low abundant or low molecular weight proteins. Liquid chromatography-mass spectrometry (LC-MS) is a powerful identification tool for proteins, but it is not suitable for analysing proteins directly (Lim and Elenitoba-Johnson, 2004). It has been suggested that the combination of SELDI-TOF, 2-D and LC-MS may provide a better solution to identify disease-associated proteomic biomarkers (Adkins et al, 2002; Sheng et al, 2006).
HCC is closely associated with chronic inflammation, which produces a repertoire of cytokines and chemokines at low molecular weights. We hypothesise that these low molecular peptides may provide a clue for HCC early detection. The aim of this study was to identify low molecular weight serum protein biomarkers in HCC. The analysis was performed by using SELDI-TOF-MS technology to screen potential protein patterns specific for HCC. Candidate protein peaks were then separated by SDS–PAGE followed by trypsin digestion and identified by LC-MS analysis.
Materials and methods
Collection and preparation of samples for SELDI-TOF-MS analysis
Samples for SELDI-TOF-MS analysis
With patient consent, serum samples were obtained from 80 HCC patients (60 male, 20 female; mean age, 65 years; range, 35–82 years) with HBV/cirrhosis from the Tianjin Medical University Cancer Institute and Hospital, China from January 2005 to November 2007. All HCC patients were diagnosed according to the combined clinical criteria, including imaging data and serum tumour markers, and further confirmed by histopathological analysis. Samples from 50 patients with liver cirrhosis (35 male, 15 female; mean 63 years) and 30 HBV-infected patients (20 male, 10 female; mean 60 years) were kindly provided by the Department of Infection, Tianjin Medical University General Hospital, China. Diagnosis of liver cirrhosis was mainly dependent on clinical history, physical examination, laboratory findings, ultrasonography and/or computed tomography, with or without liver biopsy. Serum samples of HBV-infected patients were collected based on the presence of HBsAg (hepatitis B surface antigen), HBeAg (hepatitis B e antigen) and HBV DNA. None of the patients had received any treatment before collection of blood samples. In addition, 60 blood donors (45 male, 15 female; mean age 61 years; range 50–76 years) without liver neoplasia, alcoholic cirrhosis, hepatitis B or hepatitis C infection were recruited from routine health examination at the Tianjin Medical University Cancer Institute and Hospital, China.
Samples for CCL15 analysis
Patients with carcinoma: with patient consent and approval by the ethical committee, serum samples were obtained from the Tianjin Medical University Cancer Institute and Hospital, China. This group of 125 patients contained 55 patients with hepatitis B-related HCC (the presence of HBsAg, HBeAg and HBV DNA; mean age 58 years), 25 with hepatitis C-related HCC (the presence of HCV RNA; mean age 59 years), 15 with lung cancer (mean age 56 years), 15 with gastric cancer (mean age 60 years) and 15 carcinoma of gallbladder (mean age 57 years).
Control patients: With patient consent, serum samples were obtained from the Tianjin Medical University General Hospital, China and are as follows: benign diseases: liver cirrhosis (n=25; mean age 61 years); calculus of bile duct (n=20; mean age 60 years); B hepatitis (n=20; mean age 59 years); and 60 blood donors (mean age 61 years).
Serum samples from HCC patients were immunodepressed with CCL15 antibody, which was bought from German Roche (Berlin, Germany) and then prepared for SELDI-TOF-MS analysis. All serum samples were separated and divided into 10 μl aliquots, and stored at −80 °C until being assayed.
SELDI-TOF-MS analysis
Protein profiling of serum samples was performed using the eight-spot format WCX-2 (weak cationic exchange) Protein Chip Arrays (Ciphergen Biosystems, Fremont, CA, USA). Frozen serum samples were thawed and spun at 10 000 r.p.m. for 5 min at 4 °C. In all, 20 μl of U9 buffer was added to 10 μl aliquots of each serum sample and placed on ice for 30 min before adding 360 μl WCX-2 buffers. Arrays were prepared as follows: each array was pre-equilibrated 2 × 5 min in 200 μl WCX-2 buffer on a horizontal shaker (MSI Mini-shaker, Benchmark, Brownstown, PA, USA) before sample addition. The sample supernatant was added and incubated for 1 h on the shaker. After incubation, the sample was removed, and each spot was washed with 200 μl WCX-2 buffers for 2 × 5 min with agitation. After washing, the array was carefully separated from the bioprocessor and washed briefly with deionized water. In all, 0.5 μl sinapinic acid was deposited on the array spots and allowed to air dry.
The ProteinChip Arrays were read by SELDI-TOF-MS (ProteinChip PBS II reader, Ciphergen). This was calibrated using NP20 chips that had been bound with all-in-one standard proteins to set up the parameters. The optimal detection parameter of mass/charge size range was set between 5000 and 20 000 M/Z with a maximum of 50 000 M/Z. The laser intensity was set at 175 and detector sensitivity was set at 5. An average value of 130 spots was presented for each sample. All samples were detected with the same parameters. All the raw data was normalised with the ProteinChip Software version 3.1 (homogenisation of the total ion strength and M/Z). The M/Z sample peaks with >2000 M/Z were normalised with biomarker wizard of ProteinChip Software version 3.1 for noise filtering. The first threshold for noise filtering was set at 5, and the second was set at 2. The minimum threshold for clustering was set at 10%. Spectrum analysis was performed using the Biomarker Patterns Software (Ciphergen, Sparta, NJ, USA).
Tricine-SDS–PAGE and LC-MS analysis
Pooled serum samples (n=15) from HCC patients with high SELDI intensities at 7777.27 M/Z were selected. These samples were diluted with 9 ℳ urea, 10 mℳ Tris/HCI (pH 7.4) and applied to AKTA Purifier T-900 column system (Amersham, Piscataway, NJ, USA). After sample purification, albumin and immunoglobulin were removed from the serum by 3GA and then by Protein A. The rest of the fractions were loaded onto a Tricine–SDS–PAGE gel using ETTAN II (Amersham Pharmacia, Piscataway, NJ, USA) gel electrophoresis system. Electrophoresis was run at 20 mA for 3 h. Gels were then stained with Coomassie Brilliant Blue. The bands corresponding to the 8000 M/Z markers were excised and then destained with two washes of 50 μl deionised water, followed by 50 μl ACN/50 mM l−1 NH4HCO3 (1 : 1, v/v), and dried in a SpeedVac concentrator (FisherScientific, Pittsburgh, PA, USA). The dried gel slices were rehydrated with 10 mℳ DTT followed by 50 mℳ IAM (45 min at room temperature in the dark). After several washes with 25 mℳ NH4HCO3 and 100% CAN, 20 mg l−1 solution of trypsin was added to the gel slices and digestion was allowed to proceed at 37 °C for 12 h. The trypsin-digested sample was loaded onto a C18 reversed-phase column (5 mm × 250 μm, PepMapC18, LC Packings, Amsterdam, The Netherlands), and the peptides were separated by electrospray ionisation (ESI, Bruker Esquire 3000, Bruker Daltonik, Bremen, Germany). Proteins were identified by an automated searching algorithm against the SWISS-Protand NCBI protein database.
Immunohistochemistry (IHC) assay
Normal (n=50), adjacent liver tissues (n=80) and HCC tissues (n=80) were processed according to the standard approaches. The anti-CCL15 serum (1 : 1600, Immunechem Pharmaceuticals INC, Beijing, China) was applied to the slides of three groups and incubated in a moist chamber at 4 °C overnight. In all, 0.01 ml PBS was used as the negative control in all the experiments. Slides cut in parallel to the IHC-treated sections were stained by hematoxylin and eosin for better identification of the different tissue areas. To avoid interindividual bias of IHC staining differentiations, all slides were determined by an experienced pathologist.
Western blotting analysis in serum
The serum samples from mixture of HCC and mixture of LC were used in western blotting analysis. The protein concentrations of these pooled samples were measured by DC protein assay (BioRad, Chicago, IL, USA). Fifteen micrograms of total protein of each sample was separated in 4–20% Tris-glycine SDS–PAGE gel (Invitrogen, Carlsbad, CA, USA) and transferred to PVDF membranes. Membranes were blocked by 5% nonfat milk in PBS-0.1% Tween 20 buffer (PBS-T) for 1 h at room temperature and then incubated overnight at 4 °C with mAb of CCL15, followed by HRP-conjugated secondary antibody. Signal was developed using ECL plus Western Blotting Detection Reagents (Amersham Bioscience, Piscataway, NJ). The PVDF membrane was dried and stained with SimplyBlue Safe Stain (Invitrogen) after western blot transferring.
Enzyme-linked immunosorbent assay (ELISA) analysis
Patients’ samples were collected as serum, centrifuged for 15 min at 1500 g and then stored until analysis at −80 °C. CCL15 was determined using a commercially available sandwich enzyme-linked immunosorbent assay (ELISA, Schebo Tech, Giessen, German). Serum determination of AFP was performed by means of an enzyme immunoassay with a commercially available kit only for hepatitis B-related HCC. The measurements of two tumour markers were performed blindly.
Cell culture and reagents
Three human HCC cell lines, including HepG2 (American Type Culture Collection) and Huh-7, HLE (Human Sciences Research Resources Bank, Tokyo, Japan), were used in this study. HepG2 was cultured in MEM media containing 10% foetal bovine serum (FBS), 1% non-essential amino acids and 1% sodium pyruvate (Thermo Scientific Hyclone, Logan, VT, USA), whereas Huh-7 and HLE were cultured in DMEM media with 10% FBS and RPMI1640 with 10% FBS, respectively. Chemotaxis chambers and membranes were from Neuroprobe (Gaithersburg, MD, USA). Recombinant Human CCL15/68 and antibody to CCL15 were from RD Systems (USA). Antibody to CCR1 was from Abcam Inc. (Cambridge, MA, USA). Antibody to β-actin, goat anti-mouse IgG-FITC and donkey anti-goat IgG-FITC were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA).
Gene transfection
2 × 105 cells were cultured in 12-well plates 1 day before transfection in MEM containing 10% FBS, 1% non-essential amino acids and 1% sodium pyruvate. After starvation for 3 h, transfection was performed by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. To establish stable cell lines, the cells were enriched using 0.6 mg ml−1 G418 sulphate (Gibco/Invitrogen, Grand Island, NY, USA) or 0.4 mg ml−1 Hygromycin B from BD Biosciences (Bedford, MA, USA). After the cells grew up, single cell expressing GFP was picked out and cultured into 96-well plates, and then each clone was validated by RT–PCR and western blotting analysis.
RT–PCR
Total RNA from cells was extracted using Trizol (Invitrogen). Then the RNA was transcribed using the RT–PCR Kit from TaKaRa Biotechnology Co. Ltd. (Dalian, China). Specific primers for GAPDH (forward, 5′-GCACCGTCAAGGCTGAGAAC-3′ reverse, 5′-TGGTGAAGACGCCAGTGGA-3′), CCL15 (forward, 5′-CGCGAATTCATGAAGGTCTCCGTGGCTG-3′ reverse, 5′-GCGCTCGAGTTATATTGAGTAGGGCTTCAG-3′), and CCR1 (forward, 5′-CTCTTCCTGTTCACGCTTCC-3′ reverse, 5′-GCCTGAAACAGCTTCCACTC-3′) were from Sangon (Shanghai, China).
Western blotting assay
Western blotting assay was performed as described bySun et al (2005). Cells and clones were washed twice with ice cold PBS pH 7.4 and then lysed by 1 × SDS sample buffer (Tris-HCl, pH 6.8, 62.5 mℳ, 2% SDS, 10% glycerol.). Equal amounts of cell lysates (20 μg total protein per lane) were loaded onto 10% or 14% SDS–PAGE gels, transferred onto PVDF membranes (Millipore, Billerica, MA, USA), probed with anti-CCR1 or anti-CCL15 primary antibody, followed by HRP-conjugated secondary antibody and visualised by enhanced chemiluminescence reagents ECL (Pierce, Rockford, IL, USA). Western blotting analysis of β-actin was used as loading control.
Wound-healing assay
Cells were seeded in 35 mm dishes for 2 days, starved in serum-free medium overnight, after which, scrape wounds were created with a 10-μl sterile plastic pipette tip in the confluent cell monolayers. After washing with PBS, serum-free medium (to prevent cell proliferation) was added. The widths of the wounds were recorded at different time points (0, 3, 6, 9, 12 and 24 h) using the number of grids in the ocular of our microscope. Ten grids represented 1 mm in length, and we could therefore calculate the migration distances by the changes of the grid number. Three measurements at different positions along each scratching line were recorded at each time points. And in order to test the effect of CCL15 on the motility of hepatocarcinoma cells, the serum-free medium containing CCL15 (1 μℳ) was added to the scraped cells and cultured for 24 h.
Chemotaxis assay
Chemotaxis assays were performed essentially as previously described using Boyden chambers equipped with 8 μm porosity polyvinylpyrrolidone-free polycarbonate filters(Carloni et al, 1998). Briefly, polycarbonate filters were coated on the lower surface with 20 μg ml−1 human type I collagen (Collaborative Biomedical Products; Bedford, MA, USA) for 30 min at 37 °C and placed between the lower chamber and upper chamber. The lower chamber was filled with MEM media containing CCL15 as chemoattractants. Serum-deprived HepG2, Huh-7 and HLE cells were separately washed, trypsinized, resuspended in serum-free medium containing 1% albumin at a concentration of 3 × 105 cells ml−1 and placed in the upper chamber. The chambers were incubated at 37 °C in a cell-culture incubator for 6 h. Then the filter membrane was washed, and cells attached were fixed and stained. The number of migrating cells was counted in six randomly chosen fields ( × 400) by light microscope, and the counts were averaged (means±s.d.). Chemotaxis index=the migrating cell number in a chemoattractant gradient/the migrating cell number in a medium control.
Matrigel invasion assay
For invasion assays, 105 cells in 400 μl serum-free MEM medium were plated in the top chamber of Transwells with a Matrigel-coated membrane (Corning Incorporated, Corning, NY, USA) containing 8-μm diameter pores. The inserts were placed into the bottom chamber wells of a 24-well plate containing MEM with CCL15 as a chemoattractant. After 36 h of incubation, cells remaining on the insert top layers were removed by cotton swab scrubbing, as recommended by the manufacturer. Cells on the lower surface of the membrane were fixed and stained. Inserts were washed for several times in distilled water before air drying. The membranes were photographed, and cell counts were determined after totalling five random fields.
Statistical analysis
All statistical analyses were carried out with the use of SPSS software Version 13.0 (Barbecana, Houston, TX, USA). Data were expressed as the mean±s.e. from at least three separate experiments. Statistical significance of the results was analysed by two-way ANOVA and P <0.05 was considered to be statistically significant.
Results
Identification of CCL15 from serum samples of hepatoma patient
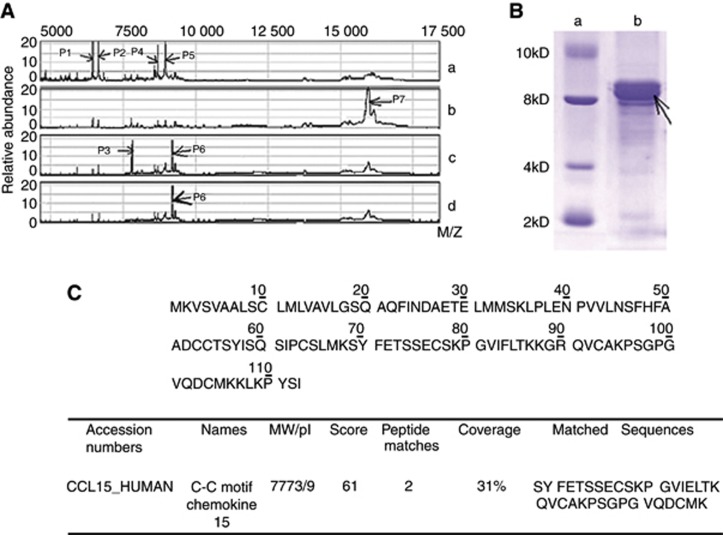
SELDI-MS was used to investigate the expression of low molecular weight peptides in serum samples of HCC patients. As shown in Figure 1A, four peptides, designated as P1 at 6489 Dalton, P2 at 6662 Dalton, P4 at 8593 Dalton, and P5 at 8720 Dalton, were detected in the pooled serum from healthy controls (Figure 1A, Lane a), but not in the serum from patients with benign diseases (Lane b), or HCC (Lane c and d, Table 1). P7 at 16 200 Dalton only appeared in serum from patients with benign diseases, including liver cirrhosis and HBV-carriers (Figure 1A, Lane b). Two bands, P3 at 7777 Dalton and P6 at 9250 Dalton, were detected only in HCC patients (Figure 1A, Lane c).
Figure 1.
Identification of CCL15 in HCC patient serum. (A) Differential expression of the SELDI peaks in the comparisons of four groups. a: the sample from mixture of healthy controls; b: the sample from mixture of benign diseases (liver cirrhosis or hepatitis); c: the sample from mixture of HBV-HCC; and d: the sample from mixture of HBV-HCC after immunodepression assay using CCL15 antibody. P1-P7 shows different peptides or proteins. (B) Isolation of the 7777.27 M/Z peak. Arrow indicates the band of 7777.27 M/Z protein. a: marker; b: sample from mixture of HCC. (C) The 7777.27 M/Z protein-matched CCL15 protein (MW=7773.10 M/Z, pI=9) with two consistent sequences (Score=61).
Table 1. Comparative content of seven different proteins at M/Z in WCX-2.
| Peptides/M/Z (Da) | Comparative content of proteins (%) | |||
|---|---|---|---|---|
|
Proteins
|
Healthy control | Benign diseases | HCC | |
| P1 | 6489.48 | 19.77±3.54a | 6.46±2.18 | 6.87±2.30 |
| P2 | 6662.34 | 22.82±4.15a | 9.96±2.93 | 8.06±2.47 |
| P3 | 7777.27 | 4.09±1.14 | 6.87±2.18 | 15.87±4.30a |
| P4 | 8593.75 | 11.45±2.73a | 2.13±0.89 | 3.21±1.21 |
| P5 | 8720.23 | 13.69±2.30a | 4.57±2.12 | 4.32±2.89 |
| P6 | 9250.00 | 4.51±1.32 | 4.87±1.82 | 23.15±3.81a |
| P7 | 16200.17 | 8.83±2.76 | 26.72±4.51a | 7.63±2.48 |
Abbreviations: HCC=hepatocellular carcinoma; WCX-2=weak cationic exchange.
Benign diseases include liver cirrhosis and hepatitis B.
This value was significantly higher than that in the other two groups (P<0.01).
To isolate the proteins of interest and to determine candidate protein identities, 15 serum samples from HCC patients containing high SELDI intensity of 7777.27 M/Z were pooled and separated by Tricine-SDS–PAGE (Figure 1B). The band at 8 kDa was excised, trypsinized and analysed by LC-MS/MS. As shown in Figure 1C, two peptide sequences were identified, which matched with human CCL15, at a 31% of coverage, suggesting that expression of CCL15 was enhanced in serum from HCC patients. CCL15 antibody was used to treat serums from HCC patients, and the band P3 disappeared after immuno-depletion, showing that P3 at 7777 Dalton, not P6, was CCL15 (Figure1A, Lane d). Taken together, MS revealed that expression of CCL15 was elevated in HCC patients.
CCL15 as a biomarker for HCC
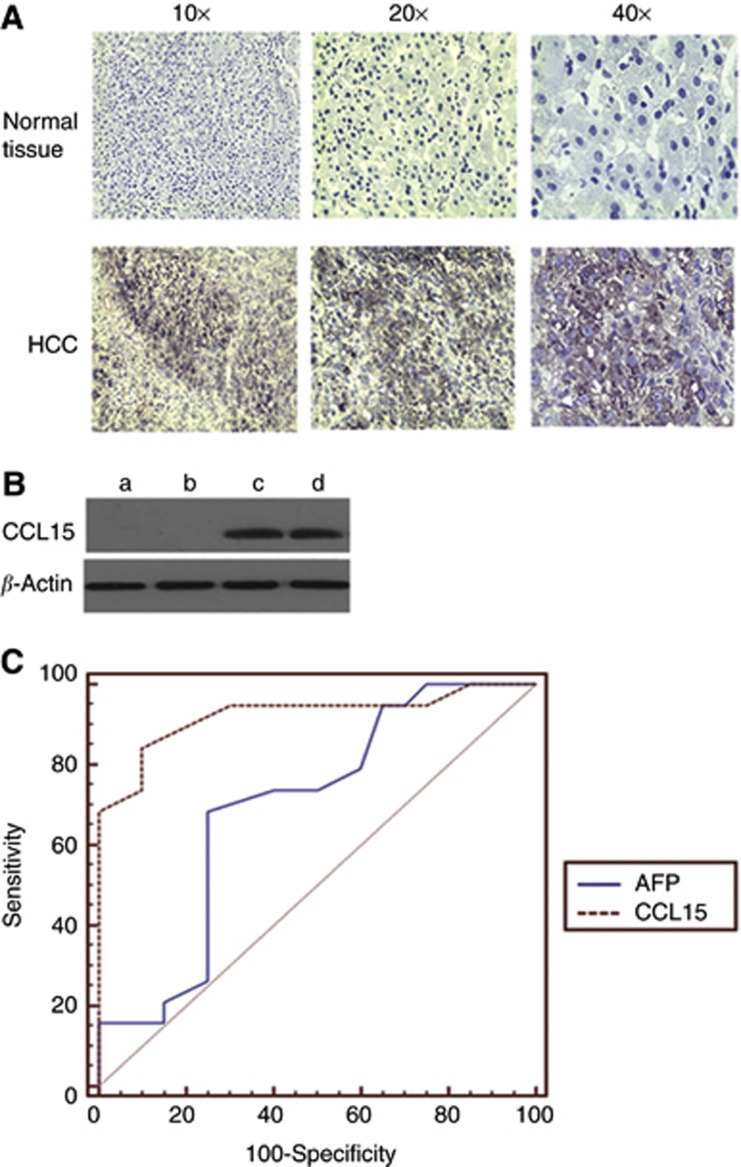
Immunohistochemistry analysis was used to assess CCL15 expression in liver tissues. As shown in Figure 2A, CCL15 were detected on tumour tissues from HCC patients but not in tissues from healthy samples. Among 80 tumour samples, CCL15 were detected in 64 tumour tissues and only in 16 tissues adjacent to tumour (Table 2). Only 1 out of 50 samples from healthy tissues showed positive staining with CCL15. CCL15 expression in liver tumour sera and liver cirrhosis sera was analysed by western blotting. Interestingly, strong staining signals were found in HCC sera (Figure 2B and c), whereas no CCL15 staining was found in liver cirrhosis sera (Figure 2B and a). ELISA was used to assess serum CCL15 levels in HCC patients (Table 3), in other cancer patients and controls (benign patients and healthy controls; Table 4). A significant increase in serum CCL15 was detected in HCC patients and treatment reduced serum levels of CCL15.
Figure 2.
The expression of CCL15 in liver tissues and sera, and its comparison with AFP as a proteomic biomarker. (A) Immunohistochemical staining of CCL15 expression in HCC tissues and adjacent liver tissues. The positive signals for CCL15 were observed in brown. No positive signal of CCL15 was detected in adjacent normal liver tissues. (B) Western blotting analysis of CCL15 expression in HBV-HCC sera and liver cirrhosis sera. Arrow indicates that CCL15 were observed in HCC. a, b: sample from two batches of mixture of liver cirrhosis; c,d; sample from two batches of mixture of HCC; (C) Receiver operation characteristic (ROC) curves in patients with B-related HCC and B-hepatitis.
Table 2. The expression of CCL15 in the liver tissues.
| Liver tissue | Samples | Positive rate (%) | Score |
|---|---|---|---|
| Normal | 50 | 1 (2.00) | 0.12±0.13a |
| Adjacent cancer | 80 | 16 (20.00) | 0.55±1.23b |
| HCC | 80 | 64 (80.00) | 2.25±1.29c |
Abbreviation: CCL15=C-C motif chemokine 15.
Normal vs hepatocellular carcinoma (HCC); P<0.05.
Adjacent cancer vs normal, P>0.05.
Adjacent cancer vs HCC, P<0.05.
Table 3. The relationship between clinicopathological data of patients with HCC and the serum levels of CCL15.
| HCC | CCL15 (pg ml−1) |
|---|---|
| Total (before treatment) (55) | 26.6±1.2 |
|
Subtype
| |
| Poorly differentiated (15) | 30.3±2.2a |
| Moderately differentiated (20) | 24.3±0.6 |
| Well differentiated (20) | 19.1±0.6 |
|
Tumour size
| |
| ⩾3 cm (35) | 27.6±1.2b |
| <3 cm (20) | 19.6±2.1 |
|
Metastasis
| |
| Yes (30) | 27.9±3.6c |
| No (25) | 20.6±1.1 |
| After treatment | 9.7 ±1.2d |
| Operation (25) | 9.9±0.8 |
| Arterial embolization (20) | 10.0±1.4 |
| Percutaneous tumour ablation (10) | 9.0±1.6 |
| Healthy controls (60) | 1.2±0.1 |
Abbreviations: CCL15=C-C motif chemokine 15; HCC=hepatocellular carcinoma.
Showed P<0.05 between poorly differentiated and well differentiated.
Showed P<0.05 between tumour size ⩾3 and <3 cm.
Showed P<0.05 between patients with metastasis and without metastasis.
Showed P<0.05 between patients before and after treatments.
Table 4. Differential levels of CCL15 in various groups.
| Groups | CCL15 (pg ml−1) |
|---|---|
|
Cancer
| |
| B-related HCC (55) | 26.6±1.2 |
| C-related HCC (25) | 23.9±3.1a |
| Lung cancer (15) | 4.3±1.7b |
| Gastric cancer (15) | 4.1±0.5c |
| Carcinoma of gallbladder (15) | 3.6±1.5d |
| Benign diseases (65) | 2.0±0.9e,f |
| Healthy controls (60) | 1.2±0.1g |
Abbreviations: CCL15=C-C motif chemokine 15; HCC=hepatocellular carcinoma.
Showed P>0.05 between B-related HCC and C-related HCC.
Showed P<0.05 between B-related HCC and lung cancer.
Showed P<0.05 between B-related HCC and gastric cancer.
Showed P<0.05 between B-related HCC and carcinoma of gallbladder.
Showed P<0.05 between B-related HCC and benign diseases.
Showed P>0.05 between benign diseases and healthy controls.
Showed P<0.05 between B-related HCC and healthy controls.
Receiver operation characteristic curves were plotted to assess the specificity and sensitivity of CCL15 as a biomarker for HCC in distinction to chronic hepatitis B (Figure 2C). The AUC (95% CI) was 0.964 (0.812–0.989), 0.723(0.555–0.861) for CCL15 and AFP, respectively. A significant difference was noted between the AUC for CCL15 and those for AFP (P<0.05), suggesting that CCL15 was a better biomarker to AFP. Based on the maximisation of the Youden index, the optimal cutoff value was 16 ng ml−1 for CCL15 (sensitivity, 88.2% specificity, 93%) and 25 μg l−1 for AFP (sensitivity, 72.4% specificity, 81%).
CCL15 promoted HCC cell migration and invasion
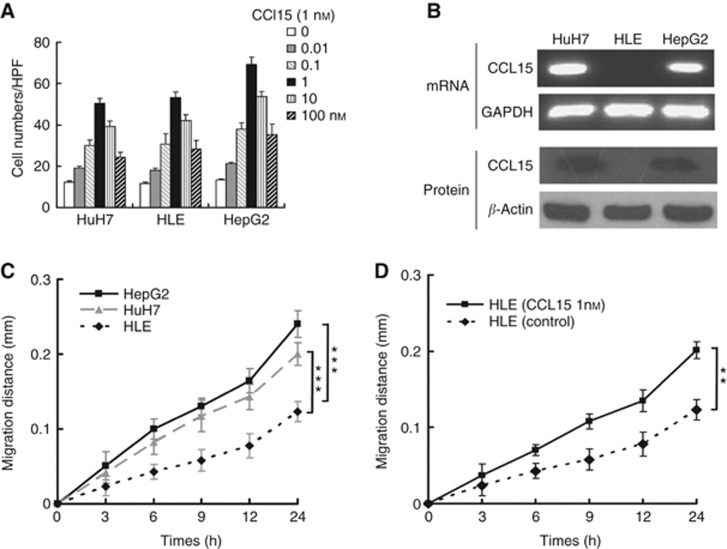
CCL15, a chemokine, induces cell migration by binding to CCR1, a receptor expressed in most liver cells (Brown et al, 2007). We hypothesise that elevated CCL15 promoted HCC cell migration and invasion. To investigate the role of CCL15, a micro-Boyden chamber assay was performed on HCC cells. CCL15 induced robust chemotaxis of Huh7, HLE and HepG2 cells, indicating that CCL15 mediated HCC cell migration (Figure 3A). Immunohistochemistry results suggested that CCL15 was expressed by HCC cells (Figure 2A). To confirm this result, we examined the expression of CCL15 in HCC cell lines. CCL15 mRNA and protein were detected in both Huh7 and HepG2, two HCC cell lines, but not in HLE, less malignant cells (Figure 3B). A scratch assay was used to further assess HCC migration. As shown in Figure 3C, both HepG2 and Huh7 showed a stronger migration capacity than HLE cells. Treatment with CCL15 induced a marked increase in HLE cell migration (Figure 3D). Taken together, the results clearly showed that CCL15 promoted HCC cell migration.
Figure 3.
Effects of CCL15 on hepatocarcinoma cells chemotaxis and migration. (A) Chemotaxis assay of three kinds of hepatocarcinoma cells with indicated amount of CCL15 as chemoattractant for 6 h. (B) RT–PCR and western blotting analysis of CCL15 in Huh-7, HLE and HepG2 cells. (C) Wound-healing assay of the three kind of hepatocarcinoma cells. Wound widths were recorded at indicated time points. ***P<0.0005. (D) Wound-healing assay of HLE cells with or without CCL15 (1 nℳ) treatment. ***P<0.005.
Knockdown CCL15 impaired HCC cell invasion
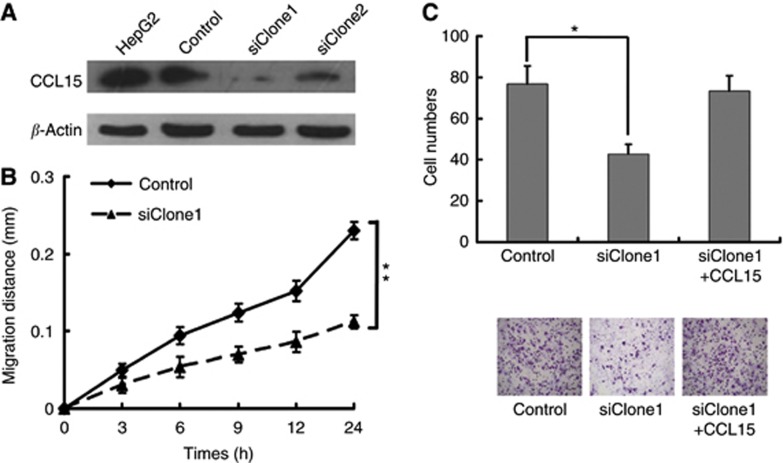
CCL15 may be secreted from two major sources, tumour infiltrated leucocytes and HCC cells. CCL15 from HCC may be sufficient to promote tumour invasion and micrometastasis. To further confirm that CCL15 from HCC had an active role in invasion, expression of CCL15 was knocked down by small RNA interference (Figure 4A). A scratch assay showed a decrease in HepG2 migration, suggesting that CCL15 from HepG2 functioned in an autocrine way to promote HepG2 migration (Figure 4B). A Matrigel analysis showed that knockdown of CCL15 impaired HepG2 cell invasion, while supplement with CCL15 restored the invasiveness (Figure 4C). Thus, the results suggested that CCL15 from HCC was sufficient to promote tumour invasion in an autocrine mode.
Figure 4.
Downregulation of CCL15 impaired HepG2 cells migration and invasion. (A) Western blotting analysis of HepG2 cells and its clones with downregulation of CCL15. (B) Wound-healing assay of HepG2 cells and siCCL15/HepG2 cells at indicated time points. (C) Invasion assays of HepG2 cells and siCCL15/HepG2 cells. CCL15 coated in Matrigel-enhanced HepG2/siClone2 cells invasive ability, with pictures shown ( × 200; *P<0.05, **P<0.005).
Discussion
SELDI has been successfully applied in defining early detection biomarkers in multiple cancers, including ovarian, prostate and breast cancers (Zhang et al, 2004; Hu et al, 2005; Le et al, 2005). However, it is rather a challenge to identify the molecular identity of these novel biomarkers. Our results revealed CCL15 as a HCC biomarker by using SELDI in combination with SDS–PAGE and LC-MS/MS. SELDI analysis showed two molecules at 7.8 and 9.3 kD specifically expressed in serum from HCC patients. SDS–PAGE and LC-MS/MS analysis showed that CCL15 was specifically expressed in serum of HCC patient. Treatment with anti-CCL15 antibody eliminated SELDI signal at 7.8 kD, confirming that the CCL15 was specifically expressed in serum of HCC patients. Taken together, both SELDI and SDS–PAGE/LC-MS/MS results suggest that CCL15 is a specific serum biomarker for HCC patients.
Micrometastasis is a major cause of failure in HCC treatments. Identification of CCL15 from patient tumour tissues provides a new insight into tumorigenesis and micrometastasis process of HCC. CCL15 is a peptide in chemokine family, which has a pivotal role in mediating leucocyte trafficking and essential for inflammatory responses (Richter et al, 2005; Kwon et al, 2008). Receptors of CCL15, CCR1, were expressed in liver (Nomiyama et al, 2001). Our results clearly showed that CCL15 enhanced migration and invasion of HCC cells. Furthermore, it has been reported that CCL15 also increased the expression of matrix metalloproteinase expression and induced angiogenesis (Hwang et al, 2004; Kwon et al, 2008). Thus, CCL15/CCR1 may have a critical role in the micrometastasis of HCC and can be used as a target for anti-metastasis treatment.
The most effective treatment of HCC is surgery, which is typically applied to small HCC (<3 cm). Thus, the key to improve survival rate is to detect tumour at early stage. Current serum biomarker, AFP, has its limitation in sensitivity and approximately 30% HCC patients do not show an elevation in AFP. Our preliminary investigations suggest that CCL15 may be a promising candidate, which shows a better AUC (95%) at 0.964 by testing 55 HCC patients and 60 health donors. Although, this investigation focused on HBV-HCC patients, based on our studies of HCC cell lines, we speculate that CCL15 may also have an important role in other types of HCC. Further clinical investigation by using larger sample sizes is required to determine the diagnosis value of CCL15. It is also possible that a combination of AFP and CCL15 will be a more sensitive and specific way to detect HCC at an early stage.
Acknowledgments
This work was supported by research grants from 973 programme grants (No. 2011CB933100 and No. 2010CB933900), National Scientific Foundation of China (NSFC No. 81201646, No. 81072160, No. 81101754) and Changjiang Scholars and Innovative Research Team in University in China (Grant IRT1076).
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL, Pounds JG (2002) Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics 1(12): 947–955 [DOI] [PubMed] [Google Scholar]
- Aivado M, Spentzos D, Germing U, Alterovitz G, Meng XY, Grall F, Giagounidis AA, Klement G, Steidl U, Otu HH, Czibere A, Prall WC, Iking-Konert C, Shayne M, Ramoni MF, Gattermann N, Haas R, Mitsiades CS, Fung ET, Libermann TA (2007) Serum proteome profiling detects myelodysplastic syndromes and identifies CXC chemokine ligands 4 and 7 as markers for advanced disease. Proc Natl Acad Sci USA 104(4): 1307–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc JF, Lalanne C, Plomion C, Schmitter JM, Bathany K, Gion JM, Bioulac-Sage P, Balabaud C, Bonneu M, Rosenbaum J (2005) Proteomic analysis of differentially expressed proteins in hepatocellular carcinoma developed in patients with chronic viral hepatitis C. Proteomics 5(14): 3778–3789 [DOI] [PubMed] [Google Scholar]
- Brown MF, Bahnck KB, Blumberg LC, Brissette WH, Burrell SA, Driscoll JP, Fedeles F, Fisher MB, Foti RS, Gladue RP, Guzman-Martinez A, Hayward MM, Lira PD, Lillie BM, Lu Y, Lundquist GD, McElroy EB, McGlynn MA, Paradis TJ, Poss CS, Roache JH, Shavnya A, Shepard RM, Trevena KA, Tylaska LA (2007) Piperazinyl CCR1 antagonists--optimization of human liver microsome stability. Bioorg Med Chem Lett 17(11): 3109–3112 [DOI] [PubMed] [Google Scholar]
- Carloni V, Romanelli RG, Mercurio AM, Pinzani M, Laffi G, Cotrozzi G, Gentilini P (1998) Knockout of alpha6 beta1-integrin expression reverses the transformed phenotype of hepatocarcinoma cells. Gastroenterology 115(2): 433–442 [DOI] [PubMed] [Google Scholar]
- Fujiyama S, Tanaka M, Maeda S, Ashihara H, Hirata R, Tomita K (2002) Tumor markers in early diagnosis, follow-up and management of patients with hepatocellular carcinoma. Oncology 62(Suppl 1): 57–63 [DOI] [PubMed] [Google Scholar]
- Grazi GL, Mazziotti A, Legnani C, Jovine E, Miniero R, Gallucci A, Palareti G, Gozzetti G (1995) The role of tumor markers in the diagnosis of hepatocellular carcinoma, with special reference to the des-gamma-carboxy prothrombin. Liver Transpl Surg 1(4): 249–255 [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhang S, Yu J, Liu J, Zheng S (2005) SELDI-TOF-MS: the proteomics and bioinformatics approaches in the diagnosis of breast cancer. Breast 14(4): 250–255 [DOI] [PubMed] [Google Scholar]
- Hwang J, Kim CW, Son KN, Han KY, Lee KH, Kleinman HK, Ko J, Na DS, Kwon BS, Gho YS, Kim J (2004) Angiogenic activity of human CC chemokine CCL15 in vitro and in vivo. FEBS Lett 570(1-3): 47–51 [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2): 69–90 [DOI] [PubMed] [Google Scholar]
- Jiang J, Wu C, Shen Y, Xu B, Zheng X, Li X, Xu N (2011) Clinical application of determining serum AFP-IgM complexes for diagnosis of small hepatocellular carcinoma. Anticancer Res 31(2): 687–691 [PubMed] [Google Scholar]
- Johnson PJ (2001) The role of serum alpha-fetoprotein estimation in the diagnosis and management of hepatocellular carcinoma. Clin Liver Dis 5(1): 145–159 [DOI] [PubMed] [Google Scholar]
- Kwon SH, Ju SA, Kang JH, Kim CS, Yoo HM, Yu R (2008) Chemokine Lkn-1/CCL15 enhances matrix metalloproteinase-9 release from human macrophages and macrophage-derived foam cells. Nutr Res Pract 2(2): 134–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le L, Chi K, Tyldesley S, Flibotte S, Diamond DL, Kuzyk MA, Sadar MD (2005) Identification of serum amyloid A as a biomarker to distinguish prostate cancer patients with bone lesions. Clin Chem 51(4): 695–707 [DOI] [PubMed] [Google Scholar]
- Lee IN, Chen CH, Sheu JC, Lee HS, Huang GT, Chen DS, Yu CY, Wen CL, Lu FJ, Chow LP (2006) Identification of complement C3a as a candidate biomarker in human chronic hepatitis C and HCV-related hepatocellular carcinoma using a proteomics approach. Proteomics 6(9): 2865–2873 [DOI] [PubMed] [Google Scholar]
- Lim MS, Elenitoba-Johnson KS (2004) Proteomics in pathology research. Lab Invest 84(10): 1227–1244 [DOI] [PubMed] [Google Scholar]
- Marrero JA, Lok AS (2004) Newer markers for hepatocellular carcinoma. Gastroenterology 127(5 Suppl 1): S113–S119 [DOI] [PubMed] [Google Scholar]
- Nomiyama H, Hieshima K, Nakayama T, Sakaguchi T, Fujisawa R, Tanase S, Nishiura H, Matsuno K, Takamori H, Tabira Y, Yamamoto T, Miura R, Yoshie O (2001) Human CC chemokine liver-expressed chemokine/CCL16 is a functional ligand for CCR1, CCR2 and CCR5, and constitutively expressed by hepatocytes. Int Immunol 13(8): 1021–1029 [DOI] [PubMed] [Google Scholar]
- Paradis V, Degos F, Dargere D, Pham N, Belghiti J, Degott C, Janeau JL, Bezeaud A, Delforge D, Cubizolles M, Laurendeau I, Bedossa P (2005) Identification of a new marker of hepatocellular carcinoma by serum protein profiling of patients with chronic liver diseases. Hepatology 41(1): 40–47 [DOI] [PubMed] [Google Scholar]
- Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, Liotta LA (2002) Use of proteomic patterns in serum to identify ovarian cancer. Lancet 359(9306): 572–577 [DOI] [PubMed] [Google Scholar]
- Richter R, Bistrian R, Escher S, Forssmann WG, Vakili J, Henschler R, Spodsberg N, Frimpong-Boateng A, Forssmann U (2005) Quantum proteolytic activation of chemokine CCL15 by neutrophil granulocytes modulates mononuclear cell adhesiveness. J Immunol 175(3): 1599–1608 [DOI] [PubMed] [Google Scholar]
- Rocken C, Ebert MP, Roessner A (2004) Proteomics in pathology, research and practice. Pathol Res Pract 200(2): 69–82 [DOI] [PubMed] [Google Scholar]
- Seow TK, Ong SE, Liang RC, Ren EC, Chan L, Ou K, Chung MC (2000) Two-dimensional electrophoresis map of the human hepatocellular carcinoma cell line, HCC-M, and identification of the separated proteins by mass spectrometry. Electrophoresis 21(9): 1787–1813 [DOI] [PubMed] [Google Scholar]
- Sheng S, Chen D, Van Eyk JE (2006) Multidimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phase of the human serum proteome: optimization and protein database. Mol Cell Proteomics 5(1): 26–34 [DOI] [PubMed] [Google Scholar]
- Soga K, Watanabe T, Aikawa K, Toshima M, Shibasaki K, Aoyagi Y (1998) Serum des-gamma-carboxyprothrombin level by a modified enzyme immunoassay method in hepatocellular carcinoma: clinical significance in small hepatocellular carcinoma. Hepatogastroenterology 45(23): 1737–1741 [PubMed] [Google Scholar]
- Sun R, Gao P, Chen L, Ma D, Wang J, Oppenheim JJ, Zhang N (2005) Protein kinase C zeta is required for epidermal growth factor-induced chemotaxis of human breast cancer cells. Cancer Res 65(4): 1433–1441 [DOI] [PubMed] [Google Scholar]
- Ward DG, Cheng Y, N'Kontchou G, Thar TT, Barget N, Wei W, Billingham LJ, Martin A, Beaugrand M, Johnson PJ (2006) Changes in the serum proteome associated with the development of hepatocellular carcinoma in hepatitis C-related cirrhosis. Br J Cancer 94(2): 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witjes CD, de Man RA, Eskens FA, Dwarkasing RS, Zondervan PE, Verhoef C, Ijzermans JN (2010) [Hepatocellular carcinoma: the significance of cirrhosis for treatment and prognosis--retrospective study]. Ned Tijdschr Geneeskd 154: A1747. [PubMed] [Google Scholar]
- Zhang Z, Bast RC, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY, Berchuck A, Van Haaften-Day C, Hacker NF, de Bruijn HW, van der Zee AG, Jacobs IJ, Fung ET, Chan DW (2004) Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res 64(16): 5882–5890 [DOI] [PubMed] [Google Scholar]