Abstract
Background:
Aberrant mitogen/extracellular signal-regulated kinase 5 (MEK5)–extracellular signal-regulated protein kinase 5 (ERK5)-mediated signalling has been implicated in a number of tumour types including prostate cancer (CaP). The mechanism for ERK5 activation in CaP remains to be fully elucidated. Studies have recently implicated the role of microRNA (miRNA) mir143 expression in the regulation of ERK5 expression.
Methods:
We utilised a tissue microarray (TMA) of 530 CaP cores from 168 individual patients and stained for both mir143 and ERK5. These TMAs were scored by a combination of observer and automated methods.
Results:
We observed a strong inverse relation between ERK5 and mir143, which manifested itself most strongly in the subgroup of 417 cores with non-zero mir143 and ERK5 immunoreactivity, or with only one of mir143 or ERK5 being zero (cc=0.2558 and P<0.0001). Mir143 neither correlate with Gleason scores or prostate-specific antigen levels, nor was it a predictor of disease-specific survival on univariate analysis.
Conclusion:
Although the mechanism for ERK5 activation in CaP remains to be fully elucidated, we have further validated the potential role of mir143 in regulating ERK5 levels in the clinical context. In addition, we demonstrate that the automated counting method for nuclear ERK5 is a clinically useful alterative to observer counting method in patient stratification in the context of ERK5 targeting therapy.
Keywords: mir143, ERK5, prostate cancer
Prostate cancer (CaP) is a major cause of cancer morbidity and mortality in the Western world, and its incidence continues to rise worldwide with routine prostate-specific antigen (PSA) testing and aging populations (Hsing et al, 2000; Andriole et al, 2009; Schroder et al, 2009). Prostate carcinogenesis represents a complex multistep process, initiated by the accumulation of mutations in cancer genes to transform the benign prostatic epithelium to locally invasive disease, which ultimately may progress to form metastasis (Abate-Shen and Shen, 2000). CaP is heterogeneous with highly variable risk for disease progression. It is therefore important to study how key cellular signalling pathways drive the development of CaP, and to define potential and biological relevant biomarkers to identify patients with poor prognosis and/or predict tumour response to conventional or novel targeted therapies.
Recent work by our laboratory has demonstrated the importance of the MEK5/ERK5 (mitogen/extracellular signal-regulated kinase 5/extracellular signal-regulated protein kinase 5) cascade in invasive CaP (Mehta et al, 2003; McCracken et al, 2008; Ramsay et al, 2011). ERK5 is a member of the mitogen-activated protein kinase (MAPK) family, and is a specific substrate of MEK5, responding to a number of mitogens, cytokines and stresses (Ranganathan et al, 2006). Work from our research group and others have proposed that the MEK5–ERK5 pathway represents an attractive potential target for cancer therapy, with application in both untreated (primary and metastatic) CaP and castrate-resistant disease (McCracken et al, 2008; Ramsay et al, 2011).
The mechanism for ERK5 activation in CaP remains to be fully elucidated. MicroRNA (miRNA) mir143 expression has recently been implicated to regulate ERK5 expression (Clape et al, 2009; Ramsay et al, 2011). MicroRNAs are a class of evolutionary conserved small non-coding RNAs, critically involved in the regulation of mRNA function by controlling the stability and/or translation efficiency of the target transcripts (He and Hannon, 2004; Lim et al, 2005). Target recognition is mediated by the RNA-induced silenced complex that allows the matching between mature miRNAs and mRNA 3′-untranslated region. Recent evidence suggests that miRNAs control a plethora of basic cellular functions, ranging from survival to apoptosis (Bartel, 2004; Care et al, 2007; Thum et al, 2008). By targeting oncogenes or tumour suppressors, miRNA are classified as anti-oncomiRs or oncomiRs, respectively. miRNA143 represents one of the best-characterised anti-oncomiRs. miR143 is located at a fragile site, often deleted in cancers, and its expression is frequently downregulated in cancer cell lines and in clinical tumours including colon and gastric cancer and B cell lymphoma. Of particular interest to us, data from a number of independent studies have revealed that mir143 expression is consistently downregulated in CaP (Clape et al, 2009; Leite et al, 2011; Peng et al, 2011; Xu et al, 2011; Wach et al, 2012). However, whether the expression of mir143 and ERK5 are indeed inversely associated in a large clinical cohort of prostate tumours has not been previously been tested. Here, we report our findings showing a significant correlation between low mir143 and elevated ERK5 levels in primary human prostate cancers.
Materials and methods
Clinical samples
Five hundred thirty cancer cores from 168 individual patients within the entire TAPG cohort were studied (Cuzick et al, 2006; Berney et al, 2009). National approval was obtained from the Northern Multi Research Ethics Committee, MREC/99/3/40 followed by local ethics committee approval at each of the collaborating hospital trusts. In short, men were included in this study if they were under 76 years of age at diagnosis and had clinically localised CaP diagnosed between January 1990 and December 1996. Patients who had a radical prostatectomy or radiation therapy within 6 months of diagnosis, or clear evidence of metastatic disease (by bone scan, X-ray, CT scan, MRI, bone biopsy, lymph node biopsy or pelvic lymph node dissection) or clinical indications of metastatic disease (including pathologic fracture, soft tissue metastasis, spinal compression or bone pain) at or within 6 months of diagnosis, were excluded. Eligibility was established by review of patient records by registry data collection officers and trained medical staff. Clinical staging was centrally reviewed. All patients had centralised Gleason grading by a panel of genitourinary pathologists and had initial diagnostic serum PSA available. Blocks from the trans-urethral resection specimens, which were available, were identified and the corresponding haematoxylin and eosin sections marked for cancerous areas. These were microarrayed in a series of 24 blocks using 0.6-mm cylinders of tissue. Four cores were taken from different areas of tumour to account for tumour heterogeneity in each case, and areas of adjacent normal tissue were also sampled.
Immunohistochemical analysis
Immunohistochemistry for ERK5 was performed as previously described (Gnanapragasam et al, 2003). For automated image analysis for ERK5 immunoreactivity, digitised slides were accessed through the Slidepath Digital Image Hub (Dublin, Ireland) and evaluated with the program’s nuclear scoring algorithm (Mohammed et al, 2012). Individual tissue microarray (TMA) cores were identified, annotated on the scanned image and associated with TMA map entries. Individual nuclei stained with polymerised diaminobenzidine were identified by a thresholding and segmentation algorithm that outlined the individual nuclei and separated touching nuclei. A nuclear size limit unique to these samples was specified to accept or reject individual nuclei to be quantified. Staining for ERK5 in each nucleus was classified as positive or negative, based on thresholds specified by the observer. Pseudo-colours (red/blue) display these staining intensity measurements for individual nuclei, allowing thresholds to be chosen appropriately (Mohammed et al, 2012). These thresholds were chosen using a sample of TMA cores from the entire patient cohort, and once chosen they were used for analysis for the rest of the study without further adjustment.
Similarly, in situ hybridisation for mir143 mRNA was performed as previously reported (Clape et al, 2009). Stained slides were scanned using a Hamamatsu NanoZoomer (Hertfordshire, UK). Visualisation was carried out using the Slidepath Tissue IA system version 3.0. The score for staining signals were measured manually using the histoscore scale from 0 (no staining) to 300 (very strong staining), and then converted to a scale from 0 to 3 depending on the histoscore (0=0, 1=1–100, 2=101–200, 3=201–300).
Statistics
Variation between human and automated methods of nuclear ERK5 and mir143 was examined to facilitate comparisons with published studies using the interclass correlation coefficients. The inter-observer reliability analysis using the Kappa statistic (κ) was performed to determine consistency between the human and automated methods (values of κ from 0.40 to 0.59 are considered to represent at best moderate agreement; 0.60–0.79 good; and 0.80 or more, very good agreement) (Landis and Koch, 1977). Analysis was performed using STATA version 11.2 (StataCorp LP, College Station, TX, USA). Additional analysis to test for potential correlation between the expression of mir143 and Ki67 in the patient cohort studied was carried out based on previously data on Ki67 (Berney et al, 2009).
In silico analysis of mir143 gene targets
In silico prediction of gene targets for mir143 was performed using the TargetScan (www.targetscan.org, Release 6.2: June 2012) and verified using both miRanda (http://www.microrna.org, August 2010 Release) and PicTar (http://pictar.mdc-berlin.de/). The target gene list was ranked according to the PCT, the probability of conserved targeting, as described by Friedman et al (2009). Using the method, PCT scores reflect the Bayesian estimate of the probability that a site is conserved because of selective maintenance of miRNA targeting rather than by chance or any other reason not pertinent to miRNA targeting. Pathway analysis was carried out using Genego software (www.genego.com) to highlight the pathways that were over-represented. The list of the mir143 targets was uploaded to the GeneGo, and the function for ‘Enrichment by Protein Function’ was applied.
Results
Patients demographics
Immunohistochemical staining for ERK5 and mir143 was recorded for 530 tumour rich cores from 168 individual patients. Majority of the cases were represented by at least three independent tissue cores, with ⩾3 tissue cores for 116 out of 168 (70%) patients (Supplementary Figure 1).
Loss of mir143 expression correlates with upregulated ERK5 immunoreactivity but not with Ki67 level
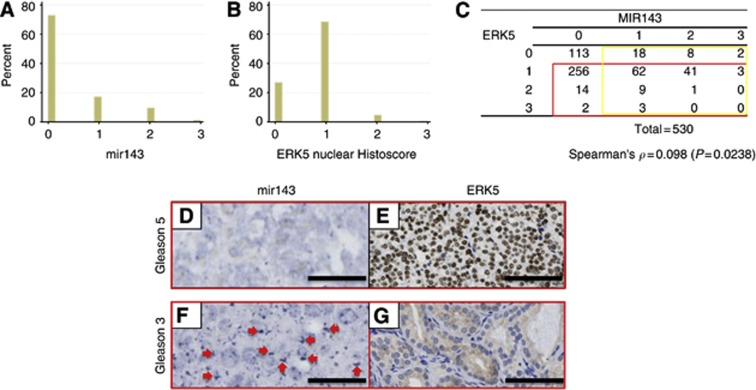
As previously mentioned a number of independent studies have revealed that mir143 expression is consistently downregulated in CaP (Clape et al, 2009; Leite et al, 2011; Peng et al, 2011; Xu et al, 2011; Wach et al, 2012) (summarised in Supplementary Table 1). In our patient cohort (530 CaP cores representing samples from 168 patients), nuclear ERK5 immunoreactivity and mir143 mRNA expression were studied. Initially, the ERK5 samples were scored using the Slidepath Image Analysis software. The data was then converted into scores ranging from 0 to 3, to facilitate correlative analysis with data from mir143 expression. We found that majority of the cores studied exhibited ERK5 score of 1 (simplified from histoscore range 0–300, median 110), whereas majority of the cases were negative for mir143 expression with score of 0 (Figures 1A and B).
Figure 1.
Bar charts summarising the distribution (in percentage) among the CaP cases with different expression scores (0–3) for mir143 (A) and ERK5 (B). (C) The correlative analysis for mir143 and ERK5 expression (Spearman’s ρ=0.098, P=0.0238). Yellow box highlights samples with scores for mir143>0, whereas red box highlights samples with scores for ERK>0. The inverse relationship between the two markers manifests itself most strongly in the subgroup of 417 cores with non-zero mir143 and ERK5 readings (yellow and red boxes combined), or with only one of mir143 or ERK5 being zero (yellow and red boxes, respectively). Representative images showing an inverse relationship between mir143 and ERK5 expression. D/E (Gleason 5) and F/G (Gleason 3) are paired images from two individual patients. For each patient’s samples, namely D/E and F/G, represent tissue from sequential sections were analysed. D shows suppressed mir143 expression, whereas F demonstrated detectable mir143 transcript expression (red arrows). E and G demonstrate upregulated and negligible nuclear ERK5 immunoreactivity, respectively. Black bar measures 200 mm ( × 20 objective).
ERK5 immunoreactivity was observed in the malignant epithelium as previously reported. Of note, a proportion of ERK5 expressing cells are also noted to have significant nuclear ERK5 localisation (Figure 2), which signifies activated ERK5. Mir143 mRNA expression was detected by in situ hybridisation and identified as punctate signals predominantly within the cytoplasm (Figure 2). Figure 1D–G illustrated the inverse relationship between ERK5 immunoreactivity and mir143 expression in sequential histology sections.
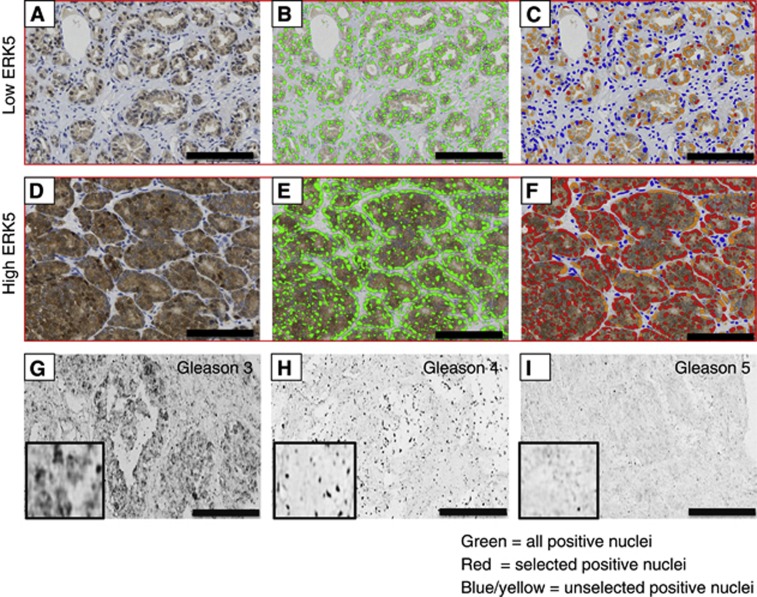
Figure 2.
Gleason pattern grade 3 (A–C) and Gleason pattern grade 5 (D–F) tumours were scored using the automated system. A and D demonstrate low and high ERK5 immunoreactivity in Gleason grade 3 and 5 tumours, respectively. B and E show all of the epithelial nuclei highlighted green, whereas C and F show the classification of ERK5 within each core as positive (red) or negative (blue) based on the adjustable input parameters (see details in Materials and Method section). Scores for sections represented in A–C were negative (0), weak (1), moderate (2) and strong (3) for nuclear immunoreactivity at 88%, 0%, 11%, 1%, respectively, giving a histoscore of 25. Score for sections in D–F were negative, weak, moderate and strong nuclei at 17%, 0%, 31%, 52%, respectively, giving a histoscore of 218. G–I represent typical in situ hybridisation images for mir143 expression: high, moderate to low for G, H and I, respectively. Black bar measures 200 mm ( × 20 objective).
As the two variables (mir143 and ERK5 expression) are not normally distributed, Spearman’s correlation was applied for the analysis. Analysing the entire 530 CaP cores, the Spearman correlation tests were significant (cc=0.0982, P=0.0238) (Figure 1C). However, if we restricted the analysis to the cancer cores (n=116) with both non-zero mir143 and non-zero ERK5 readings, we only observed an insignificant trend for inverse correlation (cc=−0.1817 and P=0.0509). Finally, if we only excluded cores with both mir143 and ERK5 readings being zero (which were considered ‘not informative’), among these 417 cores, there was a strong inverse correlation between mir143 and ERK5 expression (cc=−0.2558 and P<0.0001). Thus, the inverse relation between the two markers manifests itself most strongly in the subgroup of 417 cores with non-zero mir143 and ERK5 readings, or with only one of mir143 or ERK5 being zero.
As mir143 modulates the expression of a large number of target genes (Wach et al, 2012), we wished to test the impact of mir143 as an anti-oncomiR and test whether its expression was related to Ki67 status (Supplementary Figure 2). Corroborating against data from previous analysis Ki67 expression (Berney et al, 2009), we found no evidence of inverse correlation between mir143 and Ki67 (treated dichotomous with cutoff 5). Data on 428 tissue cores representing 133 cases were available for both mir143 expression and Ki67 immunoreactivity, with Pearson χ2=4.7353 and 4.7619 for tissue cores and individual cases, respectively (with corresponding P-values at 0.192 and 0.190), signifying the absence of overall impact on cancer cell proliferation with reduced mir143 expression.
Among the studied tumour samples, mir143 expression did not correlate with tumour Gleason scores (differentiation) (Spearman’s ρ=0.114, P=0.172) or PSA levels (Spearman’s ρ=0.139, P=0.099). In a univariate analysis, mir143 was not a predictor of disease-specific survival.
In silico analysis of mir413 target genes
Despite the significant inverse correlation between mir143 and ERK5 expression, the levels of mir143 was not found to correlative with neither tumour parameters nor patient survivals. We next performed a comprehensive survey for target genes according to consensus mir143 binding sites using three independent informatics softwares. On the basis of analyses using TargetScan, a total of 404 conserved target genes (containing 421 conserved sites and 210 poorly conserved sites) were identified to be putative mir143-targeted genes (Supplementary Table 2). Supporting data in the literature and consistent with findings in our study, ERK5 (or MAPK7) was among the highest ranked target genes based on TargetScan analysis, with an aggregate PCT score of 0.78, which is the 7th highest PCT score. Similarly, out of the 7990 predicted targets for mir143, analysis using miRanda identified ERK5 to be 6th highest ranked with mirSVR score of −0.7511. Interestingly, analysis with PicTar did not identify ERK5 as one of the targets. However, only 217 predicted targets were suggested by PicTar, and it is possible that the stringency of this analysis may result in false negative in identifying mir143 target genes. It is worth noting that kinases, in which ERK5 belongs, is among the most over-represented pathway an mir143 target (Supplementary Table 3).
Owing to the lack of correlation between mir143 expression and clinical correlates despite its association with ERK5 expression, we hypothesised that, due to the large number of putative target genes, the overall effects of mir143 on growth and tumourigenesis may be balanced out due to effects on genes with opposing function. On the basis of the gene list from TargetScan analysis, we carried out analysis utilising the ‘Enrichment by Protein Function’ tool within GeneGo software to examine enrichment of mir143 targets by objects from different protein classes. The three most enriched protein classes were kinases, transcription factors and phophatases (Table 1). Although enzymes represented the most abundant single protein class by function (at 11%) within the gene list, this protein class was however not over-represented in the mir143 target gene list (observed ratio=0.95). Kinases were over-represented with 27 of 450 objects (6%) present within the target gene list (P=0.003). Similarly, transcription factors were also significantly over-represented with 34 of 450 objects (7.56% P=0.0008). Hence, our working hypothesis is that the functional impact of mir143 loss is modulated by the overall effects of regulated kinases and transcription factors.
Table 1. Analysis of mir143 target gene set using GeneGo for enrichment of genes according to their protein function categories.
|
Enrichment by protein function
|
Percentage
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein class | Actual | n | R | N | Expected | Ratio | P-value | z-score | In data set | In protein function | Protein function in database |
| Kinases | 27 | 450 | 651 | 22771 | 12.87 | 2.099 | 2.634E−04 | 4.038 | 6.00% | 4.15% | 2.86% |
| Transcription factors | 34 | 450 | 959 | 22771 | 18.95 | 1.794 | 7.836E−04 | 3.567 | 7.56% | 3.55% | 4.21% |
| Phosphatases | 7 | 450 | 230 | 22771 | 4.545 | 1.54 | 1.719E−01 | 1.169 | 1.56% | 3.04% | 1.01% |
| Proteases | 13 | 450 | 554 | 22771 | 10.95 | 1.187 | 3.030E−01 | 0.6341 | 2.89% | 2.35% | 2.43% |
| Enzymes | 51 | 450 | 2715 | 22771 | 53.65 | 0.9505 | 3.820E−01 | −0.3899 | 11.33% | 1.88% | 11.92% |
| Receptors | 25 | 450 | 1522 | 22771 | 30.08 | 0.8312 | 1.930E−01 | −0.968 | 5.56% | 1.64% | 6.68% |
| Ligands | 8 | 450 | 512 | 22771 | 10.12 | 0.7907 | 3.147E−01 | −0.6802 | 1.78% | 1.56% | 2.25% |
| Other | 288 | 450 | 15680 | 22771 | 309.9 | 0.9294 | 1.479E−02 | −2.248 | 64.00% | 1.84% | 68.86% |
Columns have the following meaning:
Protein class, a broadly defined protein function.
Actual, number of network objects from the activated data set(s) for a given protein class.
n, number of network objects in the activated data set(s).
R, number of network objects of a given protein class in the complete database or background list.
N, total number of network objects in the complete database or background list.
Expected, mean value for hypergeometric distribution (n*R/N).
Ratio, connectivity ratio (Actual/expected).
z-score, z-score ((actual-expected)/sqrt (variance)).
P-value, probability to have the given value of Actual or higher (or lower for negative z-score).
In data set, fraction of network objects with a selected function in the activated data set.
In protein function, fraction of network with a selected function in the activated data set among network objects with this function in the complete database or background list.
Protein function in database, fraction of network objects with a selected function in the complete database or background list.
Discussion
In this study, we demonstrate an inverse correlation between the miRNA mir143 and the MAPK ERK5 in a large multi-centre human prostate tumour TMA resource. Indeed we observe an inverse relation between ERK5 and mir143, manifesting itself most strongly in the subgroup of 417 cores with non-zero mir143 and ERK5 readings, or with only one of mir143 or ERK5 being zero. We have previously observed that, in sequential sections, the pattern of reduced mir143 expression closely matched that of enhanced ERK5 expression within the malignant epithelium (and vice versa for benign tissue) (Ramsay et al, 2011). This strongly implicates a role for mir143 at least partially contributing to abnormal ERK5 expression in CaP. Clape et al (2009) have previously demonstrated that mir143 has a tumour suppressor role in CaP by controlling cell proliferation and survival through modulation of ERK5. For the first time, we have validated the proposed inverse association between mir143 and ERK5 expression. The basis of loss of mir143 expression in prostate cancer remains to be defined. Besides gene deletion, mir155 has recently been identified as a potent repressor of mir143 expression by targeting C/EBPb, a transcriptional activator for mir143 (Jiang et al, 2012). Immunoreactivity for nuclear ERK5 was used as a surrogate for activated ERK5. p-ERK5 can be detected by western blot analysis in in vitro cell models either with activated ERK via constitutively active MEK5 or following exogenous stimulation with growth factors such as EGF (Supplementary Figure 3). However, despite extensive attempt to optimise available antibodies against p-ERK5, immune reactivity for p-ERK5 suffered from high background and was not useful to analyse endogenous levels of p-ERK5 in clinical samples. In fact, besides the significant numbers of non-specific bands recognised on the western blot (Supplementary Figure 3), p-ERK5 also binds to p-ERK1/2 due to the high homology among the ERK1, 2 and 5. Hence, as a surrogate for ERK5 function, immunoreactivity for nuclear ERK5 is studied in this study. In addition, such an approach provides the basis to study translocated ERK5 in the nucleus with or without the required canonical activating phosphorylation by MEK5 (discussed below).
Amplification or mutation of the MEK5 and ERK5 genes is infrequent; within the data available at catalogue of somatic mutations in cancer (COSMIC, Sanger Institute, UK; www.sanger.ac.uk/genetics/CGP/cosmic), they are relatively uncommon events seen in the skin, prostate, liver and breast cancer. Our finding of reduced mir143 expression with corresponding enhanced ERK5 expression in the tissue sections strongly argue for a role of mir143 at least partly contributing to abnormal ERK5 expression. In support of our data, a recent report in colorectal cancer revealed correlation in 63 cancer specimens between reduced mir143 levels and upregulated ERK5 expression (Akao et al, 2010). However, downregulation of mir143 may be acting via mechanisms other than modulation of ERK5 and the exact mode of interaction has yet to be elucidated. Interestingly, recent studies in cervical carcinoma and osteosarcoma have demonstrated downregulation of mir143 expression and its impact in enhancing Bcl-2 expression. It is suggested that, in normal circumstances, mir143 downregulates the level of Bcl-2, an anti-apoptotic molecule, thus reducing cell viability and tumourigenicity by promoting apoptosis (Akao et al, 2010; Liu et al, 2012). Thus, re-expression of mir143 in HeLa cells significantly induced apoptosis and reduced proliferation. Similarly, in nude mice, increased mir143 expression suppressed the levels of Bcl-2 (Liu et al, 2012). When Bcl-2 levels were overexpressed, there was a partial reversal of the mir143-induced anti-mitotic and pro-apoptotic effects in the HeLa cells. In mesenchymal stem cells, cell cycle progression driven by co-overexpression of Akt and Ang-1 was found to be mediated by the upregulation of miR143 expression, which in turn promoted FoxO1 and Erk5 signalling (Lai et al, 2012). In contrast to our findings in prostate cancer, miR143 inhibition in mesenchymal stem cells repressed phosphorylation of Erk5 and abrogated cyclin D1 with concomitant reduction in cells entering cell cycle (Lai et al, 2012), thus further support a complex relationship between mir143 expression and its functional impact on cellular proliferation.
We found no evidence for correlation between the levels of expression between mir143 and Ki67 (a marker of proliferation). Considering its role as an anti-oncomiR, we would expect a mitogenic effect upon reduced mir143 expression. This may reflect the complex pattern of target genes regulated by mir143, including oncogenic kinases, inactivating phosphatases and transcription factors. In addition, the role of ERK5 in control of cell cycle progression is complicated by the relative role of MEK5-mediated ERK5 phosphorylation and nuclear localisation. Intriguingly, nuclear ERK5 may be induced independent of canonical MEK5–ERK5 activity via cyclin-dependent kinases (Inesta-Vaquera et al, 2010).
On the basis of GeneGo analysis, phosphastases as a protein group ranked as the third most enriched functional group, implicating seven phosphatases out of 230 genes, which is however not statistically significant. Hence, our working hypothesis is that mir143 target genes within the categories of kinases and transcription factors may in fact have opposing effects in modulating cell growth and possibly carcinogenesis. This may have implication on the potential application of miRNA as prognostic or diagnostic markers.On the basis of the complexity of miRNA targets genes, it is therefore likely that exploitation of miRNA as biomarkers will require the use of a panel of miRNAs, perhaps along with other biomarkers at mRNA, DNA and protein levels, rather than individual miRNA as isolated biomarkers. Our data from in situ hybridisation for mir143 and automated detection of (nuclear) ERK5 expression provide a model to develop future strategies in stratifying patients for targeted therapy in prostate cancer. We propose that ERK5 immunoreactivity, rather than mir143 expression, is a valid approach to guide targeted therapies aimed at the MEK5–ERK5 pathway.
Acknowledgments
Cancer Research UK funded research work in the Urology Group, Beatson Institute for Cancer Research. IA was funded by a clinical research fellowship from the Medical Research Council and Cancer Research UK. We thank Clare Orange for technical assistance, and the ‘Think Pink’ charity for the purchase of the slide scanner.
Author Contributions
Experimental design: IA and HYL. Experiments were performed by: IA, LBS and JF. Data analysis: IA, ZHY, GK and GF. Provision of reagents: CC, JC, DMB, HM and PS. Manuscript preparation: IA and HYL.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Abate-Shen C, Shen MM (2000) Molecular genetics of prostate cancer. Genes Dev 14(19): 2410–2434 [DOI] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh T, Kojima K, Nakashima R, Kitade Y, Naoe T (2010) Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther 17(6): 398–408 [DOI] [PubMed] [Google Scholar]
- Andriole GL, Crawford ED, Grubb RL, Buys SS, Chia D, Church TR, Fouad MN, Gelmann EP, Kvale PA, Reding DJ, Weissfeld JL, Yokochi LA, O’Brien B, Clapp JD, Rathmell JM, Riley TL, Hayes RB, Kramer BS, Izmirlian G, Miller AB, Pinsky PF, Prorok PC, Gohagan JK, Berg CD (2009) Mortality results from a randomized prostate-cancer screening trial. N Engl J Med 360(13): 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2): 281–297 [DOI] [PubMed] [Google Scholar]
- Berney DM, Gopalan A, Kudahetti S, Fisher G, Ambroisine L, Foster CS, Reuter V, Eastham J, Moller H, Kattan MW, Gerald W, Cooper C, Scardino P, Cuzick J (2009) Ki-67 and outcome in clinically localised prostate cancer: analysis of conservatively treated prostate cancer patients from the Trans-Atlantic Prostate Group study. Br J Cancer 100(6): 888–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G (2007) MicroRNA-133 controls cardiac hypertrophy. Nat Med 13(5): 613–618 [DOI] [PubMed] [Google Scholar]
- Clape C, Fritz V, Henriquet C, Apparailly F, Fernandez PL, Iborra F, Avances C, Villalba M, Culine S, Fajas L (2009) miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS One 4(10): e7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Fisher G, Kattan MW, Berney D, Oliver T, Foster CS, Moller H, Reuter V, Fearn P, Eastham J, Scardino P (2006) Long-term outcome among men with conservatively treated localised prostate cancer. Br J Cancer 95(9): 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1): 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanapragasam VJ, Robinson MC, Marsh C, Robson CN, Hamdy FC, Leung HY (2003) FGF8 isoform b expression in human prostate cancer. Br J Cancer 88(9): 1432–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5(7): 522–531 [DOI] [PubMed] [Google Scholar]
- Hsing AW, Tsao L, Devesa SS (2000) International trends and patterns of prostate cancer incidence and mortality. Int J Cancer 85(1): 60–67 [DOI] [PubMed] [Google Scholar]
- Inesta-Vaquera FA, Campbell DG, Tournier C, Gomez N, Lizcano JM, Cuenda A (2010) Alternative ERK5 regulation by phosphorylation during the cell cycle. Cell Signal 22(12): 1829–1837 [DOI] [PubMed] [Google Scholar]
- Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH, Liang S, Li B, Li Y, Li D, Wang ED, Liu MF (2012) A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J 31(8): 1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai VK, Ashraf M, Jiang S, Haider K (2012) MicroRNA-143 is a critical regulator of cell cycle activity in stem cells with co-overexpression of Akt and angiopoietin-1 via transcriptional regulation of Erk5/cyclin D1 signaling. Cell Cycle 11(4): 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33(1): 159–174 [PubMed] [Google Scholar]
- Leite KR, Tomiyama A, Reis ST, Sousa-Canavez JM, Sanudo A, Camara-Lopes LH, Srougi M (2011) MicroRNA expression profiles in the progression of prostate cancer-from high-grade prostate intraepithelial neoplasia to metastasis. Urol Oncol [DOI] [PubMed]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433(7027): 769–773 [DOI] [PubMed] [Google Scholar]
- Liu L, Yu X, Guo X, Tian Z, Su M, Long Y, Huang C, Zhou F, Liu M, Wu X, Wang X (2012) miR-143 is downregulated in cervical cancer and promotes apoptosis and inhibits tumor formation by targeting Bcl-2. Mol Med Rep 5(3): 753–760 [DOI] [PubMed] [Google Scholar]
- McCracken SR, Ramsay A, Heer R, Mathers ME, Jenkins BL, Edwards J, Robson CN, Marquez R, Cohen P, Leung HY (2008) Aberrant expression of extracellular signal-regulated kinase 5 in human prostate cancer. Oncogene 27(21): 2978–2988 [DOI] [PubMed] [Google Scholar]
- Mehta PB, Jenkins BL, McCarthy L, Thilak L, Robson CN, Neal DE, Leung HY (2003) MEK5 overexpression is associated with metastatic prostate cancer, and stimulates proliferation, MMP-9 expression and invasion. Oncogene 22(9): 1381–1389 [DOI] [PubMed] [Google Scholar]
- Mohammed ZM, McMillan DC, Elsberger B, Going JJ, Orange C, Mallon E, Doughty JC, Edwards J (2012) Comparison of visual and automated assessment of Ki-67 proliferative activity and their impact on outcome in primary operable invasive ductal breast cancer. Br J Cancer 106(2): 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Guo W, Liu T, Wang X, Tu X, Xiong D, Chen S, Lai Y, Du H, Chen G, Liu G, Tang Y, Huang S, Zou X (2011) Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS One 6(5): e20341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay AK, McCracken SR, Soofi M, Fleming J, Yu AX, Ahmad I, Morland R, Machesky L, Nixon C, Edwards DR, Nuttall RK, Seywright M, Marquez R, Keller E, Leung HY (2011) ERK5 signalling in prostate cancer promotes an invasive phenotype. Br J Cancer 104(4): 664–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan A, Pearson GW, Chrestensen CA, Sturgill TW, Cobb MH (2006) The MAP kinase ERK5 binds to and phosphorylates p90 RSK. Arch Biochem Biophys 449(1-2): 8–16 [DOI] [PubMed] [Google Scholar]
- Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, Denis LJ, Recker F, Berenguer A, Maattanen L, Bangma CH, Aus G, Villers A, Rebillard X, van der Kwast T, Blijenberg BG, Moss SM, de Koning HJ, Auvinen A (2009) Screening and prostate-cancer mortality in a randomized European study. N Engl J Med 360(13): 1320–1328 [DOI] [PubMed] [Google Scholar]
- Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S (2008) MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456(7224): 980–984 [DOI] [PubMed] [Google Scholar]
- Wach S, Nolte E, Szczyrba J, Stohr R, Hartmann A, Orntoft T, Dyrskjot L, Eltze E, Wieland W, Keck B, Ekici AB, Grasser F, Wullich B (2012) MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer 130(3): 611–621 [DOI] [PubMed] [Google Scholar]
- Xu B, Niu X, Zhang X, Tao J, Wu D, Wang Z, Li P, Zhang W, Wu H, Feng N, Hua L, Wang X (2011) miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem 350(1-2): 207–213 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.