Abstract
Background:
Whether methylation of the microRNA (mir)-124-3 CpG island is of relevance for the clinical course of a solid cancer and whether it shows association with clinicopathology or survival of patients with renal cell cancer (RCC) is not known as yet.
Methods:
In a cross-sectional study, relative methylation of mir-124-3 was measured in 111 RCC samples and 77 paired normal appearing tissues using quantitative methyl-specific PCR. Results were statistically compared with tumour histology, clinicopathological parameters and disease recurrence.
Results:
We found tumour-specific hypermethylation of mir-124-3 in samples of RCCs with clear cell histology (ccRCC) compared with paired normal appearing tissues (P<0.0001). Methylation was significantly increased in tumours with state of advanced disease (P<0.0001). Higher relative methylation was associated with worse recurrence-free survival in both univariate (hazard ratio=9.37; P=0.0005) as well as bivariate Cox regression analyses considering age, sex, diameter of tumours and state of advanced disease, metastasis and lymph node metastases as covariates (hazard ratios=5.9–18.2; P-values of 0.0003–0.008).
Conclusion:
We identified mir-124-3 CpG islands (CGI) methylation as a relevant epigenetic mark for ccRCC thus underlining the need for functional studies of potentially affected signalling pathways in kidney tumour models. Methylation of mir-124-3 is suggested as an independent prognosticator for ccRCC.
Keywords: renal cell carcinoma, microRNA, mir-124-3 , DNA methylation, prognosis
With over 111 000 new cases and 43 000 deaths reported in 2008, kidney cancer is among the top 10 causes of cancer death of men in the developed countries (Jemal et al, 2011). Nephron-sparing surgery and nephrectomy are still the gold standard for therapy of localised stages of renal cell carcinoma (RCC), but despite recent advances in targeted therapy, the prognosis for patients with progressive disease is still poor. Moreover, few diagnostic methods are available to predict progression of the disease. The identification of biomarkers for RCC could provide a basis for both future stratification of targeted therapy schemes and the identification of relevant signalling pathways for RCC development. The most frequent histology of RCC is clear cell RCC (ccRCC) exhibiting mutations in the von Hippel-Lindau (VHL) and polybromo 1 (PBRM1) (Varela et al, 2011) genes in at least 30% of tumours analysed. von Hippel-Lindau is involved in various cellular mechanisms such as tumour suppression, response to hypoxia, protein degradation and interaction with intra- as well as extracellular matrix proteins (Nordstrom-O'Brien et al, 2010) and PBRM1 has been identified as part of chromatin structure modifying protein complexes (Reisman et al, 2009; Thompson, 2009).
However, from other studies it has become apparent, that numerous epigenetic alterations are also involved in renal carcinogenesis (Baldewijns et al, 2008; Morris et al, 2008, 2010, 2011). Most of these alterations refer to changed DNA methylation as observed in CGI of various genes and some of them have been suggested in functional analyses as candidate tumour suppressor genes (TSG) function (Dreijerink et al, 2001; Morris et al, 2010, 2011). Changes in clinical outcome of RCC patients have been attributed so far either to multimarker methylation panels (Urakami et al, 2006) or to gene-specific CGI methylation of genes such as basonuclin 1 (BNC1), collagen type XIV α 1 (COL14A1), DAL-1/4.1B, GATA-binding protein 5 (GATA5), secreted frizzled-related protein 1 (SFRP1) and signal peptide CUB domain EGF-like 3 (SCUBE3) (Yamada et al, 2006; Morris et al, 2010; Peters et al, 2012). Interestingly, a subset of these candidate prognosticators were independent from other clinicopathological parameters including DAL-1/4.1B (independent from stage and grade of tumours) COL14A1 (size, grade and stage), gremlin 1 (GREM CGI region 1, age and sex) and SFRP1 (size, grade, stage, age and gender) (Yamada et al, 2006; Morris et al, 2010; van Vlodrop et al, 2010; Atschekzei et al, 2012).
Alteration of microRNA (miR) expression as a result of DNA methylation has recently also been described as a common mechanism in human tumourgenesis (Esquela-Kerscher and Slack, 2006). Considering that miR’s may interact with a large number of target mRNAs leading to degradation and loss or reduction of corresponding gene expression epialterations of miR, CGIs may be of great potential relevance for the prediction of cancer progression (Han et al, 2007; Selbach et al, 2008). In line with this hypothesis, miR-9 methylation has already been identified as an epigenetic mark of potential prognostic relevance for RCC. Apart from that only miR-34a and miR-34b/c CGIs were found to be methylated in RCC as yet (Hildebrandt et al, 2010; Vogt et al, 2011).
Here we asked, whether methylation of a CGI annotated to mir-124-3 and found to be relevant in many tumours (Kent et al, 2002) can be also detected in kidney tumours and is of relevance for the clinical outcome of patients with RCC. MiR-124 matures from three precursor variants located on chromosomes 8p23.1 (mir-124-1), 8q12.3 (mir-124-2) and 20q13.33 (mir-124-3). Hypermethylation of miR-124 has been initially demonstrated to occur in colon, breast and lung cancers as well as leukaemia and lymphoma (Lujambio et al, 2007) and seems to play an important role in tumour development. However, a possible clinical relevance of mir-124-3 methylation as independent prognosticator has been reported so far only for haematological malignancies (Agirre et al, 2009; Roman-Gomez et al, 2009), while corresponding information for solid tumours in general as well as for RCC in particular are not available. Here, we analysed whether methylation of the mir-124-3 locus is relevant for RCC and, moreover, whether changes are associated with the clinicopathology and disease progression of RCC patients.
Materials and methods
Cell lines
Human kidney tumour cell lines and primary renal proximal tubular epithelial cells (RPTEC) were cultured not longer as 2 months following purchase and identity control by the manufacturer as described previously (Peters et al, 2012). Human breast cancer and cervical cancer cell line DNAs were a kind gift by Thilo Dörk (Department of Gynaecology & Obstetrics, Hannover Medical School, Hannover, Germany).
Study design and patients
Cross-sectional analyses were conducted on 111 RCC samples and 77 samples from paired histologically normal appearing renal tissues. Sample collection was approved by the local ethics committee and informed consent was obtained from each patient. TNM classification of all tissues was evaluated according to the Union for International Cancer Control 2002 classification (Sobin and Compton, 2010). Two pathologists assessed all specimens considering tumour classification, grade and histological subtype. Localised RCC was defined as pT⩽2, lymph node (N) and metastasis (M) negative (N0, M0) and a grading (G) of 1 and 1–2. Advanced tumours were classified as pT⩾3 and/or lymph node positive (N+), positive for distant metastasis (M+) or G2–3 and G3. The time to disease progression corresponds to the point in time patients demonstrating either a local recurrence or a synchronous/metachronous metastasis as detected by CT scan. Clinical and histopathological characteristics of tumour specimens are summarised in Table 1.
Table 1. Tumour patients characteristics.
| All RCC | % | ccRCC | % | ccRCC surv. group | % | |
|---|---|---|---|---|---|---|
| Total cases | 111 | 80 | 37 | |||
|
Histology
| ||||||
| ccRCC | 80 | 72.1 | 80 | 100 | 37 | 100 |
| papRCC | 23 | 20.7 | 0 | 0 | 0 | 0 |
| Chrom. RCC | 4 | 3.6 | 0 | 0 | 0 | 0 |
| Not class. | 4 | 3.6 | 0 | 0 | 0 | 0 |
|
Sex
| ||||||
| Female | 38 | 34.2 | 30 | 37.5 | 15 | 40.5 |
| Male | 73 | 65.8 | 50 | 62.5 | 22 | 59.5 |
|
Age
| ||||||
| Median (years) | 65 | 64 | 65 | |||
|
Distant metastasis
| ||||||
| M0 | 86 | 77.5 | 60 | 75 | 28 | 75.7 |
| M+ | 25 | 22.5 | 20 | 25 | 9 | 24.3 |
|
Lymph node metastasis
| ||||||
| N0 | 98 | 88.3 | 73 | 91.3 | 35 | 94.6 |
| N+ | 13 | 11.7 | 7 | 8.8 | 2 | 5.4 |
|
T-classification
| ||||||
| pT1 | 11 | 9.9 | 8 | 10 | 1 | 2.7 |
| pT1a | 32 | 28.8 | 21 | 26.3 | 12 | 32.4 |
| pT1b | 19 | 17.1 | 14 | 17.5 | 7 | 18.9 |
| pT2 | 7 | 6.3 | 5 | 6.3 | 2 | 5.4 |
| pT3 | 5 | 4.5 | 2 | 2.5 | 1 | 2.7 |
| pT3a | 9 | 8.1 | 7 | 8.8 | 2 | 5.4 |
| pT3b/c | 23 | 20.7 | 21 | 26.3 | 11 | 29.7 |
| pT4 | 1 | 0.9 | 0 | 0 | 0 | 0 |
| NA | 4 | 3.6 | 2 | 2.5 | 1 | 2.7 |
|
Differentiation
| ||||||
| G1 | 22 | 19.8 | 19 | 23.8 | 5 | 13.5 |
| G1–2 | 14 | 12.6 | 9 | 11.3 | 4 | 10.8 |
| G2 | 56 | 50.5 | 38 | 47.5 | 21 | 56.8 |
| G2–3 | 8 | 7.2 | 4 | 5 | 2 | 5.4 |
| G3 | 11 | 9.9 | 10 | 12.5 | 5 | 13.5 |
|
State of disease
| ||||||
| Loc. diseasea | 59 | 53.2 | 39 | 48.8 | 17 | 45.9 |
| Adv. diseaseb | 51 | 45.9 | 41 | 51.3 | 20 | 54.1 |
| NA | 1 | 0.9 | 0 | 0 | 0 | 0 |
|
Paired samples
| ||||||
| All RCC | 77 | |||||
| ccRCC | 58 | |||||
Abbreviation: ccRCC=clear cell renal cell carcinoma.
pT⩽2, N0, M0 and G1+G1–2.
pT⩾3 and/or N+, M+ or G2–3 G3.
DNA isolation, bisulphite conversion of DNA and quality measurements
DNA isolation from frozen sections and histopathological examination of control sections were carried out as described previously (Peters et al, 2012). Tumour tissue sections represented 100% tumour tissue and included only minor fractions of normal cells. Bisulphite conversion of DNA was performed as described before (Peters et al, 2007). To control bisulphite conversion efficiency, all samples were subjected to quantitative methylation-specific PCRs (qMSP) using the concept reported by Campan et al (2009) and van der Horst et al (2004). Briefly, detection of the methylation-independent repetitive sequence ALU-C4 (QC1) and a single copy β-Actin (ACTB) (QC2) sequence served to measure the yield of conversion and to provide for controlled input of bisulphite converted template DNA into qMSP analyses, while the repetitive sequence ALU-YB8 (QC3) allows to quantify unconverted DNA. Fully methylated bisulphite converted control DNA (M) and unmethylated bisulphite converted control DNA (U) were generated as described recently (Weisenberger et al, 2008).
Quantitative methylation-specific PCR (qMSP) analysis
Methylation analyses of bisulphite-treated genomic DNA was performed by a quantitative real-time fluorimetric 5′ exonuclease PCR assay. Our analyses determined a subregion within the CpG island (CGI) annotated to the hsa-mir-124-3 locus on chromosome 20 as indicated in Figure 1A. The qMSP-specific primers 5′-GGTCGGGTCGGGTTAGTAGG-3′ (forward) and 5′-CGCAAACCGACTACGAACCG-3′ (reverse) as well as the Taqman probe 5′-FAM-CCACGAAATCCACGCTACAAACGCCCA-BHQ-3 were designed according to Weisenberger et al (2005) and using the Beacon Designer software (PREMIER Biosoft, Palo Alto, CA, USA). Real-time PCRs were carried out in duplicate on an ABI 7900HT (Life Technologies, Foster City, CA, USA) in 384-well plates. Amplification included initial incubation for 10 min at 95 °C followed by 45 cycles of denaturation for 15 s at 95 °C, 1 min annealing and elongation at 60 °C in a reaction volume of 9 μl, consisting of 1.2 μℳ of each primer, 400 nℳ probe and 1 × Taqman Universal Master Mix II no UNG. For PCR setup, the FasTrans automatic Liquid Handling System (AnalyticJena, Jena, Germany) was used. Measurements were carried out blinded against type, order and clinicopathological status of samples by the experimenter.
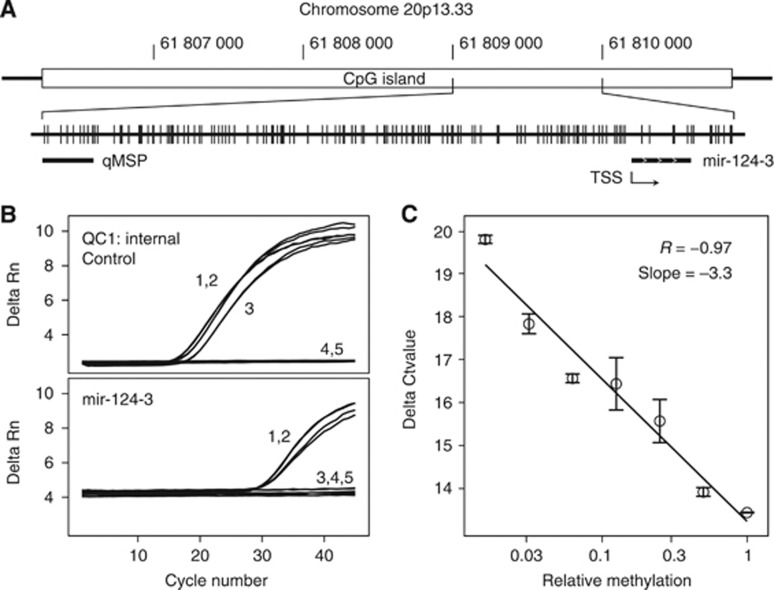
Figure 1.
(A) Structure of the mir-124-3 CGI and location of the qMSP assay relative to the mir-124-3 transcription start site (TSS). Vertical lines represent CpG sites within the CGI. Chromosomal positions refer to the GRCh37/hg19 annotation in the UCSC genome browser (Lander et al, 2001; Kent et al, 2002). (B) Exemplary primary data of quantitative methylation-specific and control PCR measurements in duplicate for A498 (1); methylated control DNA (2); unmethylated control DNA (3); unconverted DNA (4) and a blank control (5). (C) Normalised mir-124-3 assay threshold values (Ct) for a two-fold dilution series of the methylated control in non-methylated control DNA for determination of assay linearity and efficiency.
Calculation of relative methylation levels
The relative degree of methylation was determined using the method of Weisenberger et al (2005). Sample-specific total input of converted DNA in each sample was measured by detection by means of QC1 while calibrator samples M and U served as positive and negative controls, in each run. Relative methylation values were calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001). Briefly, Ct values of samples were normalised to QC1 and referenced to the positive control M, allowing calculation of ΔΔCt and the relative methylation. Notably, the resulting methylation values are related to the copy number of methylated sequences detected in the calibrator sample corresponding to a fully methylated diploid genome. Hence, both potential cancer-related alterations such as regional amplification of DNA as well as slight variation in measured Ct values may lead to the measurement of methylation values of >100%. Therefore, the methylation values were designated as ‘relative methylation level’. Samples exhibiting lower template DNA input than a QC1 Ct value of 21.1 were excluded. The analytical sensitivity of the mir-124-3 assay was estimated calculating the limit of quantitative detection (LQ) from the variance measurements of blank values as suggested by Currie (1968). First the non-methylated control U was measured 12 times and all runs were found to be undetermined. To consider the variance of ‘blank values’ present within sample group, LQ was then calculated by addition of the mean of background methylation levels as determined from 153 undetermined samples and the corresponding 10-fold standard deviation.
Statistical analyses
Explorative statistical data analyses were conducted by the use of the statistical software R 2.12 (R Development Core Team, 2011). P<0.05 were considered to indicate statistical significance. Relative methylation levels are converted to the natural logarithmic scale before conducting further statistical calculations. Linear regression analysis was performed for linearity and PCR efficiency measurements of the qMSP assay. Statistical comparison of the mean relative methylation levels observed for paired tumour and adjacent normal appearing tissue were carried out using the paired t-test. For group comparisons of independent tissue samples representing different clinicopathological classifications, univariate logistic regression analysis was conducted, providing both statistical significance and odds ratio (OR) serving as a measure of the observed effect size. Analysis of recurrence-free survival was achieved using Cox’s proportional-hazards regression model. The grade and state of lymph node metastasis were not considered in bivariate analyses due to low number of cases in subgroups following dichotomization. Note that G and N were part of the localised and advanced disease classifications (see above). Optimum threshold calculations for dichotomization of methylation levels were performed using the R package ‘maxstat’.
Results
Measurement of technical controls
We analysed the specificity of the mir-124-3 qMSP analysis by duplicate measurements of converted methylated (M), converted non-methylated (U) and non-converted DNA control samples. We exclusively observed Ct values of 45 (undetermined) for the U and non-converted DNA samples, while the M sample demonstrated Ct values of about 29 (Figure 1B). Non-converted DNA was detected neither in QC1 control PCR nor in mir-124-3-specific qMSP, thus demonstrating that only methylated and converted DNA has been measured by the mir-124-3 methylation assay. To determine PCR efficiency and linearity of the methylation detection assay, we analysed a two-fold dilution series of the M control within the U control DNA adjusting for constant total converted DNA input. Linear regression analysis revealed a slope of ΔCT=−3.3 per 10-fold dilution and a coefficient of correlation of r=0.97 (P=0.001), indicating a high efficiency and linearity of the assay (Figure 1C). Estimation of the analytical sensitivity LQ of the mir-124-3 assay according revealed a relative methylation level of 5.7E−4 (mean of blank values=3.0E−5, s.d.=5.3E−5).
mir-124-3 methylation in tumour cell lines and normal primary cells
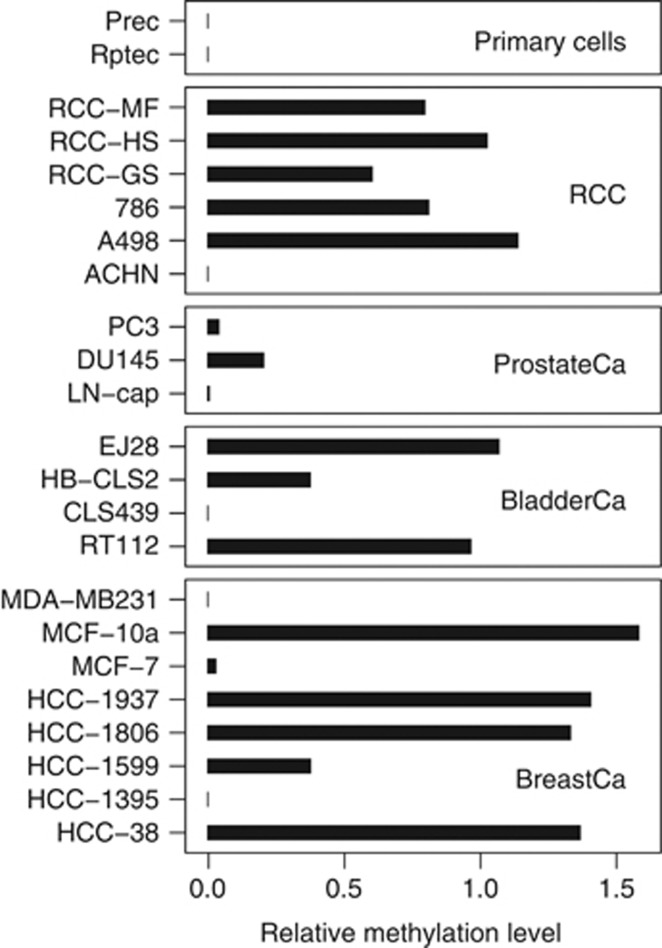
We first evaluated whether the mir-124-3 qMSP assay is capable of detecting methylation in cell lines used as surrogates for tumours with known methylation (breast cancer) as well as for normal tissues and tissues of both localised and metastatic human cancers of other origin (kidney, prostate and bladder cancer; Figure 2). As expected, methylation could be detected in five out of eight (63%) breast cancer cell lines. Interestingly, five of six (83%) of the renal cancer cell lines showed high relative methylation levels ranging from 0.006 to 1.14, while normal primary cells from kidney (RPTEC) and prostate (PreC) exhibited undetectable or low methylation, respectively. Notably, three of four (75%) bladder cancer cell lines were identified to show high relative methylation levels (Figure 2).
Figure 2.
Relative methylation levels measured for cancer cell lines and normal primary cells.
mir-124-3 is hypermethylated in RCC
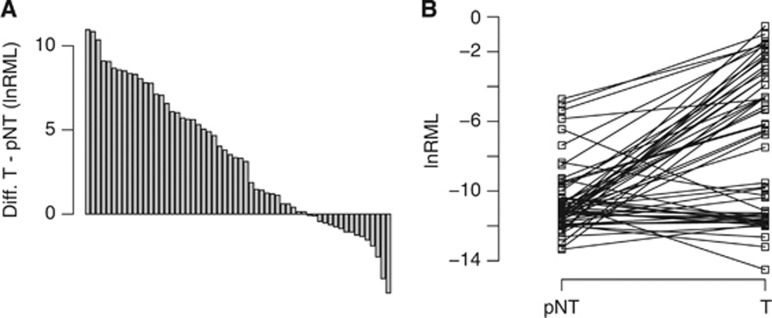
Analysis of relative methylation levels in paired tumour and adjacent normal tissues regardless of histology revealed higher values for the tumour tissues (mean=6.5E−4) compared with normal tissue samples (mean=2.5E−5; P<0.0001; paired t-test), demonstrating relative methylation levels in the range of the blank values. The corresponding analysis of the clear cell histology subset of tissues disclosed also higher relative methylation for neoplastic tissues (mean=7.1E−4 compared with paired normal tissues (2.5E−5; P<0.0001, paired t-test; Figures 3A and B). In view of previous results obtained by others and describing that methylation found in tumour tissues may be related with their normal counterparts and therefore supposed to be indicative of an epigenetic risk for tumour development, we additionally carried out a correlation analysis for the paired tissue samples. As a result, we found a weak but significant statistical association (r=0.29, P=0.026, Pearson’s correlation analysis).
Figure 3.
Analysis of paired normal appearing and tumour tissues for the ccRCC subgroup. (A) Assorted pairwise differences for natural logarithms of relative methylation levels (lnRML) observed in tumour (T) and paired normal appearing tissues (pNT). (B) Direct comparison of natural logarithms of relative methylation levels for paired normal appearing (pNT) and tumour (T) samples.
mir-124-3 methylation and statistical association with clinicopathologic parameters
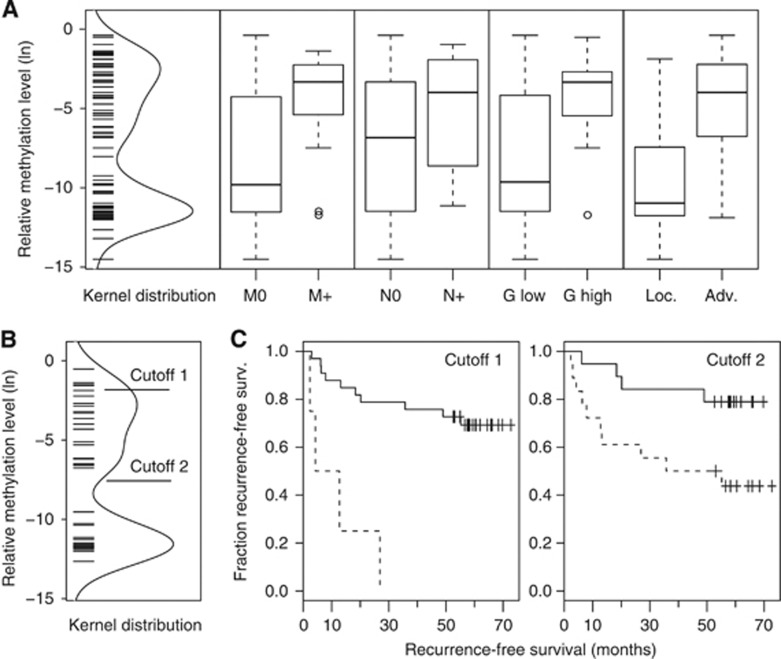
Analysis of all RCC samples revealed a significant higher relative methylation levels for tumours with clear cell histology (mean=7.1E−4) compared with papillary tumours (mean=6.5E−5, P=0.011; OR=0.843; 95% CI=0.74–0.97). Therefore, further statistical analyses were exclusively carried out for the ccRCC tissue subset. As a result, we identified significant differences in methylation levels for the parameters state of distant metastasis (P<0.0001), state of localised vs advanced disease (P<0.0001) and high- (G2–3, G3) or low-grade (G1, G1–2, G2) tumour (P=0.0063; Figure 4A; Table 2). Methylation of ccRCCs was not statistically associated with sex, age and lymph node metastasis status but showed a trend for the tumour diameter (P=0.062).
Figure 4.
(A) Distribution of methylation values detected in ccRCC and box plot analysis for subset-specific relative methylation levels of clinicopathological parameters; negative (M0) or positive (M+) for distant metastasis, negative (N0) or positive (N+) for lymph node metastasis, low (G<=2) or high (G>2) grade tumours and localised (pT⩾2, N0, M0 and G1+G1–2) or advanced (pT⩾3 and/or N+, M+ or G2–3 G3) disease. Bold horizontal lines show group medians; boxes and whiskers show the 25 and 75% and 10 and 90% quartiles, respectively. (B) Distribution of methylation values detected in the ccRCC survival subgroup and location of the statistical optimum (cutoff 1) or the limit of quantitative detection LQ (cutoff 2) cutoff values. (C) Kaplan–Meier plots showing relative survival of patients dichotomised by use of cutoff 1 or cutoff 2 values as indicated in (B). Note that some of the symbols displaying censored cases coincide due to similar follow-up periods of patients.
Table 2. Statistical association of mir-124-3 CGI methylation with clinicopathological parameters of ccRCC patients.
| RML a | |||||
|---|---|---|---|---|---|
| Median | Median | P -value b | OR | 95% CI | |
|
ccRCC
| |||||
| Dist. metastasis (M0/M+) | 5.5E–5 | 0.03 | 0.0010 | 1.30 | 1.11–1.51 |
| Lymph node met. (N0/N+) | 1.0E–3 | 0.01 | 0.2253 | 1.13 | 0.93–1.39 |
| Grade (low/high) | 6.5E–5 | 0.03 | 0.0063 | 1.28 | 1.07–1.52 |
| Diameterc | 3.4E–5 | 3.8E–3 | 0.0620 | 1.14 | 0.99–1.30 |
| Localised/advanced dis. | 1.7E–5 | 0.01 | <0.0001 | 1.36 | 1.19–1.57 |
Abbreviations: OR=odds ratio; CI=confidence interval; ccRCC=clear cell renal cell cancer; CGI=CpG island.
Relative methylation level.
Univariate logistic regression.
Dichotomised using median as cut point.Bold numbers indicate statistical significance (P<0.05).
mir-124-3 methylation is associated with worse recurrence-free survival of patients
To assess whether mir-124-3 methylation is associated with the clinical course of patients with ccRCC, we first performed a Cox regression analysis using a statistically calculated optimum cutoff value corresponding to a relative methylation level of 0.0015 for dichotomization of patients (Figure 4B). We identified the methylation status to be associated with increased risk for disease recurrence (P=0.0005; hazard ratio (HR)=9.37; CI=2.68–32.8; Table 3). The corresponding Kaplan–Meier analysis showed that in this case four out of four patients (100%) were identified with disease recurrence within <30 months and methylation above the cut point (Figure 4B,C). In contrast, 7 of 33 (21%) were detected with low methylation and disease recurrence within the same period. Comparison of HRs from univariate Cox regression analysis exhibited the values 9.37, 4.86 and 4.28 for the parameters methylation, metastatic state or advanced disease (Table 3). To investigate whether the association of methylation status and recurrence-free survival could be a statistically independent parameter, we performed pairwise bivariate Cox proportional-hazards regression considering the clinicopathological parameters distant metastasis, state of localised or advanced disease, gender, age and tumour diameter as covariates. Our analysis revealed that methylation dichotomised with the optimised cutoff value remained as a significant parameter in bivariate Cox regression models exhibiting HR values of 5.87–18.2 and P-values between 0.0003 and 0.008 (Table 4). From the clinicopathological parameters, only distant metastasis remained as a significant factor in the bivariate Cox regression models.
Table 3. Univariate statistical association of mir-124-3 CGI methylation and clinicopathological parameters with recurrence-free survival of patients.
| P- value a | HR | 95% CI | |
|---|---|---|---|
| mir-124-3 methylation | 0.0005 | 9.37 | 2.68–32.8 |
| Metastasis | 0.0032 | 4.86 | 1.7–14 |
| Lymph node status | 0.8751 | 1.18 | 0.15–9.02 |
| Localised/advanced disease | 0.0260 | 4.28 | 1.19–15.4 |
| Gender | 0.2080 | 2.11 | 0.66–32.8 |
| Ageb | 0.7058 | 0.82 | 0.28–2.35 |
| Diameterb | 0.4278 | 1.67 | 0.47–5.92 |
Abbreviations: CI=confidence interval; HR=hazard ratio; CGI=CpG island.
Univariate Cox regression analysis.
Dichotomisation by the median of parameter.Bold numbers indicate statistical significance (P<0.05).
Table 4. Association of mir-124-3 CGI methylation and clinicopathological parameters with recurrence-free survival in bivariate survival analysis.
| P- value a | HR | 95% CI | |
|---|---|---|---|
| mir-124-3 methylation | 0.0003 | 13 | 3.3–51.7 |
| Metastasis | 0.0017 | 6.05 | 1.9–18.6 |
| mir-124-3 methylation | 0.0081 | 5.87 | 1.6–21.8 |
| Localised/advanced | 0.1055 | 3.06 | 0.8–11.8 |
| mir-124-3 methylation | 0.0004 | 10.90 | 2.9–40.3 |
| Gender | 0.1410 | 2.45 | 0.74–8.07 |
| mir-124-3 methylation | 0.0005 | 18.20 | 3.6–92.4 |
| Ageb | 0.1429 | 0.35 | 0.9–1.14 |
| mir-124-3 methylation | 0.0006 | 18.20 | 3.45–95.5 |
| Diameterb | 0.1986 | 2.46 | 0.6–9.7 |
Abbreviations: CI=confidence interval; HR=hazard ratio; CGI=CpG island.
Bivariate Cox regression analyses.
Dichotomised by the use of the median of parameter values.Bold numbers indicate statistical significance (P<0.05).
Considering that statistical optimisation revealed a small subset of patients at higher risk for disease recurrence, we asked whether lowering the cutoff value thus increasing the number of positive tumours, could give also prognostic information for a greater number of patients. We chose LQ as the lowest estimated methylation level that can be reliably quantitatively measured for dichotomization, hence including the maximum number of patients into the risk group (Figure 4B and C). As a result, we found that these patients also exhibited a worse recurrence-free survival in univariate (P=0.025; HR=4.32; 95% CI=1.2–15.5) but not in the pairwise bivariate Cox regression analyses. The corresponding Kaplan–Meier plot showed that within a follow-up period of 70 months only 4 (24%) of 17 patients from the low methylation group, but 11 (55%) of 20 patients with higher methylation demonstrated disease recurrence (Figure 4C, cutoff 2).
Discussion
Our analyses demonstrated that increased methylation levels of a subregion in the mir-124-3 CGI are associated with adverse clinicopathological parameters of ccRCC patients including metastasis, grade, state of advanced disease and greater tumour diameter. Our data, therefore suggest that this epialteration could be involved in cancer progression in patients with ccRCC. This view is supported by our findings that methylation is significantly associated with recurrence-free survival in univariate and in bivariate analyses as well. Association of mir-124-3 CGI methylation and recurrence-free survival in solid tumours, to our knowledge, has not been described as yet. Previous studies reported a shortened disease-free survival for patients with acute lymphoblastic leukaemia (ALL) using a multi-miR loci approach including also mir-124-3 (Roman-Gomez et al, 2009). Methylation of the mir-124-3 locus was then identified as an independent prognosticator for both disease-free and overall survival in ALL (Agirre et al, 2009). Contribution of mir-124-3 epigenetics to tumour progression is also indicated by gradually increasing methylation levels found in a series of premalignant and malignant cervical tissues of differing biological aggressiveness (Wilting et al, 2010). Although DNA hypermethylation is a frequently observed characteristic in RCC (Baldewijns et al, 2008) only few loci currently including COL14A1 (Morris et al, 2010), GREM1 (van Vlodrop et al, 2010), SFRP1 (Atschekzei et al, 2012) and DAL-1/4.1B (Yamada et al, 2006), were associated with RCC prognosis, provided clinicopathological parameters have been considered as covariates in multi- or bivariate analyses. In view of these results, it seems remarkable that mir-124-3 methylation contributed high and fairly independent hazards for disease recurrence in our bivariate analyses accounting for the states of distant metastasis or localised/advanced disease. So, methylation-related risk values exceed those obtained for metastasis, thus suggesting mir-124-3 methylation as a promising candidate for an independent prognosticator. However, although a high sensitivity of 100% for detection of disease recurrence has been observed, only a fraction of patients subsequently demonstrating disease recurrence would have been identified at the time of surgery. Interestingly, making use of the second cutoff value, a larger group of patients of ∼38% might be identified with a low risk of disease recurrence. We therefore conclude that mir-124-3 methylation should be further evaluated in prospective studies using independent and enlarged patients groups. Moreover, it would be of interest whether this potential biomarker gives prognostic information that is independent from other DNA methylation-based potential biomarkers or can be even improved by generation of combined methylation markers. Our methylation analyses using cell lines of different tumour entities showed that mir-124-3 methylation likely also occurs in bladder cancers, a tumour that has not been reported as yet to show this epigenetic mark. In contrast, none of the cell lines deriving from prostatic metastases demonstrated a higher degree of methylation.
The methylation analysis of the mir-124-3 locus overall revealed apparently relative low methylation values in tumours. Considering that histopathological assessment of control sections confirmed that tissues submitted to DNA extraction demonstrated a tumour cell content of at least 50%, both the absolute levels of mean methylation values and the measured increases of relative methylation levels in metastasised or advanced tumours cannot be attributed to sample-specific variation of tumour cell proportions. Comparable low mean methylation values, on the other hand, may be a consequence of the qMSP method used for methylation analysis when taking into account that only sequences exhibiting a uniformly dense methylation are detected and intra-CGI heterogeneity of methylation may occur in principle as has already been reported for another gene (van Vlodrop et al, 2010). On the other hand, our methylation values may also reflect that only a subpopulation of tumour cells carries densely methylated miR-124-3 loci. A limitation of our study is therefore, that neither explanation can be supported as yet. While our statistical findings clearly suggest miR-124-3 methylation as a marker for biologically aggressive tumours, it remains to be clarified whether it represents an early or late event in the carcinogenesis of RCC.
A relative increase in methylation levels of the mir-124-3 CGI has also been reported for non-cancerous gastric tissues of Helicobacter pylori-positive individuals when compared with corresponding normal tissue samples of H. pylori-negative patients (Ando et al, 2009). This correlation with a known risk factor for gastric cancer led to the hypothesis that mir-124-3 methylation also could contribute for cancer risk. Both, analyses of methylation in paired precancerous and cancer tissues indicating epigenetic inheritance of the methylation status into tumour tissue (Shen et al, 2005) as well as a direct risk estimation by a case–control study (Ando et al, 2009; Atschekzei et al, 2012) have been applied to assess epigenetic cancer risk contribution for other loci. Thus, our observation of a weak statistical correlation of mir-124-3 methylation in normal appearing and paired tumour tissues suggest further appropriate risk studies.
Our analyses suggest mir-124-3 methylation as a biomarker for independent identification of a subset of high-risk patients and are in line with results of recent analyses of haematological malignancies. Considering that first functional analyses in Lujambio et al (2007) described mir-124-3 methylation to effect cellular proliferation and chromosome stability via cyclin D kinase 6 (CDK6), our results may point to the existence of comparable mechanisms also taking place in RCC development. Some evidence for the relevance of CDK6 signalling in RCC development is given by public mRNA expression data showing a significant increase of CDK6 mRNA levels in ccRCC tissues when compared with normal paired samples (P=0.001; paired t-test; data from Lenburg et al, 2003). This would be consistent with a hypothesised silencing of mir-124-3 expression due to CGI methylation. Moreover, from a kinase silencing screen in RCC cell lines deficient for VHL, CDK6 has been identified as one of the targets to be functionally associated with the viability of kidney cancer cells and, in addition, can be affected by a small molecule inhibitor (Bommi-Reddy et al, 2008). Hence, our clinical analysis emphasises the need for further functional analysis of the CDK6 signalling in RCC. In case of that methylation-dependent silencing of the mir-124-3 expression can be shown to be associated with concurrent activation of CDK6 signalling, the measurement of mir-124-3 methylation would provide a rationale for treatment of such RCCs with so far experimental small molecule CDK6 inhibitors. In conclusion, mir-124-3 CGI is hypermethylated in ccRCC and associated with poorer clinical outcome. Our results emphasise the need for further functional characterisation of mir-124-3 alterations in clear cell RCC such as CDK6 signalling. Methylation of the mir-124-3 locus represents a candidate biomarker for further prospective molecular stratification of patients possibly for targeted therapy using small molecule effectors.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Agirre X, Vilas-Zornoza A, Jimenez-Velasco A, Martin-Subero JI, Cordeu L, Garate L, San Jose-Eneriz E, Abizanda G, Rodriguez-Otero P, Fortes P, Rifon J, Bandres E, Calasanz MJ, Martin V, Heiniger A, Torres A, Siebert R, Roman-Gomez J, Prosper F (2009) Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res 69(10): 4443–4453 [DOI] [PubMed] [Google Scholar]
- Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, Sugiyama T, Ushijima T (2009) DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer 124(10): 2367–2374 [DOI] [PubMed] [Google Scholar]
- Atschekzei F, Hennenlotter J, Janisch S, Grosshennig A, Trankenschuh W, Waalkes S, Peters I, Dork T, Merseburger AS, Stenzl A, Kuczyk MA, Serth J (2012) SFRP1 CpG island methylation locus is associated with renal cell cancer susceptibility and disease recurrence. Epigenetics 7(5): 447–457 [DOI] [PubMed] [Google Scholar]
- Baldewijns MM, van Vlodrop IJ, Schouten LJ, Soetekouw PM, de Bruine AP, van Engeland M (2008) Genetics and epigenetics of renal cell cancer. Biochim Biophys Acta 1785(2): 133–155 [DOI] [PubMed] [Google Scholar]
- Bommi-Reddy A, Almeciga I, Sawyer J, Geisen C, Li W, Harlow E, Kaelin WG, Grueneberg DA (2008) Kinase requirements in human cells: III. Altered kinase requirements in VHL-/- cancer cells detected in a pilot synthetic lethal screen. Proc Natl Acad Sci USA 105(43): 16484–16489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campan M, Weisenberger DJ, Trinh B, Laird PW (2009) MethyLight. Methods Mol Biol 507: 325–337 [DOI] [PubMed] [Google Scholar]
- Currie LA (1968) Limits for qualitative detection and quantitative determination. Application to radiochemistry. Anal Chem 40(3): 586–593 [Google Scholar]
- Dreijerink K, Braga E, Kuzmin I, Geil L, Duh FM, Angeloni D, Zbar B, Lerman MI, Stanbridge EJ, Minna JD, Protopopov A, Li J, Kashuba V, Klein G, Zabarovsky ER (2001) The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proc Natl Acad Sci USA 98(13): 7504–7509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ (2006) Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6(4): 259–269 [DOI] [PubMed] [Google Scholar]
- Han L, Witmer PD, Casey E, Valle D, Sukumar S (2007) DNA methylation regulates MicroRNA expression. Cancer Biol Ther 6(8): 1284–1288 [DOI] [PubMed] [Google Scholar]
- Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W, Tamboli P, Wood CG, Wu X (2010) Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene 29(42): 5724–5728 [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2): 69–90 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12(6): 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W et al (2001) Initial sequencing and analysis of the human genome. Nature 409(6822): 860–921 [DOI] [PubMed] [Google Scholar]
- Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF (2003) Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer 3: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4): 402–408 [DOI] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, Spiteri I, Das PP, Caldas C, Miska E, Esteller M (2007) Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 67(4): 1424–1429 [DOI] [PubMed] [Google Scholar]
- Morris MR, Gentle D, Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Teh BT, Latif F, Maher ER (2008) Functional epigenomics approach to identify methylated candidate tumour suppressor genes in renal cell carcinoma. Br J Cancer 98(2): 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MR, Ricketts C, Gentle D, Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Latif F, Maher ER (2010) Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene 29(14): 2104–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MR, Ricketts CJ, Gentle D, McRonald F, Carli N, Khalili H, Brown M, Kishida T, Yao M, Banks RE, Clarke N, Latif F, Maher ER (2011) Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene 30(12): 1390–1401 [DOI] [PubMed] [Google Scholar]
- Nordstrom-O'Brien M, van der Luijt RB, van Rooijen E, van den Ouweland AM, Majoor-Krakauer DF, Lolkema MP, van Brussel A, Voest EE, Giles RH (2010) Genetic analysis of von Hippel-Lindau disease. Hum Mutat 31(5): 521–537 [DOI] [PubMed] [Google Scholar]
- Peters I, Eggers H, Atschekzei F, Hennenlotter J, Waalkes S, Trankenschuh W, Grosshennig A, Merseburger AS, Stenzl A, Kuczyk MA, Serth J (2012) GATA5 CpG island methylation in renal cell cancer: a potential biomarker for metastasis and disease progression. BJU Int 110(Pt 2): E144–E152 [DOI] [PubMed] [Google Scholar]
- Peters I, Rehmet K, Wilke N, Kuczyk MA, Hennenlotter J, Eilers T, Machtens S, Jonas U, Serth J (2007) RASSF1A promoter methylation and expression analysis in normal and neoplastic kidney indicates a role in early tumorigenesis. Mol Cancer 6: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2011) R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria, ISBN 3-900051-07-0, URL http://www.R-project.org/ [Google Scholar]
- Reisman D, Glaros S, Thompson EA (2009) The SWI/SNF complex and cancer. Oncogene 28(14): 1653–1668 [DOI] [PubMed] [Google Scholar]
- Roman-Gomez J, Agirre X, Jimenez-Velasco A, Arqueros V, Vilas-Zornoza A, Rodriguez-Otero P, Martin-Subero I, Garate L, Cordeu L, San Jose-Eneriz E, Martin V, Castillejo JA, Bandres E, Calasanz MJ, Siebert R, Heiniger A, Torres A, Prosper F (2009) Epigenetic regulation of microRNAs in acute lymphoblastic leukemia. J Clin Oncol 27(8): 1316–1322 [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455(7209): 58–63 [DOI] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan PS, Krouse RS, Prasad AR, Einspahr JG, Buckmeier J, Alberts DS, Hamilton SR, Issa JP (2005) MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst 97(18): 1330–1338 [DOI] [PubMed] [Google Scholar]
- Sobin LH, Compton CC (2010) TNM seventh edition. what's new, what's changed: communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer 116(22): 5336–5339 [DOI] [PubMed] [Google Scholar]
- Thompson M (2009) Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie 91(3): 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakami S, Shiina H, Enokida H, Hirata H, Kawamoto K, Kawakami T, Kikuno N, Tanaka Y, Majid S, Nakagawa M, Igawa M, Dahiya R (2006) Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res 12(23): 6989–6997 [DOI] [PubMed] [Google Scholar]
- van der Horst EH, Leupold JH, Schubbert R, Ullrich A, Allgayer H (2004) TaqMan-based quantification of invasive cells in the chick embryo metastasis assay. Biotechniques 37(6): 940–942, 944, 946 [DOI] [PubMed] [Google Scholar]
- van Vlodrop IJ, Baldewijns MM, Smits KM, Schouten LJ, van Neste L, van Criekinge W, van Poppel H, Lerut E, Schuebel KE, Ahuja N, Herman JG, de Bruine AP, van Engeland M (2010) Prognostic significance of Gremlin1 (GREM1) promoter CpG island hypermethylation in clear cell renal cell carcinoma. Am J Pathol 176(2): 575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA (2011) Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469(7331): 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M, Munding J, Gruner M, Liffers ST, Verdoodt B, Hauk J, Steinstraesser L, Tannapfel A, Hermeking H (2011) Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch 458(3): 313–322 [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW (2005) Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 33(21): 6823–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger DJ, Trinh BN, Campan M, Sharma S, Long TI, Ananthnarayan S, Liang G, Esteva FJ, Hortobagyi GN, McCormick F, Jones PA, Laird PW (2008) DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res 36(14): 4689–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, le Sage C, Agami R, Snijders PJ, Steenbergen RD (2010) Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer 9: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada D, Kikuchi S, Williams YN, Sakurai-Yageta M, Masuda M, Maruyama T, Tomita K, Gutmann DH, Kakizoe T, Kitamura T, Kanai Y, Murakami Y (2006) Promoter hypermethylation of the potential tumour suppressor DAL-1/4.1B gene in renal clear cell carcinoma. Int J Cancer 118(4): 916–923 [DOI] [PubMed] [Google Scholar]