Abstract
Human RBM25 (RNA-binding motif protein 25) is a novel splicing factor that contains a PWI domain, a newly identified RNA/DNA-binding domain, and regulates Bcl-x pre-mRNA alternative splicing. The flanking basic region has been suggested to serve as a co-operative partner of the PWI domain in the binding of nucleic acids, but the structure of this basic region is unknown. In the present paper, we report the crystal structure of the RBM25 PWI domain and its flanking basic region. The PWI domain is revealed to comprise a conserved four-helix bundle, and the flanking basic region forms two α-helices and associates with helix H4 of the PWI domain. These interactions promote directly the formation of an enlarged nucleic-acid-binding platform. Structure-guided mutagenesis reveals a positively charged nucleic-acid-binding surface in the RBM25 PWI domain that is entirely different from that in the SRm160 PWI domain. Furthermore, we show that the promotion of the pro-apoptotic Bcl-xS isoform expression by RBM25 is facilitated by the PWI domain in vivo. Thus the present study suggests that the PWI domain plays an important role in the regulation of Bcl-x pre-mRNA alternative splicing.
Keywords: alternative splicing, Bcl-x pre-mRNA, flanking basic region, Luc7A, PWI domain, RNA-binding protein motif 25 (RBM25)
Abbreviations: dsDNA, double-stranded DNA; dsRNA, double-stranded RNA; FAM, 6-carboxyfluorescein; FPA, fluorescence polarization assay; GFP, green fluorescent protein; GST, glutathione transferase; HEK, human embryonic kidney; IPTG, isopropyl β-D-thiogalactopyranoside; Luc7AC, C-terminal region of Luc7A; NF, nucleic-acid-free; NP40, Nonidet P40; RBM25, RNA-binding motif protein 25; rmsd, root mean square deviation; RRM, RNA-recognition motif; RT, reverse transcription; SAD, single-wavelength anomalous dispersion; SeMet, selenomethionine; snRNP, small nuclear ribonucleoprotein; SR, serine- and arginine-rich; ss, splicing site; ssDNA, single-stranded DNA; ssRNA, single-stranded RNA; WT, wild-type
INTRODUCTION
Alternative splicing plays a critical role in the regulation of eukaryotic gene expression, allowing a single gene to generate various protein isoforms that have different biological functions [1–3]. According to current estimates, 92–94% of human genes undergo alternative splicing [4], which often occurs during cell differentiation and developmental stages [5]. Apoptosis, or programmed cell death, a basic biological phenomenon that allows the regulated destruction and disposal of damaged or superfluous cells, is very important in maintaining homoeostasis [6]. The activation of one of several pathways presented in normal cells can induce apoptosis [7]. A significant number of genes involved in apoptosis undergo alternative splicing [8]. Bcl-x is an apoptotic factor, and the pre-mRNA of Bcl-x is alternatively spliced, yielding the pro-apoptotic Bcl-xS isoform and the anti-apoptotic Bcl-xL isoform, which have opposing functions during programmed cell death [9]. High Bcl-xL/Bcl-xS ratios are observed in a variety of cancers, indicating an important role for the longer (Bcl-xL) isoform in cancer cell survival [10–12]. A variety of signals and effectors that regulate Bcl-x alternative splicing have been identified [7]. RBM25 (RNA-binding motif protein 25), a member of the RBM (RNA binding motif) protein family, is a novel splicing factor that is localized to splicing-factor-rich nuclear speckles and is associated with multiple splicing components. RBM25 was reported to regulate the alternative splicing of the apoptotic factor Bcl-x pre-mRNA to yield the pro-apoptotic Bcl-xS isoform and the anti-apoptotic Bcl-xL isoform [13]. RBM25 activates the Bcl-xS 5′ ss (splicing site) via its interaction with the exonic splicing enhancer CGGGCA in the Bcl-x pre-mRNA and then promotes the recruitment of U1 snRNP (small nuclear ribonucleoprotein) to the weak Bcl-xS 5′ ss through an interaction with the U1 snRNP-associated factor Luc7A. Recently, the RBM25–Luc7A complex was also reported to mediate pathological SCN5A mRNA splicing in human heart failure [14].
RBM25 is an 843-residue protein that consists of three regions: the N-terminal RRM (RNA-recognition motif) domain (residues 87–164) that binds to RNA, the central RE/RD-rich domain (residues 285–664) that facilitates the localization of RBM25 to splicing-factor-rich nuclear speckles [13] and the C-terminal PWI domain (residues 734–843) that also binds to RNA [15]. Luc7A, a human homologue of yeast yLuc7p, is a protein that is part of the U1 snRNP and a newly identified SR (serine- and arginine-rich) protein [16]. Luc7A can affect 5′ ss selection by stabilizing the interactions between U1 snRNP and pre-mRNA [13]. The full-length RBM25 has been reported to associate with Luc7AC (C-terminal region of Luc7A) [13].
The PWI domain is an RNA/DNA-binding domain [15], but little is known about its function, and only two PWI domain structures are currently found in the PDB: the SRm160 PWI domain (PDB code 1MP1) and the Prp3 PWI domain (PDB code 1X4Q). The conserved flanking basic region has been suggested to serve as a co-operative partner of the PWI domain in the binding of nucleic acids. However, this region was absent from the solved structures of the SRm160 and Prp3 PWI domains. The RBM25 PWI domain and its flanking basic region remain poorly characterized. In the present study, we undertake a combined structural and functional characterization of the PWI domain and its flanking basic region of human RBM25. We report a 2.9 Å (1 Å=0.1 nm) resolution crystal structure of the PWI domain with its flanking basic region. The latter forms two α-helices and interacts with helix H4 of the PWI domain, which is the first structure of the flanking basic region of PWI domain. These interactions bring the basic residues in the flanking basic region close to the PWI domain, accordingly, an enlarged binding platform is provided for nucleic acids. We have determined the contact residues in the flanking basic region and in the PWI domain involved in RNA/DNA binding and characterized a nucleic-acid-binding surface in the RBM25 PWI domain that is entirely different from that in the SRm160 PWI domain. Furthermore, we show that the promotion of the Bcl-xS isoform expression by RBM25 is facilitated by the PWI domain in vivo. Thus our findings suggest that the PWI domain plays an important role in the regulation of Bcl-x pre-mRNA alternative splicing.
MATERIALS AND METHODS
Cloning, expression and purification of the PWI domain
The DNA fragments of the PWI domain (residues 734–843) were amplified by PCR from human bone marrow cDNA and then ligated into the NdeI/XhoI-cleaved plasmid p28, which is a modified pET-28a(+) vector with a deletion of the thrombin cleavage site, to yield plasmid p28-PWI. The PWI mutants were constructed by conventional PCR with the p28-PWI plasmid as a template using the MutanBEST kit (TaKaRa), yielding a series of point mutations (K744A, H737A+K739A, K734A+R735A+K736A, R769A+R770A, K777A+K778A, K825A+R828A, K837A+K838A, K749A, R772A, K797A and K843A) in the p28-PWI plasmids and the plasmid with a deletion of the flanking basic region, ΔHb (Δ734–745). All of the recombinant proteins were produced in Escherichia coli BL21(DE3) cells and induced at a D600 of 0.8–1.0 with 1 mM IPTG (isopropyl β-D-thiogalactopyranoside) at 16°C for 24 h. The recombinant proteins were purified using Ni2+-chelating columns (Qiagen) following the standard protocols and were also washed with a high-salt solution (1 M NaCl) before protein elution. The proteins were then purified further with HiLoad 16/60 Superdex 75 using a high-salt buffer (1 M NaCl), and the resulting proteins were dialysed into buffer A (25 mM NaH2PO4, pH 5.65, 150 mM NaCl and 1 mM EDTA) and concentrated to 10–20 mg/ml. The methionine residues of SeMet (selenomethionine)-derived proteins were replaced with SeMet, to impede the methionine biosynthesis, and PWI was expressed in the methionine-auxotrophic E. coli strain B834 (Novagen) using LR medium supplemented with SeMet and six amino acids (leucine, isoleucine, valine, phenylalanine, lysine and threonine) [17,18]. The SeMet-derived proteins were purified using the same procedure described above.
Crystallization of the PWI domain and data collection
Crystals of both the native and SeMet-derived proteins were grown using the hanging-drop vapour-diffusion method at 281 K. The crystals suitable for X-ray diffraction of the native protein were grown in 0.2 M ammonium sulfate, 0.1 M Hepes (pH 7.5) and 25% (w/v) poly(ethylene glycol) 3350, with a protein concentration of 0.5 mM in buffer A, and the crystals of the SeMet-derived protein were grown in 0.1 M ammonium sulfate, 0.1 M Hepes (pH 7.5), and 15% (w/v) poly(ethylene glycol) 3350, with the same protein concentration in buffer A. The crystals were soaked in a cryoprotectant (70% reservoir solution supplemented with 30% glycerol) and flash-cooled to 100 K in a liquid nitrogen stream. The native crystal dataset was collected at a radiation wavelength of 0.9194 Å on a beamline BL17U at 100 K with an MX-225 CCD (charge-coupled device) (Marresearch, Germany) at the Shanghai Synchrotron Radiation Facility (SSRF). The SAD (single-wavelength anomalous dispersion) dataset was collected at one wavelength (λpeak=0.9790 Å), and both native and SAD data were processed using HKL2000 [19] and programs in the CCP4 package [20].
Structure determination and refinement
Phases were solved using the phenix.autosol wizard [21] with the SAD data, and the initial model was constructed likewise. The automated model-building program BUCCANEER [20] was used to further build the model, which was elaborated further and refined at 2.9 Å resolution using coot [22], phenix.refine [23] and Refmac5 [24] in the CCP4 package, NCS (non-crystallographic symmetry) restraints were used during model refinement. The final crystallography Rfactor and Rfree values were 26.4% and 29.8% respectively. TLS (Translation–Libration–Screw-rotation) refinement [25] was executed in Refmac5 at the final stage. The stereochemistry of the structure was verified by MolProbity [26]. The data collection and refinement statistics are listed in Table 1. The Figures were prepared using PyMOL (http://www.pymol.org).
Table 1. Crystallographic statistics.
The values in parentheses are for the highest resolution shell. Rmerge =ΣhklΣi|Ii(hkl)−<I(hkl)>|/ΣhklΣi|Ii(hkl)|, where Ii(hkl) is the intensity of an observation and <I(hkl)> is the mean value for its unique reflection; summations are over all reflections. Rfactor=Σh|Fo(h)|−|Fc(h)|/Σh|Fo(h)|, where Fo and Fc are the observed and calculated structure factor amplitudes respectively. Rfree was calculated with 5% of the data excluded from the refinement.
| Parameter | Native | SeMet |
|---|---|---|
| Data collection | ||
| Space group | C2221 | C2221 |
| Cell dimensions | ||
| a, b, c (Å) | 81.58, 86.17, 197.17 | 81.34, 86.57, 199.85 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Molecules per asymmetric unit | 5 | 5 |
| Peak | ||
| Wavelength (Å) | 0.9194 | 0.9790 |
| Resolution (Å) | 50.00–2.80 (2.85–2.80) | 50.00–3.00 (3.11–3.00) |
| Rmerge (%) | 6.7 (57.8) | 8.2 (78.4) |
| I/σI | 21.9 (1.9) | 18.1 (1.9) |
| Completeness (%) | 97.0 (84.7) | 96.9 (92.4) |
| Redundancy | 5.9 (4.0) | 5.7 (4.6) |
| Refinement | ||
| Resolution (Å) | 49.30–2.90 (2.98–2.90) | |
| Number of reflections | 14732 | |
| Rfactor/Rfree (%) | 26.4/29.8 | |
| Number of atoms of protein | 4150 | |
| Average B-factors | 81.65 | |
| Rmsd | ||
| Bond lengths (Å) | 0.0066 | |
| Bond angles (°) | 0.9728 | |
| Ramachandran plot (%) | ||
| Favoured (98%) regions | 97.32% (509/523) | |
| Allowed (>99.8%) regions | 100% (523/523) | |
| PDB code | 3V53 |
FPAs (fluorescence polarization assays)
FPAs were performed in buffer B (25 mM Tris/HCl, pH 8.0, 150 mM NaCl and 1 mM EDTA) at 298 K using a SpectraMax M5 Microplate Reader system to investigate the PWI domain–nucleic acid interactions. The fluorescence excitation and emission wavelengths were 490 nm and 530 nm respectively. The single- and double-stranded nucleic acids were labelled at their 5′ ends using 5′ FAM (6-carboxyfluorescein) [27]. We used 100 nM fluorescently labelled DNA probes and 40 nM fluorescently labelled RNA probes as ligands in 96-well plates respectively, and mixed these ligands with various amounts of PWI or PWI mutants in a final volume of 200 μl. Nucleic-acid-free controls (PWI or PWI mutants only) were included in each assay. The fluorescence polarization, P (in mP units), was calculated using the equation [28]:
| (1) |
The fluorescence polarization change, ΔP (in mP units), was fitted to the equation [29]:
| (2) |
The sequences of the 5′ FAM-labelled nucleic acid probes are listed in Table 2.
Table 2. Sequences of 5′ FAM-labelled DNA/RNA probes.
| 5′ FAM-labelled DNA/RNA probe | Sequence |
|---|---|
| ssDNA | 5′-FAM-CCTTACAGCAAA-3′ |
| dsDNA | 5′-FAM-CCTTACAGCAAA-3′ |
| 5′-FAM-TTTGCTGTAAGG-3′ | |
| ssRNA | 5′-FAM-AUCGGGCA-3′ |
| dsRNA | 5′-FAM-AUCGGGCA-3′ |
| 5′-FAM-UGCCCGAU-3′ |
CD measurements
The CD spectra of PWI and PWI mutants were carried out on a Jasco-810 spectropolarimeter at 298 K. The spectra were recorded at wavelength between 190 and 250 nm using a 0.1 cm pathlength cell. A buffer-only sample was used as reference.
Vector construction, cell culture and transfection
The plasmid pCMV6-XL5-RBM25, containing the cloned full-length human RBM25 cDNA, was purchased from OriGene. The coding sequence was subcloned into the p3×FLAG-Myc-CMV-24 vector, yielding plasmid 3×FLAG-RBM25, and the RBM25 mutants were created by conventional PCR using the MutanBEST kit (TaKaRa) with the 3×FLAG-RBM25 plasmid as a template. The 3×FLAG-RBM25ΔPWI nomenclature indicates that the PWI domain and the flanking basic region (residues 734–843) were deleted from RBM25, 3×FLAG-PWI indicates that the mutant contains only the PWI domain and the flanking basic region. HEK (human embryonic kidney)-293 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum using standard approaches. Transient transfection was performed with Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions.
RT (reverse transcription)–PCR and Western blotting
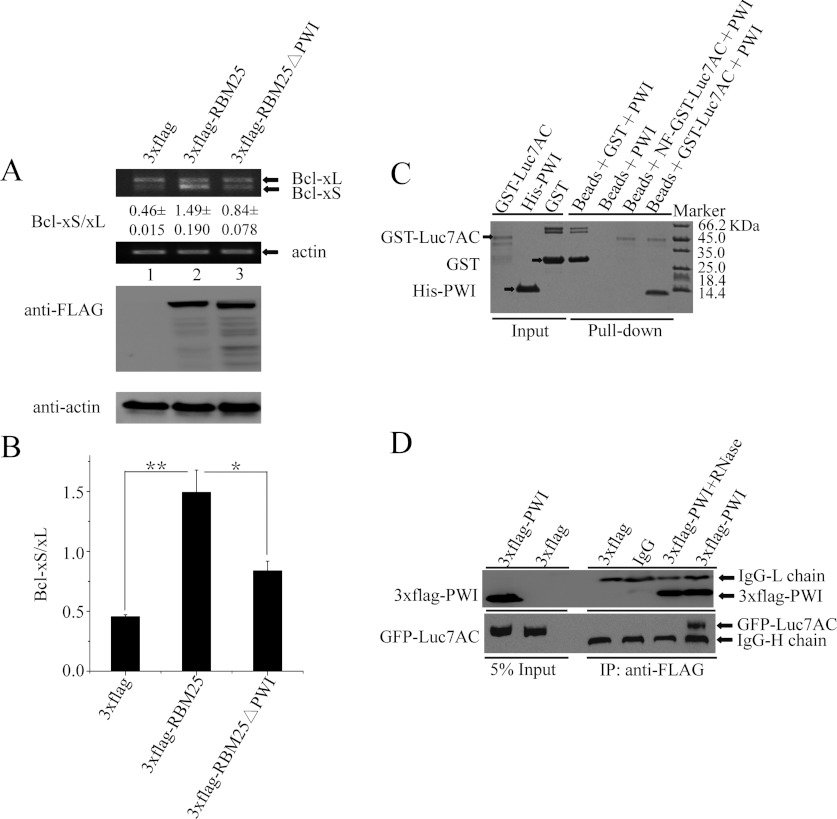
Semi-quantitative RT–PCR analysis of the alternative splicing products was performed as reported previously [30]. HEK-293 cells were harvested 36 h after transfection for RT–PCR and Western blotting. RNAs were isolated with TRIzol® reagent (Invitrogen) from cells transfected with the various vectors and reverse-transcribed with oligo(dT) primer. For the Bcl-x splicing analyses, PCR was performed with the primers Bcl-S, 5′-ATGTCTCAGAGCAACCGGGAGCTG-3′, and Bcl-A, 5′-TCATTTCCGACTGAAGAGTGAGCC-3′. Each experiment was repeated in triplicate. Western blot analysis was performed as described previously [31] using anti-FLAG and anti-actin antibodies. All values of RT–PCR are presented as means±S.D. Statistical analysis of the data was performed using Student's t test to evaluate the statistical significance of differences. P<0.05 (*) was considered significant and P<0.01 (**) was considered highly significant.
Co-immunoprecipitation and Western blot analysis
The C-terminal half of human U1 snRNP-associated factor Luc7A (residues 225–432) was cloned into the pEGFP-N1 vector to yield the GFP-Luc7AC plasmid. In the co-immunoprecipitation assay, two 6-cm-diameter dishes of HEK-293T cells (transiently transfected with 3×FLAG/GFP-Luc7AC control plasmids and 3×FLAG-PWI/GFP-Luc7AC plasmids respectively) were harvested [32], cells were washed with PBS and lysed in NP40 (Nonidet P40) lysis buffer containing 50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 1% (v/v) NP40 and a protease inhibitor cocktail (Roche). Briefly, after centrifugation, total proteins were then incubated overnight at 10°C with anti-FLAG antibodies (Sigma) and Protein A/G Plus–agarose beads (Santa Cruz Biotechnology), and immunoprecipitation complexes were washed twice in NP40 lysis buffer. Protein complexes were eluted by boiling in 2×SDS/PAGE loading buffer and subjected to Western blot analysis. Western blot analysis was performed as described previously [31] using anti-FLAG and anti-GFP (green fluorescent protein) antibodies.
GST (glutathione transferase) pull-down assay
The C-terminal half of human U1 snRNP-associated factor Luc7A (residues 225–432) was cloned into the pGEX4T vector to yield the GST-Luc7AC plasmid. GST–Luc7AC expression was induced at a D600 of 0.8–1.0 with 0.1 mM IPTG at 16°C for 24 h. The recombinant proteins were purified using glutathione–Sepharose 4B beads (GE Healthcare) following standard protocols. The resulting proteins were purified further with HiLoad 16/60 Superdex 200 using a high-salt buffer (1 M NaCl), yielding the NF (nucleic-acid-free) proteins. In pull-down assay, GST-tagged proteins and GST (control) were bound to glutathione–Sepharose 4B beads in buffer C (25 mM sodium phosphate, pH 6.5, 150 mM NaCl, 0.5% NP40 and 1 mM EDTA) by incubation for 1 h at 8°C in an Eppendorf tube followed by washing five times with 500 μl of buffer C and resuspension in 500 μl of buffer C. Next, the same amount of PWI was added to the suspensions, which were then incubated for 2 h at 8°C for the binding reactions. Finally, the beads were each washed five times with 500 μl of buffer C and then mixed with 30 μl of 2×SDS/PAGE loading buffer and heated for 10 min at 100°C. All samples were analysed by SDS/PAGE using Coomassie Brilliant Blue staining.
RESULTS
Crystal structure of the PWI domain and its flanking basic region
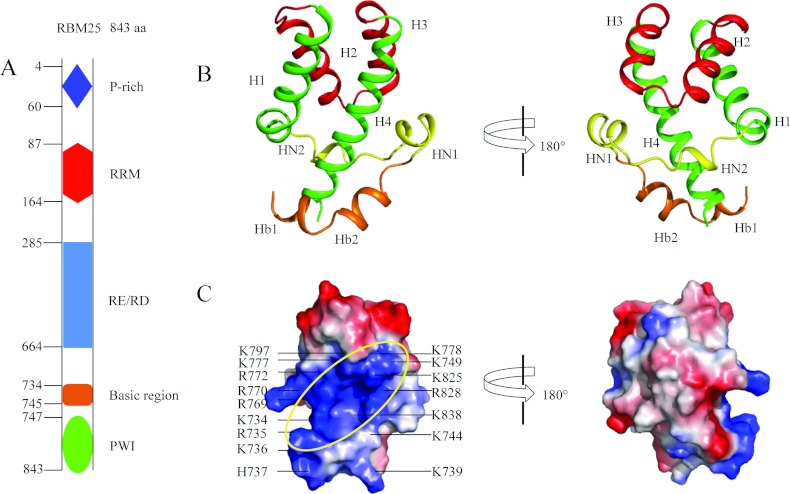
The human RBM25 consists of a P (proline)-rich region and an N-terminal RRM domain, a central RE/RD-rich domain and a C-terminal PWI domain (Figure 1A). The crystal structure of the PWI domain and its flanking basic region (residues 734–843) was determined by SAD phasing; the crystallographic statistics are summarized in Table 1. There are five molecules in each asymmetric unit; overall alignment of the backbones of the five molecules is shown in Supplementary Figure S1(A) at http://www.biochemj.org/bj/450/bj4500085add.htm. The superimposition was generalized using PyMOL software. The overall structures of the five molecules in each asymmetric unit are similar to each other, which adopt almost identical conformations. The calculated Cα rmsd (root mean square deviation) values are shown in Supplementary Figure S1(B); the mean value of pairwise rmsd is ~0.48 Å. The structure shows that the PWI domain comprises a conserved four-helix bundle (H1, H2, H3 and H4) and two structured N-terminal elements (HN1 and HN2). HN1 (residues 749–754) forms an α-helix, and HN2 (residues 759–761) forms a 310-helix (Figure 1B). HN2 is composed of Trp759, Ser760 and Ile761 (Figure 2). The core helix, H2, parallels helix H3, and the H1 helix is almost parallel with H4. The flanking basic region (residues 734–745) forms two α-helices (Hb1 and Hb2) (Figure 1B) near H4 of the PWI domain.
Figure 1. Crystal structure of the RBM25 PWI domain and its flanking basic region.
(A) Schematic diagram of the structural organization of RBM25 and its mutants. RBM25 consists of a proline (P)-rich region and an N-terminal RRM domain, a central RE/RD-rich domain and a C-terminal PWI domain. Amino acid positions are indicated on the left. (B) The structure of the PWI domain and its flanking basic region. The basic region contains two helices (orange). The two views differ by a 180° rotation around the vertical axis. (C) The electrostatic potential surface of the PWI domain and its flanking basic region. The ellipse highlights the surface region rich in positive charge; the black lines mark the basic residues. Single-letter code is used for amino acids. The orientations of (C, left) and (B, left) are the same.
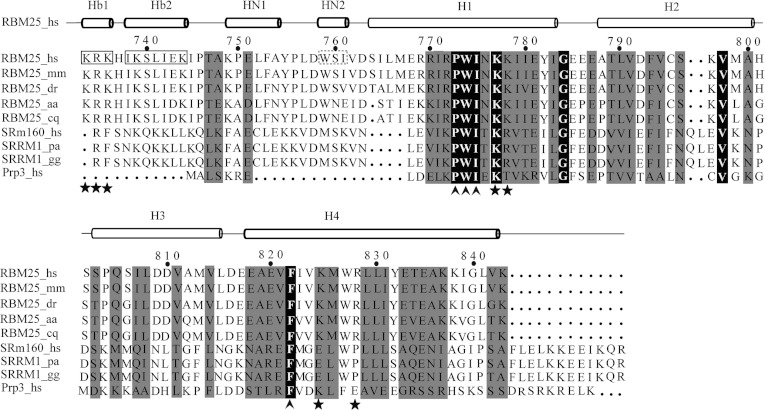
Figure 2. Sequence alignment of the RBM25 PWI domain with its homologues.
The following sequences were aligned using ClustalW2 (http://www.genome.jp/tools/clustalw): Homo sapiens (hs) RBM25 (NP_067062.1, NM_021239.2), Mus musculus (mm) RBM25 (NP_081625.3, NM_027349.3), Danio rerio (dr) RBM25 (NP_956084.1, NM_199790.1), Aedes aegypti (aa) RBM25 (XP_001649953.1, XM_001649903.1), Culex quinquefasciatus (cq) RBM25 (XP_001850266.1, XM_001850214.1), Homo sapiens (hs) SRm160 (NP_005830.2, NM_005839.3), Pongo abelii (pa) SRRM1 (NP_001126712.1, NM_001133240.1), Gallus gallus (gg) SRRM1 (NP_001026665.1, NM_001031494.1) and Homo sapiens (hs) Prp3 (NP_004689.1, NM_004698.2). The box highlights the flanking basic regions Hb1 and Hb2. The broken box highlights the 310-helix (HN2). The highly conserved residues Pro-Trp-Ile and phenylalanine are labelled with an upwards-pointing chevron. The contact residues for RNA/DNA binding are labelled with a star.
The electrostatic potential surface shows a positively charged region stretching across one side of PWI domain and the flanking basic region (Figure 1C). Twelve positively charged residues in the PWI domain (Lys749, Arg769, Arg770, Arg772, Lys777, Lys778, Lys797, Lys825, Arg828, Lys837, Lys838 and Lys843) and six positively charged residues in the basic region (Lys734, Arg735, Lys736, His737, Lys739 and Lys744) make up the positively charged surface (Figure 1C), and the PWI domain probably binds nucleic acids using this positively charged region. In fact, together with the biochemical results (details below), we confirm that the RBM25 PWI domain binds nucleic acids via this positively charged surface.
Structural comparison of the PWI domain of RBM25 and the PWI domains of other proteins
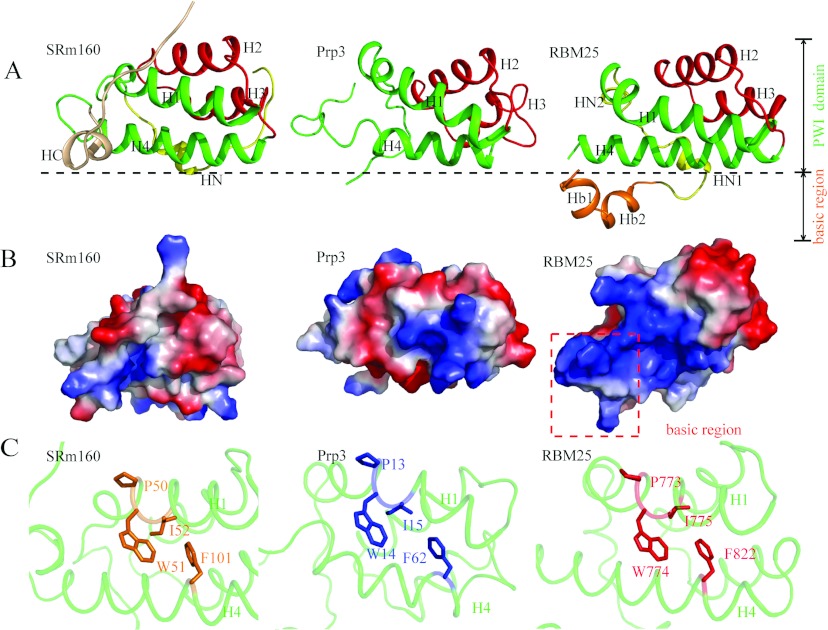
To date, only two PWI domain structures have been deposited in the PDB: the SRm160 PWI domain (PDB code 1MP1) and the Prp3 PWI domain (PDB code 1X4Q). Their overall sequence homologies with the RBM25 PWI domain are 22% and 19% respectively. All of the PWI domains have a highly conserved four-helix bundle, but only the SRm160 PWI domain has a C-terminal element (HC), which is orthogonal to the core helices [15]. The Prp3 PWI domain has no C- or N-terminal elements. The RBM25 PWI domain has two N-terminal elements (Figure 3A). HN1 folds into an α-helix, and HN2 folds into a 310-helix.
Figure 3. Structural comparison of RBM25 PWI with the SRm160 and Prp3 PWI domains.
(A) Structural comparisons of RBM25 PWI with the SRm160 and Prp3 PWI domains. Only our structure shows the structure of the flanking basic region. H1 and H4 are shown in green, H2 and H3 are shown in red, the N-elements (HN) are shown in yellow, the C-element (HC) is shown in wheat, and the flanking basic region is shown in orange. (B) Comparison of surface electrostatic potentials of RBM25 PWI domain with the SRm160 and Prp3 PWI domains. The red broken box encloses the flanking basic region. (C) The highly conserved phenylalanine residue in H4 packs against the tryptophan and isoleucine residues in the highly conserved signature Pro-Trp-Ile sequence of H1. Single-letter code is used for amino acids.
The conserved flanking basic region has been suggested to act as a co-operative partner with the PWI domain in the binding of nucleic acids [15]. However, this region was absent from the solved structures of the SRm160 and the Prp3 PWI domains [15]. In the present paper, we report the crystal structure of the flanking basic region of RBM25. We have shown that the flanking basic region forms two α-helices (Figure 1B) and interacts with H4 of the PWI domain via a hydrogen bond and hydrophobic and ionic interactions (details below), which were analysed using the Protein Interactions Calculator (http://pic.mbu.iisc.ernet.in/job.html).
Inspection of the surface properties of the RBM25 PWI domain reveals that there is a surface region rich in basic residues, but there are no positively charged regions in the overall electrostatic potential surface of SRm160 PWI domain and Prp3 PWI domain (Figure 3B). These differences in surface properties indicate that PWI domains may have diversified binding surfaces for nucleic acids.
The PWI domain is named after an almost invariant signature Pro-Trp-Ile sequence [33] (Figure 2). This motif localizes to H1 and is primarily involved in hydrophobic interactions within the core helices [15], where there is a highly conserved phenylalanine residue in H4 packed against the Trp-Ile pair, similar to the arrangement of the SRm160 PWI domain and the PRP3 PWI domain (Figure 3C), the highly conserved Pro-Trp-Ile motif resides in H1 of the RBM25 PWI domain and the Trp774 and Ile775 residues packed against the highly conserved Phe822 domain in H4 (Figure 3C). These highly conserved residues probably stabilize the four-helix bundle through the formation of a hydrophobic core.
The flanking basic region is necessary for the nucleic-acid-binding activity of the PWI domain
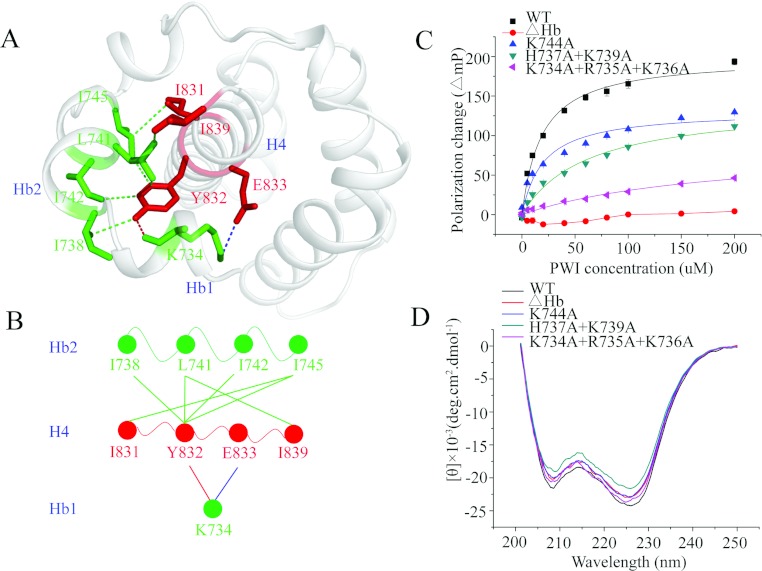
The crystal structure of the PWI domain and its flanking basic region reveals that the flanking basic region interacts with H4 of the PWI domain. The hydroxy group of Tyr832 in H4 was demonstrated to form a hydrogen bond with the carbonyl group of Lys734 in Hb1. Moreover, Lys734 interacts with the Glu833 region in H4 through ionic interactions, and Tyr832 interacts with Ile738, Leu741, Ile742 and Ile745 in Hb2 through hydrophobic interactions. In addition, Leu741 and Ile745 also interact with Ile839 and Ile831 respectively in H4, through hydrophobic interactions (Figures 4A and 4B). These interactions bring the residues in the flanking basic region close to the PWI domain, promoting directly the formation of an enlarged basic-residue-rich region.
Figure 4. The flanking basic region acts as a co-operative partner with the PWI domain in the binding of nucleic acids.
(A) The interactions between the flanking basic region and helix H4 of the PWI domain. The blue broken line represents the ionic interaction between Glu833 in H4 and Lys734 in Hb1; the red broken line represents the hydrogen bond between Tyr832 in H4 and Lys734 in Hb1; and the green broken lines represent hydrophobic interactions. Single-letter code is used for the amino acids. (B) A brief model of the interactions between the flanking basic region and helix H4 of the PWI domain. The colour code is the same as in (A). (C) The FPAs of the PWI domain and its mutants K744A, H737A+K739A, K734A+R735A+K736A and ΔHb: interactions with 5′ FAM-labelled ssRNA. (D) CD spectra of the PWI domain and its mutants.
The flanking basic region of the SRm160 PWI domain has been proposed to act as a co-operative partner in the binding of nucleic acids [15]. There are six positively charged residues in the flanking basic region of the RBM25 PWI domain; to investigate whether the flanking basic region contributes in nucleic acid binding, the six positively charged residues were mutated to alanine. We performed three mutants to test the RNA/DNA-binding activity of these basic residues using FPAs: one single mutant and two combined mutants (the residues are adjacent). The binding affinities (reported as values of the dissociation constant, Kd) for the single mutant K744A binding to ssRNA (single-stranded RNA) and dsDNA (double-stranded DNA) were ~19.9 μM (Figure 4C and Table 3) and ~23.6 μM (Supplementary Figure S2A at http://www.biochemj.org/bj/450/bj4500085add.htm) respectively; no significant changes were observed in the Kd values compared with the WT (wild-type), for which Kd values for ssRNA and dsDNA were ~11.3 μM and ~6.7 μM respectively. The Kd values for the combined mutant H737A+K739A binding to ssRNA and dsDNA were ~61.6 μM (Figure 4C and Table 3) and ~57.0 μM (Supplementary Figure S2A) respectively, slightly increased values relative to the WT, indicating that the contribution of these residues to RNA/DNA binding is only minor. The Kd values for the combined mutant K734A+R735A+K736A binding to ssRNA and dsDNA were ~182.9 μM (Figure 4C and Table 3) and ~193.5 μM (Supplementary Figure S2A) respectively, representing significant increases relative to the WT. These mutations did not appear to perturb the secondary structure as examined by CD (Figure 4D). These results reveal that Lys734, Arg735 and Lys736 in the flanking basic region are the contact residues for RNA/DNA binding (Figures 2 and 5D) and confirm that the flanking basic region indeed contributes to nucleic acid binding. We also tested the RNA/DNA-binding activity of a mutant with deletion of this flanking basic region (ΔHb, Δ734–745) using FPAs. Similarly to the basic region of the SRm160 PWI domain, the RBM25 PWI domain without the flanking basic region was unable to bind RNA/DNA (Figure 4C and Supplementary Figure S2A). These results reveal that the flanking basic region is essential for the nucleic-acid-binding activity of the PWI domain, indicating that this flanking basic region acts as a co-operative partner to facilitate the stable binding of the PWI domain to nucleic acids.
Table 3. Kd values for PWI domain or PWI mutants binding to a 5′ FAM-labelled RNA probe.
The probe used was 5′-FAM-AUCGGGCA-3′. ND, interaction not detectable under the experimental conditions.
| Protein | Kd value (μM) |
|---|---|
| WT | 11.3±1.0 |
| ΔHb | ND |
| K744A | 19.9±4.4 |
| H737A+K739A | 61.6±9.3 |
| K734A+R735A+K736A | 182.9±44.7 |
| R769A+R770A | 32.1±1.4 |
| K777A+K778A | 155.9±29.4 |
| K825A+R828A | 106.1±17.3 |
| K837A+K838A | 15.7+2.1 |
Figure 5. The PWI domain interacts with nucleic acids.
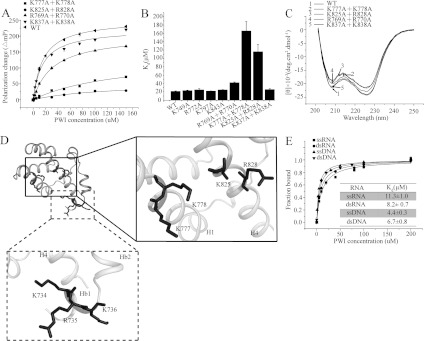
(A) The FPAs of the PWI and the double mutants K777+K778, R769A+R770A, K825A+R828A and K837A+K838A interactions with 5′ FAM-labelled ssRNA. Results are means±S.D. for three experiments. (B) Comparison of the Kd values for the binding of PWI mutants to ssRNA with the PWI domain. Values are means±S.D. for three experiments. (C) CD spectra of PWI and its mutants. (D) Structure-guided mutations reveal the contact amino acids for nucleic acid binding. The box indicates the contact residues in the PWI domain, and the broken box encloses the contact residues in the flanking basic region. Single-letter code is used for amino acids. (E) The FPAs of the PWI domain interactions with single- and double-stranded nucleic acids. The PWI domain exhibits similar binding affinities with single- and double-stranded nucleic acids. Results are means±S.D. for three experiments.
Structure-guided mutagenesis reveals important contact residues for RNA/DNA binding
The PWI domain has been reported to function as a novel nucleic-acid-binding domain [15]. In RBM25, there are 12 positively charged residues in the PWI domain and six positively charged residues in the basic region. These residues form a positively charged surface. Similarly to its flanking basic region, the PWI domain probably binds nucleic acids using its positively charged region. To verify this hypothesis, the 12 basic residues in the PWI domain were mutated to alanine. We constructed eight mutants to test the RNA/DNA-binding affinities of these basic residues using FPAs: four double mutants, each with two adjacent basic residues, and four single mutants. The Kd values of the double mutants K837A+K838A and R769A+R770A were ~15.7 μM and ~32.1 μM respectively for ssRNA (Figure 5A and Table 3) and ~40.4 μM and ~46.0 μM for dsDNA (Supplementary Figure S2B); slightly increased values relative to the WT, for which Kd values for ssRNA and dsDNA were ~11.3 μM and ~6.7 μM respectively. Similarly, the single mutants K749A, R772A, K797A and K843A bound to ssRNA and dsDNA with similar Kd values of approximately 12–25 μM (Figure 5B and Supplementary Figures S2C and S2D), indicating that the contribution of these basic residues to RNA/DNA binding is only minor. The Kd values for the double mutants K777A+K778A and K825A+R828A (these two residues are adjacent in the secondary structure rather than the primary sequence) were ~155.9 μM and ~106.1 μM respectively for ssRNA (Figure 5A and Table 3), and ~159.7 μM and ~105.5 μM for dsDNA (Supplementary Figure S2B), representing significant increases relative to the WT. The CD spectra indicate that the secondary structure has not been perturbed significantly (Figure 5C). These results indicate that Lys777 and Lys778 in helix H1 and Lys825 and Arg828 in helix H4 are the contact residues for RNA/DNA binding (Figures 2 and 5D) and confirm that the PWI domain uses the same surface to bind both RNA and DNA.
The PWI domain has an equal preference for binding to single- or double-stranded nucleic acids
The SRm160 PWI domain has an equal preference for single- and double-stranded nucleic acids [15]. To assess the binding affinity of the RBM25 PWI domain for single- and double-stranded nucleic acids, we performed binding assays with ssRNA, dsRNA (double-stranded RNA), ssDNA (single-stranded DNA) and dsDNA using FPAs. The Kd values for the PWI domain when binding to ssRNA, dsRNA, ssDNA and dsDNA oligonucleotides were similar at ~11.3 μM, ~8.2 μM, ~4.4 μM and ~6.7 μM respectively (Figure 5E). These results reveal that the RBM25 PWI domain has an equal preference for binding to either single- or double-stranded nucleic acids.
The modulation of Bcl-x pre-mRNA alternative splicing by RBM25 is facilitated by the PWI domain
RBM25 regulates Bcl-x pre-mRNA 5′ ss selection through interactions with Luc7A [13]; to investigate whether the PWI domain facilitates the recruitment of U1 snRNP by RBM25 to the weak Bcl-xS 5′ ss in vivo, we examined changes in the expression of the Bcl-x isoform with overexpression of 3×FLAG-RBM25ΔPWI, 3×FLAG-RBM25 and 3×FLAG, with the last-named being the control construct. All the vectors were transfected into HEK-293 cells, and the cells were harvested 36 h after transfection. The expression of RBM25 and the mutant constructs was validated by Western blot assay using anti-FLAG antibodies. RNA was isolated from cells transfected with the vectors and analysed for the alternative splicing patterns of the Bcl-x gene by RT–PCR. The empty vector (3×FLAG) produced two splice variants of the Bcl-x transcripts, with a Bcl-xS/Bcl-xL ratio of ~0.46 in HEK-293 cells, meaning that the Bcl-xL transcripts were significantly more numerous than the Bcl-xS transcripts (Figures 6A, lane 1, and 6B). The overexpression of RBM25 (3×FLAG-RBM25) significantly increased the ratio of Bcl-xS to Bcl-xL to ~1.49 (Figures 6A, lane 2, and 6B) (**P<0.01), indicating that increased recruitment of U1 snRNP stimulates the usage of the weak Bcl-xS 5′ ss when RBM25 is overexpressed. However, the effect of RBM25 was significantly inhibited when the PWI domain was deleted from RBM25 (3×FLAG-RBM25ΔPWI), resulting in a marked decrease in the expression of the Bcl-xS isoform, yielding a Bcl-xS/Bcl-xL ratio of ~0.84 (Figures 6A, lane 3, and 6B) (*P<0.05). These results demonstrate that the modulation of Bcl-x pre-mRNA alternative splicing by RBM25 is facilitated by the PWI domain in vivo.
Figure 6. Effects of the PWI domain on the alternative splicing of the Bcl-x gene.
(A) Deletion of the PWI domain decreased the expression of Bcl-xS isoforms. (B) Means±S.D. densitometric determination of the ratios of Bcl-xS to Bcl-xL in the three trials. All statistical analyses were performed using Student's t test (*P<0.05; **P<0.01). (C) The GST pull-down assay shows that the PWI domain was pulled down only with GST–Luc7AC, but not with NF-GST–Luc7AC. (D) The co-immunoprecipitation assay shows that GFP–Luc7AC was found only in the immunoprecipitate (IP) complexes without prior RNase treatment, but not with prior RNase treatment. 3×FLAG-PWI+RNase denotes RNase treatment of the 3×FLAG-PWI cell lysates.
RNA molecules serve as a bridge between the PWI domain and Luc7AC
RBM25 can associate with Luc7AC [13]. To examine whether the PWI domain interacts with Luc7AC, we investigated the interactions of PWI with Luc7AC (residues 225–432) using GST pull-down and co-immunoprecipitation assays. Because both the PWI domain and Luc7AC can bind to RNA, we performed a GST pull-down experiment using NF-GST–Luc7AC to examine whether RNA molecules serve as a bridge between the PWI domain and Luc7AC. The results show that the PWI domain was pulled down only with the GST–Luc7AC, but not with the NF-GST–Luc7AC (Figure 6C). We also performed overexpression-based co-immunoprecipitation assay in the HEK-293T cell line with co-expressed FLAG-epitope-tagged PWI domain and GFP-epitope-tagged Luc7AC. The cell lysates were immunoprecipitated with anti-FLAG monoclonal antibodies, and the co-precipitated GFP–Luc7AC was detected by Western blot analysis with anti-GFP antibodies. Similarly, we performed a control sample in which the cell lysates were treated with RNase. GFP–Luc7AC was found only in the immunoprecipitate complexes without prior RNase treatment, but not with prior RNase treatment (Figure 6D). These results indicate that RNA molecules can serve as a bridge between the PWI domain and Luc7AC.
DISCUSSION
At the time of writing, there were only two structures for PWI domains in the PDB: SRm160 PWI (PDB code 1MP1) and Prp3 PWI (PDB code 1X4Q). The conserved flanking basic region has been suggested to serve as a co-operative partner for the PWI domain during its binding to nucleic acids [15]. However, this region is absent from the solved structures of the SRm160 and Prp3 PWI domains [15]. Thus the structure of the flanking basic region and how this region affects the RNA/DNA-binding activity of the PWI domain remain unknown. For the first time, we report the crystal structure of the RBM25 PWI domain together with its flanking basic region (Figure 1B). This structure demonstrates that the flanking basic domain forms two α-helices that are intimately associated with H4 of the PWI domain through hydrogen bond and hydrophobic and ionic interactions (Figure 4A). These interactions bring the basic residues in the flanking basic region close to the PWI domain, promoting directly the formation of an enlarged basic-residue-rich region. Accordingly, an enlarged binding platform is provided for nucleic acids, which facilitates the stabilization of the interactions between the PWI domain and nucleic acids. Structure-guided mutagenesis revealed that Lys734, Arg735 and Lys736 in the flanking basic region are the contact residues for both RNA and DNA binding, indicating that the flanking basic region indeed contributes to nucleic acid binding. After the deletion of the flanking basic region, the RBM25 PWI domain was unable to bind to RNA/DNA (Figure 4C and Supplementary Figure S2A), meaning that the flanking basic region is essential for RNA/DNA binding. The enlarged binding platform may be the main mechanism by which the conserved flanking basic region acts as a co-operative partner with the PWI domain in the binding of nucleic acids.
The PWI domain is a nucleic-acid-binding domain. Structure-guided mutagenesis revealed that the positively charged surface composed of the basic residues of the RBM25 PWI domain is crucial for both RNA and DNA binding (Figure 5A and Supplementary Figure S2B). The most important residues are Lys777 and Lys778 in H1 and Lys825 and Arg828 in H4. This nucleic-acid-binding surface is entirely different from that of the SRm160 PWI domain. In the latter, the nucleic-acid-binding sites are located on the turns between helices 1 and 2 and helices 3 and 4 [15]. The reason for these differences may be that the SRm160 PWI domain has no region rich in basic residues. A similar situation has been also shown to exist for RRM domains, which provide highly plastic platforms for nucleic acid binding. In order to achieve high affinity and specificity, RRM domains show remarkable adaptability in RNA recognition [34]: some RRM proteins use loop 1, 3 or 5, whereas others use the C- and N-termini or the β4 and β2 strands [34]. In addition, the β-sheet surface of an RRM can be modulated by using only one or up to four β-strands for RNA binding [34]. Similarly to RRM domains, the differences in RNA recognition between the RBM25 PWI domain and SRm160 PWI domain indicate that PWI domains also provide diversified binding surfaces for nucleic acids. The RNA/DNA-binding properties indicate that the PWI domain probably binds primarily to the sugar-phosphate backbones of nucleic acids through electrostatic interactions.
Several RNA-binding domains are known to play significant roles in gene expression [35], including RRM [36], KH (K homology) [37], dsRBD (double-stranded RNA-binding domain) [38], zinc-finger motifs [39] and several basic-residue-rich motifs [40]. The PWI domain is a newly identified RNA/DNA-binding domain. However, little is known about the roles of PWI domains, except that the SRm160 PWI domain is involved in stimulating 3′ end formation [15]. The PWI domain may also play an important role in the assembly of splicing complexes in the presence of the SR domain [33], but the underlying mechanism remains unclear. In the present study, deletion of the PWI domain decreases the expression of Bcl-xS significantly (Figures 6A and 6B) in HEK-293 cells, indicating that the PWI domain plays an important role in modulating Bcl-x pre-mRNA alternative splicing in vivo. RBM25 activates the Bcl-xS 5′ ss via its interaction with the exonic splicing enhancer CGGGCA in the Bcl-x pre-mRNA and then promotes the recruitment of U1 snRNP to the weak Bcl-xS 5′ ss through an interaction with the U1 snRNP-associated factor Luc7A [13]. GST pull-down and co-immunoprecipitation assays indicate that RNA molecules can serve as a bridge between the PWI domain and Luc7AC, but it is unclear whether this complex plays a functional role in the Bcl-x pre-mRNA alternative splicing in vivo; further investigation will be needed to explain the significance of this finding. In addition, further experiments will be required to elucidate the details of the interactions between RBM25 and Luc7A.
Online data
AUTHOR CONTRIBUTION
Deshun Gong and Yunyu Shi designed the research. Deshun Gong performed the biochemical experiments and wrote the paper. Deshun Gong, Fudong Li and Minhao Wu conducted the structural determination and refinement. Fan Yang performed the RT–PCR assay. Dandan Qian, Zhenhua Shao, Mian Wu, Jihui Wu and Yunyu Shi helped with editing the paper before submission.
ACKNOWLEDGEMENTS
Single-crystal X-ray diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) using beamline BL17U. We thank Dr Lei Liu, Dr Yu Qiu, Dr Zhiyong Zhang, Dr Weiwei Wang and Dr Zhenwei Song for their valuable suggestions throughout this project.
FUNDING
This work was supported by the National Basic Research Program of China [grant numbers 2012CB917201, 2011CB966302 and 2011CB911104], and the Chinese National Natural Science Foundation [grant numbers 30830031 and 31170693].
References
- 1.Black D. L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis T., Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 3.Stetefeld J., Ruegg M. A. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem. Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luco R. F., Allo M., Schor I. E., Kornblihtt A. R., Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwerk C., Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 7.David C. J., Manley J. L. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C. Y., Chu J. Y., Yu J. K., Huang X. Q., Liu X. J., Shi L., Che Y. C., Xie J. Y. Regulation of alternative splicing of Bcl-x by IL-6, GM-CSF and TPA. Cell Res. 2004;14:473–479. doi: 10.1038/sj.cr.7290250. [DOI] [PubMed] [Google Scholar]
- 9.Boise L. H., Gonzalezgarcia M., Postema C. E., Ding L. Y., Lindsten T., Turka L. A., Mao X. H., Nuñez G., Thompson C. B. bcl-X, a Bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 10.Olopade O. I., Adeyanju M. O., Safa A. R., Hagos F., Mick R., Thompson C. B., Recant W. M. Overexpression of BCL-x protein in primary breast cancer is associated with high tumor grade and nodal metastases. Cancer J. Sci. Am. 1997;3:230–237. [PubMed] [Google Scholar]
- 11.Takehara T., Liu X., Fujimoto J., Friedman S. L., Takahashi H. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology. 2001;34:55–61. doi: 10.1053/jhep.2001.25387. [DOI] [PubMed] [Google Scholar]
- 12.Xerri L., Parc P., Brousset P., Schlaifer D., Hassoun J., Reed J. C., Krajewski S., Birnbaum D. Predominant expression of the long isoform of Bcl-x (Bcl-xL) in human lymphomas. Br. J. Haematol. 1996;92:900–906. doi: 10.1046/j.1365-2141.1996.423958.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou A., Ou A. C., Cho A., Benz E. J., Jr, Huang S. C. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol. Cell. Biol. 2008;28:5924–5936. doi: 10.1128/MCB.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao G., Xie A., Huang S. C., Zhou A., Zhang J., Herman A. M., Ghassemzadeh S., Jeong E. M., Kasturirangan S., Raicu M. Role of RBM25/LUC7L3 in abnormal cardiac sodium channel splicing regulation in human heart failure. Circulation. 2011;124:1124–1131. doi: 10.1161/CIRCULATIONAHA.111.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szymczyna B. R., Bowman J., McCracken S., Pineda-Lucena A., Lu Y., Cox B., Lambermon M., Graveley B. R., Arrowsmith C. H., Blencowe B. J. Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing. Genes Dev. 2003;17:461–475. doi: 10.1101/gad.1060403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puig O., Bragado-Nilsson E., Koski T., Seraphin B. The U1 snRNP-associated factor Luc7p affects 5′ splice site selection in yeast and human. Nucleic Acids Res. 2007;35:5874–5885. doi: 10.1093/nar/gkm505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickson W. A., Horton J. R., LeMaster D. M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 19.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005;61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winn M. D., Murshudov G. N., Papiz M. Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 26.Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. Structure validation by Cα geometry: Φ,Ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y., Huang H., Zhou B. O., Wang S. S., Hu Y., Li X., Liu J., Zang J., Niu L., Wu J., et al. Structural analysis of Rtt106p reveals a DNA binding role required for heterochromatin silencing. J. Biol. Chem. 2010;285:4251–4262. doi: 10.1074/jbc.M109.055996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X., Levine L., Kwok P. Y. Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res. 1999;9:492–498. [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L., Qin S., Zhang J., Ji P., Shi Y., Wu J. Solution structure of an atypical PHD finger in BRPF2 and its interaction with DNA. J. Struct. Biol. 2012;180:165–173. doi: 10.1016/j.jsb.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 30.Yang G., Huang S. C., Wu J. Y., Benz E. J. An erythroid differentiation-specific splicing switch in protein 4.1R mediated by the interaction of SF2/ASF with an exonic splicing enhancer. Blood. 2005;105:2146–2153. doi: 10.1182/blood-2004-05-1757. [DOI] [PubMed] [Google Scholar]
- 31.Yao Z., Duan S., Hou D., Heese K., Wu M. Death effector domain DEDa, a self-cleaved product of caspase-8/Mch5, translocates to the nucleus by binding to ERK1/2 and upregulates procaspase-8 expression via a p53-dependent mechanism. EMBO J. 2007;26:1068–1080. doi: 10.1038/sj.emboj.7601571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu Y., Liu L., Zhao C., Han C., Li F., Zhang J., Wang Y., Li G., Mei Y., Wu M., et al. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 2012;26:1376–1391. doi: 10.1101/gad.188359.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blencowe B. J., Ouzounis C. A. The PWI motif: a new protein domain in splicing factors. Trends Biochem. Sci. 1999;24:179–180. doi: 10.1016/s0968-0004(99)01387-0. [DOI] [PubMed] [Google Scholar]
- 34.Maris C., Dominguez C., Allain F. H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 35.Dreyfuss G., Kim V. N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 36.Query C. C., Bentley R. C., Keene J. D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- 37.Siomi H., Matunis M. J., Michael W. M., Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St Johnston D., Brown N. H., Gall J. G., Jantsch M. A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malim M. H., Tiley L. S., McCarn D. F., Rusche J. R., Hauber J., Cullen B. R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.