Abstract
Invasion and angiogenesis are two major pathophysiological features of malignant gliomas. Anti-angiogenic treatment lead to enhanced tumor cell invasion and metastasis. In the current study, we tested invasion and angiogenesis related mRNA expression profiles of glioma cells via RT2Profiler PCR Array by employing an in vivo 9L homograft glioma tumor animal model and an in vitro hypoxic cell culture model. The miRNA profile was also obtained via miRNA array. Genes with mRNA expression that changed significantly in the mRNA array were selected to predict possible miRNAs that regulate mRNA expression using the TargetScan database, and were then matched with miRNA array results. Based on these criteria, NRP-2 with the matching miRNA miR-15b, and MMP-3 with the matching miRNA miR-152 were selected for further study, and to determine whether they regulate tumor microenviroment changes and affect glioma angiogenesis and invasion. The protein expression of NRP-2 and MMP-3 were verified in 9L glioma cells and were negatively correlated to miR-15b and miR-152 level, respectively. Rat astrocytes (primary and cell line), when co-cultured with 9L glioma cells, showed significantly elevated NRP-2, MMP-3 expression and reduced miR-15b, miR-152 expression compared to non co-cultured astrocytes. Luciferase activity assay confirmed that miR-15b and miR-152 attenuate expression of NRP-2 and MMP-3 protein by binding to NRP-2 and MMP-3 transcript, respectively. In vitro invasion assay data showed that miR-15b and miR-152 significantly decreased 9L cell invasiveness. Anti-miR-15b and anti-miR-152 inhibitors counteracted the inhibition of invasion caused by miR-15b and miR-152. In vitro tube formation assay data showed that miR-15b, but not miR-152, reduced tube formation in cultured endothelial cells, and anti-miR-15b inhibitor counteracted the inhibition of tube formation caused by miR-15b. A preliminary pathway study indicated that miR-15b and miR-152 deactivated the MEK-ERK pathway via NRP-2 and MMP-3 in 9L cells, respectively. In conclusion, our current study indicates that miR-15b reduces invasion of glioma cells and angiogenesis via NRP-2, and miR-152 reduces invasion of glioma cells through MMP-3.
Keywords: Glioma, tumor model, angiogenesis, invasion, miR-15b, miR-152, NRP-2, MMP-3
1. Introduction
Gliomas are both highly vascularized and invasive, and characterized by high incidence of recurrence and poor prognosis [1]. Tumor cells that have migrated from the primary site of malignant gliomas result in the nearly inevitable recurrence and tumor progression seen clinically [2; 3]. Rapid dissemination of single tumor cells throughout the brain underlies a great propensity for tumor recurrence, often rendering gliomas incurable by surgical removal, even when combined with adjuvant radiation and chemotherapy. Marked increase in blood vessel formation (angiogenesis) is another key characteristic of malignant gliomas. Glioma cells clearly need the vasculature for the delivery of nutrients and oxygen, which is crucial for tumor growth and colonization in the brain [4; 5]. Glioma blood vessels show endothelial cell proliferation which is a key feature of high grade gliomas in the WHO grading system [5; 6; 7].
Systemic therapy with anti-angiogenic treatment can modulate patterns of tumor invasion [8; 9; 10]. Antiangiogenic therapy can lead to enhanced tumor cell invasion and metastasis [8; 9; 10; 11; 12; 13]. Glioblastoma Multiforme (GBM), for example, when targeted with anti-VEGF agents, becomes more invasive [8; 11]. Orthotopic glioma models showed that antagonization of neovascularization could cause increased tumor cell migration, preferentially along preexisting host vessels [8; 14]. Although the exact mechanisms responsible for this increased invasiveness are unknown, it has been speculated that a decreased supply of oxygen and nutrients may act as a stimulus for tumor cell migration [13]
The formation of abnormal tumor vasculature and glioma cell invasion along white matter tracts are believed to be the major factors responsible for the resistance of these tumors to treatment. Therefore, investigation of both angiogenesis and invasion in glioblastoma is essential for the development of a curative therapy.
miRNAs are short single-stranded RNA molecules that function as master regulators of gene expression by post-transcriptional modifications of target mRNAs [15]. The pattern of regulation of gene expression is sequence-specific. MiRNAs bind to 3′ untranslated regions (3′-UTRs) of mRNAs and then reduce the translation and/or stability of that mRNA, leading to a reduction in protein levels. Based on the unique feature of their targeting, miRNAs may have many targets [16], and, thus, control a large number of proteins. miRNAs are integral to many biological processes. In tumor cells, miRNAs may serve as either oncogenes or tumor suppressors [17; 18]. Dysregulation of miRNAs promotes malignancy of glioblastoma and contributes to cell proliferation, invasion, and angiogenesis and glioma stem cell multipotency and survival [19; 20].
2. Material and Methods
2.1 Cell line and cell culture
The rat glioma cell line 9L was obtained from the American Type Culture Collection (Manassas, VA). The cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin. The 9L cells were maintained in a humidified 37°C incubator with 5% CO2, fed every 3 days with complete medium, and subcultured when confluence was reached.
2.2 miRNA and anti-miRNA inhibitor transfection
Transfection of the miR-15b, miR-152, anti-miR15b inhibitor, anti-miR-152 inhibitor (Applied Biosystems), and inactive (scrambled) control cel-mir-67 (Thermo Scientific Dharmacon, IL, USA), or pMIR-Report vectors was performed using Lipofectamine 2000 transfection reagent (Invitrogen, CA, USA) with 300 nmol of miRNA or 1 μg/ml DNA plasmid, respectively.
2.3 In vitro invasion assay
Matrigel invasion assays were used to assess the ability of the 9L cells to penetrate the Extra Cellular Matrix (ECM) in the presence or absence of miRNA and inhibitor. Invasion of cells through Matrigel was determined using 24-well invasion chambers (8.0μm pore size with polycarbonate membrane; BD Biosciences, Cowley, UK) in accordance with the manufacturer’s instructions with the following modifications [21]. BD invasion chambers were pre-hydrated with serum-free DMEM (500μl/well) for 2h of incubation at 37°C in 5% CO2. After trypsinization, 9L cells with different treatment were suspended in medium without serum (500μl) in a concentration of 1×105 cells/well and immediately placed onto the upper compartment of the plates. Subsequently, the lower compartment was filled with complete medium (750μl). The cells were incubated for 24 h. Following incubation, the non-invading cells were removed from the upper surface of the membrane by wiping with cotton-tipped swabs. Cells on the lower surface of the membrane were stained with CellTracker Green (Molecular Probes, Eugene, OR) for 45 minutes and fixed in 4% paraformaldehyde. Five fields of adherent cells were randomly counted in each well under a fluorescent microscope at 4X magnification, and the results were numerically averaged and counted. The invasion index for each treatment was calculated by dividing the number of invading cells by the number of cells that invaded under the control group. Invasion index was expressed as percentage of control value.
2.4 Capillary-like tube formation assay
Briefly, 0.1ml growth factor reduced Matrigel (Becton Dickinson) was added per well, and mouse brain endothelial cells (MBECs) (2×104 cells) were added and incubated in media from different treatments. All assays were performed in n=6/group. For quantitative measurements of capillary tube formation, matrigel wells were digitized under a 4x objective (Olympus 1×71) for measurement of total tube length of capillary tube formation using a video camera (Roper scientific) interfaced with the MCID image analysis system (Imaging Research, St. Catharines, Canada) at 6 hours [24]. Tracks of endothelial cells organized into networks of cellular cords (tubes) were counted and averaged in randomly selected 3 microscopic fields. The total tube length was calculated by MCID in each field. The tube formation index was expressed as tube length (mm) per mm2 area.
2.5 Western blot
Cultures were rinsed with PBS and proteins were extracted in 500μl RIPA lysis buffer. Equal amounts of protein, as determined using the BCA (bicinchoninic acid) protocol (Pierce, Rockford, IL), were loaded on 10% Bis-Tris Gels (Invitrogen, Carlsbad, CA) after being denatured. The proteins were then transferred to Invitrogen PVDF membranes (Invitrogen, Carlsbad, CA). The membranes were blocked at RT for 1h with 5% BSA in TBS-T (10mM Tris-HCl, pH 7.6 and 150mM NaCl, 0.1% Tween-20). Afterward the membranes were incubated with primary antibodies in 3% BSA at 4 °C overnight. The membranes were washed with TBS-T and incubated for 1h at RT with horseradish peroxidase (HRP)-conjugated secondary antibodies (Bio-Rad Laboratories, Hercules, CA). Following washing, the immunoblots were detected using a SuperSignal West Pico Chemiluminescent Substrate kit (Pierce, Rockford, IL). The experiment was repeated in triplicate. β-actin was used as the internal control.
2.6 Laser-capture microdissection of 9L glioma model in Fischer rat
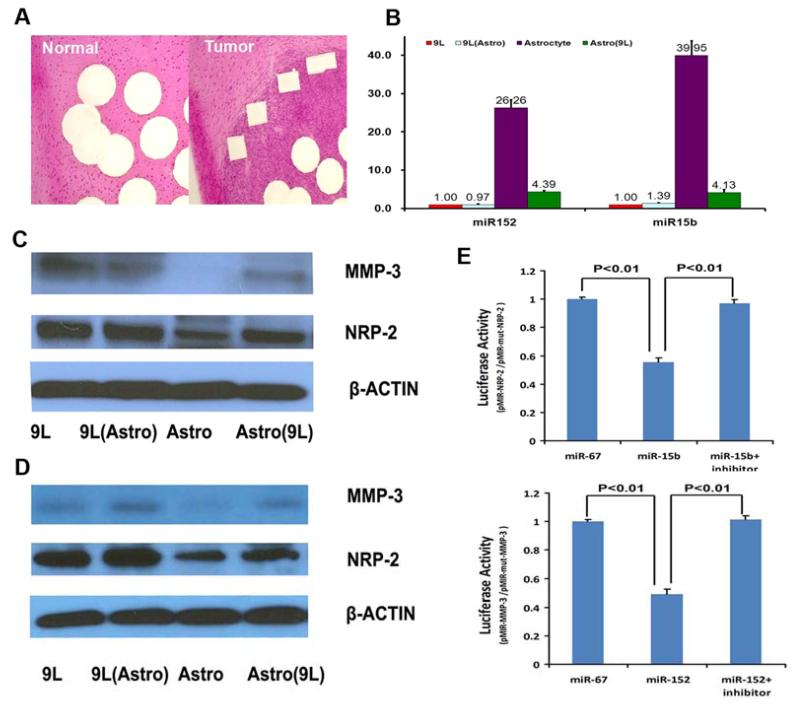
Fischer rats were anesthetized with ketamine (80 mg/kg) and xylazine (13 mg/kg), fixed in a stereotaxic device, and 5×104 9L cells were injected using a 10μL Hamilton syringe (5 μl volume). The craniectomy was covered with a film of polyvinyl chloride glued to the surrounding intact bone. Animals were sacrificed 4 weeks post-implantation. Brain coronal cryosections (10μm) obtained from rats subjected to 9L tumor implantations were mounted on LCM membrane slides (Leica Microsystems, Wetzlar, Germany). To identify tumor rat brain slice were stained with H&E, approximately 100,000 tumor cells were collected by an LMD 6000 system (Leica Microsystems). The same number of non-tumor cells collected from the homologous tissue in the contralateral hemisphere was used as control (Figure 1A). Cells were lysed in Qiazol reagent (Qiagen, Valencia, CA); total RNA was isolated immediately after cells were collected.
Figure. 1.
A. Laser Capture Microdissection. B. miRNA expression in 9L and primary rat astrocyte coculture. C. Protein expression in co-cultured 9L and primary rat astrocyte. D. protein expression in co-cultured 9L and rat astrocyte cell line. E. Luciferase activity assay. miR-152 significantly decreased luciferase activity in Luc-NRP-2-UTR (wild-type) which suggest that miR-15b directly targets the 3′-UTR of NRP-2 mRNA. miR-152b suppressed >50% activity in Luc-MMP-3-UTR (wild-type) which suggest that miR-152 directly targets the 3′-UTR of MMP-3 mRNA. 9L: 9L glioma cells. 9L(Astro): 9L glioma cells co-cultured with rat astrocyte. Astro: Rat astrocyte Astro (9L): Rat astrocyte co-cultured with 9L cells.
2.7 MiRNA luciferase assay
Position 1798-1804 of the NRP-2 3′-UTR is a predicted interaction position of miR-15b. We cloned an 80-bp sequence containing the predicted binding site or a scrambled sequence downstream of the pMIR luciferase reporter to generate pMIR-NRP2 and pMIR-mut-NRP2 vectors, respectively. Luciferase assays were carried out in 9L cells. First, cells were co-transfected with appropriate plasmids with either miR-15b miRNA mimic or control cel-mir-67 mimic in 24-well plates. Anti-miR miRNA inhibitor was also employed to transfected into 9L cells with miRNA mimic. Position 121-127 of the MMP-3 3′-UTR is a predicted interaction position of miR-152. We cloned an 80-bp sequence containing the predicted binding site or a scrambled sequence downstream of the pMIR luciferase reporter to generate pMIR-NRP-2 and pMIR-mut-NRP-2 vectors, respectively. Luciferase assays were carried out in 9L cells. First, cells were co-transfected with appropriate plasmids with either miR-152 miRNA mimic or control cel-mir-67 mimic in 24-well plates. Anti-miR miRNA inhibitors were also transfected into 9L cells. Then, the cells were harvested and lysed for luciferase assay 24 h after transfection.
2.8 Isolation of total RNA and real-time RT-PCR
For miRNA analysis, cells were lysed in Qiazol reagent and total RNA was isolated using the miRNeasy Mini kit (Qiagen). miRNA was reverse transcribed with the miRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) and amplified with TaqMan miRNA assay (Applied Biosystems), which is specific for mature miRNA sequences. For analysis of mRNA, RNA was reverse transcribed with M-MLV reverse transcriptase (Invitrogen), and amplified by SYBR Green reporter (Applied Biosystems) using custom primers (Invitrogen). Analysis of gene expression was carried out by the 2-ΔΔCt method [22].
2.9 RT2 Profiler PCR Array
RT2 Profiler PCR Array was used as a method of combining real-time PCR performance with a simultaneous analysis of a panel of genes using Angiogenesis PCR Array and Tumor Metastasis PCR Array (array PARN-024Z and PARN-028Z) The total RNA was isolated from the cultured 9L gloma cells or cell from LCM using the Rneasy Mini Kit (Qiagen). Preparation and analysis of samples were carried out in accordance with the manufacturer’s instruction. cDNA was generated from 1 μg total RNA using the RT2 qPCR Array First Strand Kit in accordance with the manual. The template was combined with RT2 SYBR Green/Fluorescein PCR master mix. Equal amounts of this mixture (25 μl) were added to each well of the RT2 qPCR profiler plate containing the predispensed gene-specific primer sets. Arrays were run on a Vii A7 PCR system (Life Technologies, Carlsbad, California). Threshold cycle values were analyzed using the ΔΔCt method with the aid of an Excel (Microsoft Excel; Microsoft, Redmond, WA). Reverse transcriptase control, cDNA control, and positive PCR control were within the accepted range.
2.10 Statistical analysis
In all in vitro studies, three unique experiments were performed employing cells from different transfection experiments. Data are presented as mean and standard error (mean ± SEM). Student’s t-test was used for comparing means between two groups. Statistical significance was analyzed by one-way ANOVA in three or more groups using SPSS software. In all experiments, a p-value of 0.05 or less was considered statistically significant.
3 Results
3.1 mRNA profiling
RT2 Profiler PCR Array was used as a method of combining real-time PCR performance with a simultaneous analysis of a panel of genes using Angiogenesis PCR Array and Tumor Metastasis PCR Array. Samples from LCM were analyzed by RT2 Profiler PCR Array. The array experiment was performed in duplicate. A comparison was performed on data to assess the gene expression of tumor tissue and normal tissue (Table 1). The differences in gene expression between tumor tissue and normal tissue were studied. More than fourfold difference was considered significant. Compared to normal tissue, mRNA expression increase in tumor tissue is labeled green, and mRNA expression decrease is labeled red in table 1.
Table 1.
mRNAs expression changes in tumor compared to normal tissue. Only mRNAs expression increase or decrease with fold-change greater than 4 compared to normal tissue are listed. The raw data, i.e., the mean ΔΔCt values of the genes, were normalized to the housekeeping gene GAPDH.
| Fold- Change |
Gene Symbol |
Gene Title | |
|---|---|---|---|
| Increased | |||
| 1 | 53.08 | PLAU | Plasminogen activator, urokinase |
| 2 | 36.25 | MMP12 | Matrix metallopeptidase 12 |
| 3 | 32.67 | VEGFA | Vascular endothelial growth factor A |
| 4 | 31.34 | TNC | Tenascin C |
| 5 | 28.25 | SPP1 | Secreted phosphoprotein 1 |
| 6 | 24.08 | CDH1 | Cadherin 1 |
| 7 | 22.01 | NRP-2 | Neuropilin 2 |
| 8 | 17.88 | SELP | Selectin, platelet |
| 9 | 15.14 | ADAMTS13 | ADAM metallopeptidase thrombospondin type 1 motif, 13 |
| 10 | 13.74 | PDGFa | Platelet-derived growth factor alpha polypeptide |
| 11 | 9.25 | MMP3 | Matrix metallopeptidase 3 |
| 12 | 7.41 | ITGA5 | Integrin alpha 5 (fibronectin receptor alpha) |
| 13 | 7.36 | VTN | Vitronectin |
| 14 | 6.36 | SPARC | Secreted protein, acidic, cysteine-rich (osteonectin) |
| 15 | 5.24 | TGFBi | Transforming growth factor, beta induced |
| 16 | 4.32 | CCL2 | chemokine (C-C motif) ligand 2 |
| Decreased | |||
| 1 | 17.75 | FN1 | Fibronectin 1 |
| 2 | 15.14 | THBS2 | Thrombospondin 2 |
| 3 | 10.41 | Col4a3 | Collagen, type IV, alpha 3 |
| 4 | 9.85 | LAMA3 | Laminin, beta 3 |
| 5 | 7.01 | FIGF | C-fos induced growth factor |
| 6 | 6.28 | TIMP-3 | TIMP metallopeptidase inhibitor 3 |
| 7 | 5.03 | TNF | Tumor necrosis factor (TNF superfamily, member 2) |
To overcome hypoxic conditions, tumor cells secrete angiogenic growth factors and invasive factors that enhance neovascularization and invasion[23]. Indeed, hypoxic tumors have been reported to have a predilection for tissue invasion and angiogenesis[24]. Therefore, in addition to testing the gene expression profile of glioma in an animal model directly, in vitro cancer cells cultured in hypoxic condition also provide another tool to investigate changes in the tumor microenvironment. Total RNA samples from cultured 9L cells were analyzed by RT2 Profiler PCR Array. The array experiment was performed in duplicate. A comparison was performed on data to assess the gene expression of hypoxia and normoxia (Table 2). The differences in gene expression between tumor tissue and normal tissue were studied. More than eightfold difference for increased gene and fourfold for decreased gene was considered significant. Compared to normoxia, mRNA expressions increased in hypoxic sample are labeled green and mRNA expression decreases are labeled red in table 2
Table 2.
mRNA expression changes in hypoxic tumor cells compared to normoxic tumor cells, mRNA expression with fold-change greater than 8 compared to normoxic tumor cells are listed in table 2 with green color. Decreased mRNA expression in hypoxic tumor cells compared to normoxic tumor cells. mRNA expression with fold-change greater than 4 compared to normoxic tumor cells are listed in table 2 with red color. The raw data, i.e., the mean ΔΔCt values of the genes, were normalized to the housekeeping gene GAPDH.
| Fold- Change |
Gene Symbol |
Gene Title | |
|---|---|---|---|
| Increased | |||
| 1 | 196.72 | VCAM1 | Vascular cell adhesion molecule 1 |
| 2 | 121.94 | ADAMTS13 | ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| 3 | 102.54 | VEGFA | Vascular endothelial growth factor A |
| 4 | 57.68 | MMP3 | Matrix metallopeptidase 3 |
| 5 | 48.84 | FGF6 | Fibroblast growth factor 6 |
| 6 | 38.85 | NRP-2 | Neuropilin 2 |
| 7 | 31.12 | CD44 | Cd44 molecule |
| 8 | 28.84 | PECAM1 | Platelet/endothelial cell adhesion molecule 1 |
| 9 | 23.26 | EFNA1 | Ephrin A1 |
| 10 | 21.41 | CCL2 | chemokine (C-C motif) ligand 2 |
| 11 | 14.83 | ID3 | Inhibitor of DNA binding 3 |
| 12 | 12.91 | MMP9 | Matrix metallopeptidase 9 |
| 13 | 11.39 | VTN | Vitronectin |
| 14 | 10.27 | SPP1 | Secreted phosphoprotein 1 |
| 15 | 9.65 | CDH5 | Cadherin 5 |
| 16 | 9.25 | PDGFA | Platelet-derived growth factor alpha polypeptide |
| 17 | 8.36 | SERPINf1 | Serine peptidase inhibitor, clade F, member 1 |
| Decreased | |||
| 1 | 58.49 | TIMP-3 | TIMP metallopeptidase inhibitor 3 |
| 2 | 27.67 | Col4a3 | Collagen, type IV, alpha 3 |
| 3 | 23.10 | THBS1 | Thrombospondin 1 |
| 4 | 17.27 | HGF | Hepatocyte growth factor |
| 5 | 15.24 | FN1 | Fibronectin 1 |
| 6 | 12.82 | Coll8a1 | Collagen type VIII alpha 1 |
| 7 | 9.45 | PDGFb | Platelet-derived growth factor beta polypeptide |
| 8 | 5.94 | LAMA2 | Laminin, alpha 2 |
| 9 | 4.44 | ITGB4 | Integrin beta 4 |
Each individual assay showed substantial numbers of genes with significantly changed expression. In order to focus on the genes that changed dramatically under different tumor progression circumstances, we reviewed the mRNA expression changes in both the in vivo animal model and the in vitro hypoxia model and selected those genes that changed in both models for further study (Table 3).
Table 3.
Expression changes both in vivo and in vitro study (green : increase; red: decreased)
| Gene Symbol | Gene Title | |
|---|---|---|
| Increased | ||
|
| ||
| 1 | ADAMTS13 | ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| 2 | VEGFA | Vascular endothelial growth factor A |
| 3 | MMP3 | Matrix metallopeptidase 3 |
| 4 | NRP-2 | Neuropilin 2 |
| 5 | CCL2 | chemokine (C-C motif) ligand 2 |
| 6 | VTN | Vitronectin |
| 7 | SPP1 | Secreted phosphoprotein 1 |
| 8 | PDGFA | Platelet-derived growth factor alpha polypeptide |
| Decreased | ||
| 9 | TIMP-3 | TIMP metallopeptidase inhibitor 3 |
| 10 | Col4a3 | Collagen, type IV, alpha 3 |
| 11 | FN1 | Fibronectin 1 |
3.2 miRNA Profiling
Using the same in vivo 9L rat xenograft tumor model, cells collected by LCM (Fig. 1A) were lysed in Qiazol reagent and total RNA was isolated using the miRNeasy Mini kit (Qiagen). miRNA profile was then detected by LC Science (Houston TX) (Table 4). Genes with mRNAs expression that changed in both in vivo tissue and in vitro hypoxia were chosen to predict possible miRNAs that regulate mRNA expression through TargetScan database. miRNA predicted in TargetScan database was then matched with miRNA array results. miRNA levels that changed significantly and that were also predicted in the database to target mRNAs that we observed to be altered were chosen for further study. These criteria were met by two mRNA-miRNA pairs. Our match results show that NRP2 and the predicted miRNA miR-15b and miR-16 exhibited significant change in both in vitro and in vivo studies, and MMP-3 and the predicted miRNA miR-152 changed significantly in both in vitro and in vivo studies. (Table 5)
Table 4. miRNA expression changes in tumor cells compared to normal cells.
Compared to normal tissue, miRNAs with significant increase (Log2(T/N)>0.5 ) in tumor tissue are listed in table4 with green color. MiRNAs with significant decrease (Log2(T/N)<-0.5) in tumor tissue are listed in table 4 with red color.
| No. | Reporter Name | p-value | Normal Mean |
Tumor Mean |
Log2 (T/N) |
|---|---|---|---|---|---|
| Increased | |||||
| 581 | rno-miR-568 | 4.55E-05 | 4 | 327 | 6.18 |
| 524 | rno-miR-466c* | 9.30E-06 | 65 | 1,526 | 4.56 |
| 521 | rno-miR-466b-1* | 3.91E-05 | 180 | 2,897 | 4.01 |
| 529 | rno-miR-483* | 6.86E-06 | 98 | 454 | 2.21 |
| 43 | rno-miR-1224 | 2.56E-05 | 1,248 | 5,105 | 2.03 |
| 308 | rno-miR-320 | 2.14E-04 | 386 | 1,065 | 1.46 |
| 6 | rno-let-7c | 2.28E-03 | 15,601 | 41,527 | 1.41 |
| 4 | rno-let-7b | 2.17E-03 | 13,720 | 34,209 | 1.32 |
| 211 | rno-miR-214 | 1.79E-04 | 238 | 566 | 1.25 |
| 230 | rno-miR-222 | 4.95E-06 | 340 | 675 | 0.99 |
| 240 | rno-miR-24 | 3.61E-04 | 498 | 971 | 0.96 |
| 1 | rno-let-7a | 3.95E-03 | 16,000 | 30,771 | 0.94 |
| Decreased | |||||
| 247 | rno-miR-26b | 1.63E-03 | 510 | 47 | −3.43 |
| 457 | rno-miR-15b | 3.90E-05 | 1197 | 135 | −3.15 |
| 674 | rno-miR-98 | 6.63E-05 | 1,188 | 158 | −2.91 |
| 48 | rno-miR-125a-5p | 1.92E-07 | 728 | 98 | −2.89 |
| 519 | rno-miR-16 | 4.98E-06 | 539 | 75 | −2.85 |
| 271 | rno-miR-152 | 2.86E-05 | 502 | 71 | −2.82 |
| 203 | rno-miR-21 | 2.61E-04 | 14,452 | 2,078 | −2.8 |
| 367 | rno-miR-352 | 8.67E-04 | 513 | 112 | −2.2 |
| 275 | rno-miR-29a | 1.14E-03 | 609 | 139 | −2.13 |
| 51 | rno-miR-125b-5p | 3.04E-04 | 7,857 | 1,826 | −2.11 |
| 245 | rno-miR-26a | 1.17E-03 | 3,547 | 2,363 | −0.59 |
| 236 | rno-miR-23a | 3.23E-03 | 3,082 | 2,066 | −0.58 |
Table 5. Matched mRNA and miRNA which changed significantly in both in vitro and in vivo studies.
| Gene Symbol | Gene Title | Predicted microRNA |
|---|---|---|
| NRP2 | Neuropilin 2 | rno-miR-15b |
| MMP3 | Matrix metallopeptidase 3 | rno-miR-152 |
Based on preliminary screening assays, NRP2 and miR-15b as well as MMP-3 and miR-152 were then selected for our detailed study to test whether and how these two miRNAs regulate tumor microenvironment changes and promote tumor angiogenesis and invasion.
3.3 miR-15b and miR-152 expression were reduced in tumor cells compared to normal cells. Co-culture with tumor cells significantly decreased miR-15b and miR-152 levels in rat astrocytes
Normal primary astrocytes and 9L glioma cells were initially employed to verify the expression of miR-15b and miRNA-152. Also, co-culture of 9L and astrocytes were tested. Our results showed 9L cells have much lower miR-15b and miR-152 expression than rat astrocytes. Co-culture of rat astrocytes with 9L cells significantly decreased miR-15b and miR-152 expression by 83.3±4.9% and 89.7±6.8%, respectively, in rat astrocytes. (Figure 1B)
3.4 NRP-2 and MMP-3 expression were increased in tumor cells compared to normal cells. Co-culture with tumor cells significantly increased NRP-2 and MMP-3 expression levels in rat astrocytes
Primary rat astrocytes and a rat astrocyte cell line were employed to verify the NRP-2 and MMP-3 protein expression. Co-culture of astrocytes with 9L were also tested. Compared to 9L glioma, primary rat astrocytes showed significantly lower NRP-2 and MMP-3 level. Co-culture of astrocytes with 9L cells significantly increased NRP-2 and MMP-3 expression in primary rat astrocytes. Rat astrocyte cell line showed the same trend of NRP-2 and MMP-3 expression as in primary rat astrocytes. (Figure 1 C,D)
3.5 miR-15b and miR-152 attenuate expression of NRP-2 and MMP-3 protein by binding to NRP-2 and MMP-3 transcripts, respectively
Luciferase activity assays were employed to further confirm that the expression of NRP-2 can be ascribed to the specific interaction between miR-15b and the binding sites for miR-15b in the 3′-UTR of NRP-2, and the expression of MMP-3 can be ascribed to the specific interaction between mir-152 and the binding sites in the 3′-UTR of MMP-3. Luciferase activities in pMIR-NRP-2 and pMIR-mut-NRP-2 vectors transfected with cel-miR-67 were set for control of miR-15b and anti-miR-15b inhibitor; luciferase activities in pMIR-MMP-3 and pMIR-mut-MMP-3 vectors transfected with cel-miR-67 were set for control of miR-152 and anti-miR-152.
Our data show that the miR-15b mimic targets the predicted binding site within the NRP-2 mRNA 3′-UTR, as it decreased the luciferase in 9L cells transfected with a luciferase reporter vector containing the predicted binding site (pMIR-NRP-2) by 44.7±2.6%. Anti-miR miRNA inhibitor co-transfected with miR-15b mimic reversed the inhibition of luciferase activity caused by miR-15b. Compared to miR-15b alone, co-transfection of anti-miR miRNA inhibitor and miR-15b mimic increased luciferase activity by 75.3±4.8%. Similarly, we determined that the miR-152 mimic targets the predicted binding site within the MMP-3 mRNA 3′-UTR, as it decreased luciferase activity in 9L cells transfected with a luciferase reporter vector containing the predicted binding site (pMIR-MMP-3) by 51.0±3.4%. Anti-miR-152 miRNA inhibitor co-transfected with miR-152 mimic reversed the inhibition of luciferase activity caused by miR-152. Compared to miR-152 transfection, co-transfection of anti-miR miRNA inhibitor and miR-15b mimic increased luciferase activity by 107.1±7.0%. (Figure 1E) These data show that miR-15b and miR-152 directly bind to their respective predicted binding sites in the NRP-2 and MMP-3.
3.6 miR-15b and miR-152 decreased NRP-2 and MMP-3 expression, respectively, and deactivated MEK-ERK pathway in 9L cells
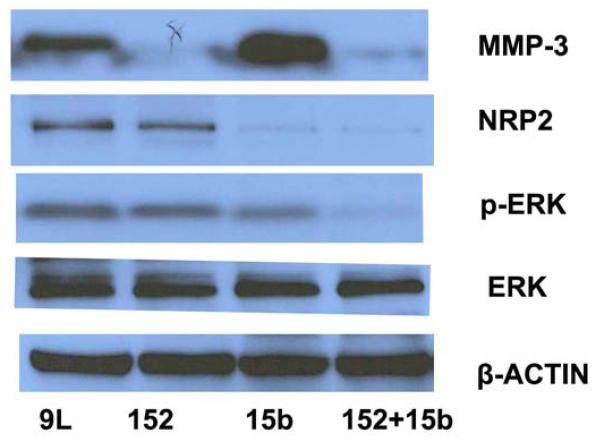
Employing a ‘scrambled’ cel-mir-67 or miR-15b and miR-152 mimic, Western blot analysis revealed that NRP-2 expression was significantly decreased by miR-15b mimic transfection, and MMP-3 expression was significantly decreased by miR-152 mimic transfection. Co-transfection of miR-15b and miR-152 mimic significantly decreased both MMP-3 and NRP-2 expression. P-MEK1/2 and p-ERK expression was significantly decreased by overexpression of miR-15b or miR-152. However, overexpression of miR-15b or miR-152 did not change mRNA expression of MEK and ERK. (Figure 2)
Figure. 2. miR-15b and miR152 decreased NRP-2 and MMP-3 expression and deactivated MEK-ERK pathway in 9L cells.
NRP-2 expression was significantly decreased by miR-15b mimic transfection; MMP-3 expression was significantly decreased by miR-152 mimic transfection. Co-transfection of miR-15b and miR-152 mimic significantly decreased both MMP-3 and NRP-2 expression. P-ERK expression was significantly decreased by overexpression of miR-15b or miR-152.
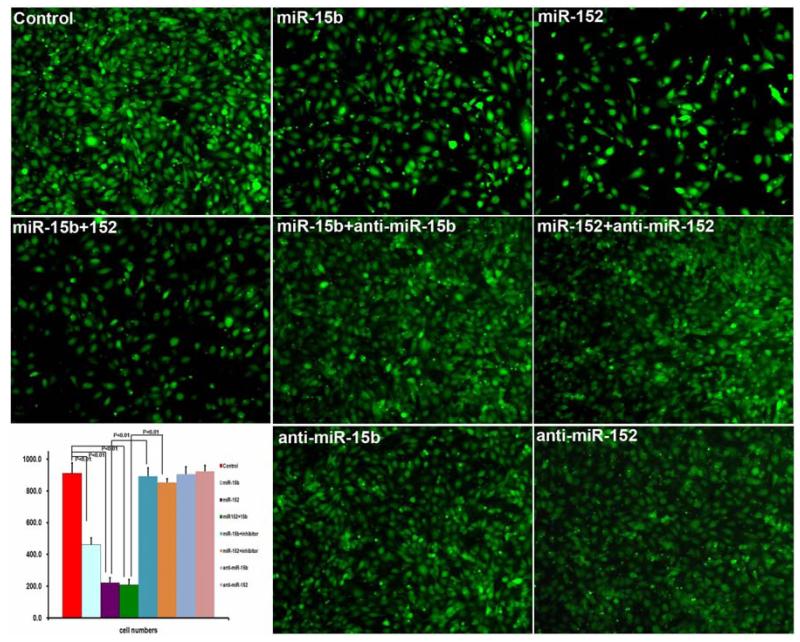
3.7 miR-15b and miR-152 decreased 9L cell invasiveness
Forty-eight hours after transfection with either miR-15b, miR-152, Cel-miR-67, or anti-miR miRNA inhibitor 9L cells were seeded into the upper chamber of invasion chamber. After 24 hours, cells that invaded through the extracellular matrix were imaged and counted. Compared to Cel-miR-67, miR-15b and miR-152 significantly decreased the number of invaded cells. Anti-miR miRNA inhibitor co-transfected with miRNA mimic reversed the inhibition of invasion caused by miR-15b or miR-152 mimic. Compared to Cel-miR-67 control, miR15b and miR-152 inhibited 9L cell invasion by 49.5±3.7% and 75.8±3.5%, respectively. Combination of miR-15b and miR-152 significantly decreased 9L cell invasion by 78.0±3.8%.
Compared to miR-15b, co-transfection of anti-miR-15b miRNA inhibitor and miR-15b mimic increased invasion by 3.04±0.23 fold. Compared to miR-152, co-transfection of anti-miR-152 miRNA inhibitor and miR-152 mimic increased invasion by 2.85±0.12 fold. (Figure 3)
Figure 3. miR-15b and miR 152 decreased 9L cell invasiveness.
miR-15b and miR-152 significantly decreased the number of invaded cells. Anti-miR miRNA inhibitor co-transfected with miRNA mimic reversed the inhibition of invasion caused by miR-15b or miR152 mimic. Compared to Cel-miR-67 control, miR15b and miR-152 inhibited 9L cell invasion by 49.5±3.7% and 75.8±3.5%. Combination of miR-15b and miR-152 significantly decreased 9L cell invasion by 78.0±3.8%. Compared to miR-15b, co-transfection of anti-miR-15b miRNA inhibitor and miR-15b mimic increased invasion by 3.04±0.23 fold. Compared to miR-152, co-transfection of anti-miR-152 miRNA inhibitor and miR-152 mimic increased invasion by 2.85±0.12 fold.
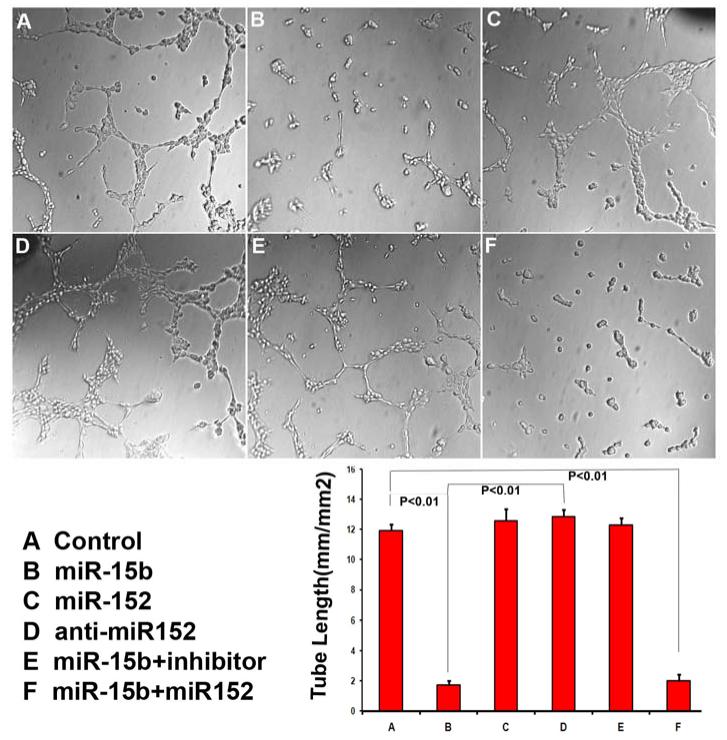
3.8 miR-15b reduces tube formation in cultured endothelial cell and anti-miR-15b inhibitor reversed the inhibition of tube formation caused by miR-15b
The effects of miR-15b and miR-152 mimic transfected 9L glioma cells on MBECs were tested using tube formation assay. Culture medium from miR-15b mimic transfected cells decreased MBECs capillary tube formation by 85.7±7.2% compared to control medium (cel-mir-67 mimic). Conversely, miR-152 mimic did not change capillary tube formation in MBECs. Cotransfection of miR-15b and miR-152 mimic significantly decreased capillary tube formation by 83.4.9±3.8% when compared to control medium. Compared to control medium, transfection of anti-miR-15b miRNA inhibitor with miR-15b mimic reversed the inhibition of tube formation in MBECs caused by miR-15b mimic. Compared to miR-15b mimic transfection, the tube formation of miR-15b mimic and inhibitor cotransfection increased by 6.24±0.29 fold. Transfection of anti-miR-15b miRNA inhibitor alone slightly increased tube formation. (Figure 4)
Figure. 4. miR-15b reduced tube formation in cultured endothelial cells and anti-miR-15b inhibitor reversed the inhibition of tube formation caused by miR-15b.
Culture medium from miR-15b mimic transfected cells decreased MBEC capillary tube formation by 85.7±7.2%. MiR-152 mimic did not change capillary tube formation in MBECs. Cotransfection of miR-15b and miR-152 mimic significantly decreased capillary tube formation by 83.4.9±3.8%. Anti-miR-15b miRNA inhibitor reversed the inhibition of tube formation in MBECs caused by miR-15b mimic. Compared to miR-15b mimic transfection, the tube formation of miR145 mimic and inhibitor cotransfection increased by 6.24±0.29 fold.
4. Discussion
The major function of miRNA is to negatively regulate mRNA target genes, and miRNA expression has been found to be deregulated in all human cancers, where miRNAs play critical roles in tumorigenesis, functioning either as tumor suppressors or as oncogenes.[25] Since the identification of miRNAs in 1993, the amount of research into their function--particularly how they contribute to malignancy--has greatly increased. This class of small RNA molecules control gene expression and provide a previously unknown control mechanism for protein synthesis. As such, it is not surprising that miRNAs are now known to play an essential part in malignancy, functioning as tumor suppressors and oncogenes. [26]
Inhibition of angiogenesis is emerging as a promising therapy for recurrent and newly diagnosed tumors. However, data from animal studies and clinical practice suggest that inhibiting angiogenesis may promote an invasive phenotype in brain tumor [8; 11; 13]. This may represent an important mechanism of resistance to anti-angiogenic therapies. Combination therapy with anti-angiogenic and novel anti-invasion agents is a promising approach that may produce a synergistic antitumor effect and a survival benefit for patients with these devastating tumors.[27]. By employing in vivo 9L homograft glioma tumor animal model and RT2Profiler PCR Array targeting invasion and angiogenesis, we identified gene expression changes in tumor tissue which may contribute to brain tumor invasion and angiogenesis. Hypoxia is a significant parameter in the progression and treatment-response of glioma. Hypoxia initiates cellular invasive processes which occur under both physiologic and pathologic conditions [28]. In vitro, cancer cells cultured under hypoxic conditions also provide another tool to investigate tumor microenvironment change. Total RNA samples from cell culture under hypoxic conditions were analyzed by RT2 Profiler PCR Array. Invasion and angiogenesis related gene expression changes under hypoxic conditions were obtained. We speculate that genes with mRNA expression changes in both in vivo animal models and in vitro hypoxia models are more likely to contribute to brain tumor progression.
By employing the same in vivo 9L rat homograft tumor model, the miRNA profile was also obtained via miRNA array. Genes with mRNA expression that changed significantly in the mRNA array were selected to predict possible miRNAs that regulate mRNA expression using the TargetScan database, and then matched with microRNA array results. After prediction and comparison, NRP-2 with matching miRNA miR-15b, and MMP-3 with matching miRNA miR152 were selected for further study on how miRNAs regulate tumor microenvironment change and promote glioma angiogenesis and invasion. Our mRNA, protein, and miRNA expression assay verified that the expression level of miRNA-15b and miRNA-152 are negatively correlated to mRNA and protein expression of NRP-2 and MMP-3, respectively. Luciferase activity assay then confirmed that miR-15b and miR-152 attenuate expression of NRP-2 and MMP-3 protein by binding to NRP-2 and MMP-3 transcript, respectively.
We used matrigel invasion assays to assess the ability of the 9L to penetrate the ECM under miRNA treatment. Our data showed that the invasion potential of 9L was significantly decreased by miR-15b and miR-152. In addition, we demonstrate that two miRNAs inhibitors significantly reversed the invasive ability of 9L reduced by miRNAs. MMP-3 and NRP-2 contribute to tumor invasion and metastasis [29; 30; 31]. Our results indicate miRNA-15b and miR-152 decreased brain tumor cell invasion by downregulating NRP-2 and MMP-3 expression.
Angiogenesis is essential for embryo development, tissue repair, inflammatory diseases, and tumor growth and metastasis[32]. Tumor growth and metastasis require angiogenesis when the tumor reaches 1-2 mm in diameter [32; 33]. Tumor angiogenesis can be triggered by extracellular signals such as growth factors and by genetic alterations such as activation of oncogenes. To address the angiogenetic function of miR-15b and miR-152 on brain tumor, we further tested tube formation in cultured endothelial cells with culture supernatant from 9L cells treated with miR-15b, miR-152 and inhibitors. Our results showed miR-15b significantly reduced capillary-like tube formation. This reduced tube formation was reversed by miR-15b inhibition. miR-152 did not significantly affect endothelial cell tube formation. It is now well established that NRP2, first described as a mediator of neuronal guidance, also mediates angiogenesis and tumor progression. NRPs are receptors for the class-3 semaphorin (SEMA) family of axon guidance molecules and also for the vascular endothelial growth factor (VEGF) family of angiogenic factors. VEGF-NRP interactions promote developmental angiogenesis and metastases. [34]. Our data indicate that miR-15b decreases angiogenesis via NRP-2 downregulation.
We further investigated a possible signal pathway involved in the miR-15b and miR-152 regulated brain tumor progression. Our protein expression assay showed NRP-2 and MMP-3 expression was significantly decreased by miR-15b and miR-152, respectively. P-MEK1/2 and P-ERK expression was significantly decreased by overexpression of miR-15b or miR-152. These data suggest that miR-15b and miR-152 inhibit brain tumor invasion and angiogenesis through activation of MEK/ERK pathway.
Our current study investigated the miRNA and gene expression profile of 9L glioma cells. By comparison with the TargetScan database, we predicted that miR-15b and miR-152 regulate glioma angiogenesis and invasion in glioma. In vitro invasion and angiogenesis study were then employed to test the function of miR-15b and miR-152 on glioma progression. Our results support the hypothesis that miR-15b inhibit brain tumor invasion and angiogenesis via NRP-2 , through deactivation of MEK/ERK pathway, and miR-152, inhibit brain tumor invasion via MMP-3 , through deactivation of MEK/ERK pathway. Hence, the development of miR-15b and miR-152 as treatment targets for brain tumor invasion and angiogenesis warrants further study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
None of the authors has declared any conflict of interest which may arise from being named as an author on the manuscript
References
- [1].Nicholas MK, Prados MD, Larson DA, Black PM, Loeffler J. Malignant Astrocytomas in Cancer of the Nervous System. Blackwell Publishers; Oxford: 1997. [Google Scholar]
- [2].Burger PC, Dubois PJ, Schold SC, Jr., Smith KR, Jr., Odom GL, Crafts DC, Giangaspero F. Computerized tomographic and pathologic studies of the untreated, quiescent, and recurrent glioblastoma multiforme. Journal of neurosurgery. 1983;58:159–169. doi: 10.3171/jns.1983.58.2.0159. [DOI] [PubMed] [Google Scholar]
- [3].Daumas-Duport C, Scheithauer BW, Kelly PJ. A histologic and cytologic method for the spatial definition of gliomas. Mayo Clin Proc. 1987;62:435–449. doi: 10.1016/s0025-6196(12)65469-x. [DOI] [PubMed] [Google Scholar]
- [4].Fischer I, Gagner JP, Law M, Newcomb EW, Zagzag D. Angiogenesis in gliomas: biology and molecular pathophysiology. Brain Pathol. 2005;15:297–310. doi: 10.1111/j.1750-3639.2005.tb00115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wurdinger T, Tannous BA. Glioma angiogenesis: Towards novel RNA therapeutics. Cell adhesion & migration. 2009;3:230–235. doi: 10.4161/cam.3.2.7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Brem S, Cotran R, Folkman J. Tumor angiogenesis: a quantitative method for histologic grading. Journal of the National Cancer Institute. 1972;48:347–356. [PubMed] [Google Scholar]
- [7].Folkerth RD. Descriptive analysis and quantification of angiogenesis in human brain tumors. Journal of neuro-oncology. 2000;50:165–172. doi: 10.1023/a:1006499824379. [DOI] [PubMed] [Google Scholar]
- [8].Rubenstein JL, Kim J, Ozawa T, Zhang M, Westphal M, Deen DF, Shuman MA. Anti-VEGF antibody treatment of glioblastoma prolongs survival but results in increased vascular cooption. Neoplasia. 2000;2:306–314. doi: 10.1038/sj.neo.7900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kim ES, Serur A, Huang J, Manley CA, McCrudden KW, Frischer JS, Soffer SZ, Ring L, New T, Zabski S, Rudge JS, Holash J, Yancopoulos GD, Kandel JJ, Yamashiro DJ. Potent VEGF blockade causes regression of coopted vessels in a model of neuroblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11399–11404. doi: 10.1073/pnas.172398399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Blouw B, Song H, Tihan T, Bosze J, Ferrara N, Gerber HP, Johnson RS, Bergers G. The hypoxic response of tumors is dependent on their microenvironment. Cancer cell. 2003;4:133–146. doi: 10.1016/s1535-6108(03)00194-6. [DOI] [PubMed] [Google Scholar]
- [11].Steeg PS. Angiogenesis inhibitors: motivators of metastasis? Nature medicine. 2003;9:822–823. doi: 10.1038/nm0703-822. [DOI] [PubMed] [Google Scholar]
- [12].Bottaro DP, Liotta LA. Cancer: Out of air is not out of action. Nature. 2003;423:593–595. doi: 10.1038/423593a. [DOI] [PubMed] [Google Scholar]
- [13].Lamszus K, Brockmann MA, Eckerich C, Bohlen P, May C, Mangold U, Fillbrandt R, Westphal M. Inhibition of glioblastoma angiogenesis and invasion by combined treatments directed against vascular endothelial growth factor receptor-2, epidermal growth factor receptor, and vascular endothelial-cadherin. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:4934–4940. doi: 10.1158/1078-0432.CCR-04-2270. [DOI] [PubMed] [Google Scholar]
- [14].Kunkel P, Ulbricht U, Bohlen P, Brockmann MA, Fillbrandt R, Stavrou D, Westphal M, Lamszus K. Inhibition of glioma angiogenesis and growth in vivo by systemic treatment with a monoclonal antibody against vascular endothelial growth factor receptor-2. Cancer research. 2001;61:6624–6628. [PubMed] [Google Scholar]
- [15].Yi R, Fuchs E. MicroRNAs and their roles in mammalian stem cells. Journal of cell science. 2011;124:1775–1783. doi: 10.1242/jcs.069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gusev Y. Computational methods for analysis of cellular functions and pathways collectively targeted by differentially expressed microRNA. Methods. 2008;44:61–72. doi: 10.1016/j.ymeth.2007.10.005. [DOI] [PubMed] [Google Scholar]
- [17].Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- [18].Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer metastasis reviews. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- [19].Novakova J, Slaby O, Vyzula R, Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochemical and biophysical research communications. 2009;386:1–5. doi: 10.1016/j.bbrc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- [20].Lawler S, Chiocca EA. Emerging functions of microRNAs in glioblastoma. Journal of neuro-oncology. 2009;92:297–306. doi: 10.1007/s11060-009-9843-2. [DOI] [PubMed] [Google Scholar]
- [21].Skinner HD, Zheng JZ, Fang J, Agani F, Jiang BH. Vascular endothelial growth factor transcriptional activation is mediated by hypoxia-inducible factor 1alpha, HDM2, and p70S6K1 in response to phosphatidylinositol 3-kinase/AKT signaling. The Journal of biological chemistry. 2004;279:45643–45651. doi: 10.1074/jbc.M404097200. [DOI] [PubMed] [Google Scholar]
- [22].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [23].Zheng X, Jiang F, Katakowski M, Kalkanis SN, Hong X, Zhang X, Zhang ZG, Yang H, Chopp M. Inhibition of ADAM17 reduces hypoxia-induced brain tumor cell invasiveness. Cancer science. 2007;98:674–684. doi: 10.1111/j.1349-7006.2007.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yoon SO, Shin S, Mercurio AM. Hypoxia stimulates carcinoma invasion by stabilizing microtubules and promoting the Rab11 trafficking of the alpha6beta4 integrin. Cancer research. 2005;65:2761–2769. doi: 10.1158/0008-5472.CAN-04-4122. [DOI] [PubMed] [Google Scholar]
- [25].Di Leva G, Briskin D, Croce CM. MicroRNA in cancer: new hopes for antineoplastic chemotherapy. Upsala journal of medical sciences. 2012;117:202–216. doi: 10.3109/03009734.2012.660551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. The lancet oncology. 2012;13:e249–258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- [27].Chi A, Norden AD, Wen PY. Inhibition of angiogenesis and invasion in malignant gliomas. Expert review of anticancer therapy. 2007;7:1537–1560. doi: 10.1586/14737140.7.11.1537. [DOI] [PubMed] [Google Scholar]
- [28].Canning MT, Postovit LM, Clarke SH, Graham CH. Oxygen-mediated regulation of gelatinase and tissue inhibitor of metalloproteinases-1 expression by invasive cells. Experimental cell research. 2001;267:88–94. doi: 10.1006/excr.2001.5243. [DOI] [PubMed] [Google Scholar]
- [29].Rushing EC, Stine MJ, Hahn SJ, Shea S, Eller MS, Naif A, Khanna S, Westra WH, Jungbluth AA, Busam KJ, Mahalingam M, Alani RM. Neuropilin-2: a novel biomarker for malignant melanoma? Human pathology. 2012;43:381–389. doi: 10.1016/j.humpath.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sternlicht MD, Bissell MJ, Werb Z. The matrix metalloproteinase stromelysin-1 acts as a natural mammary tumor promoter. Oncogene. 2000;19:1102–1113. doi: 10.1038/sj.onc.1203347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sternlicht MD, Lochter A, Sympson CJ, Huey B, Rougier JP, Gray JW, Pinkel D, Bissell MJ, Werb Z. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–146. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Folkman J. Angiogenesis. Annu Rev Med. 2006;57:1–18. doi: 10.1146/annurev.med.57.121304.131306. [DOI] [PubMed] [Google Scholar]
- [33].Jiang BH, Liu LZ. PI3K/PTEN signaling in tumorigenesis and angiogenesis. Biochimica et biophysica acta. 2008;1784:150–158. doi: 10.1016/j.bbapap.2007.09.008. [DOI] [PubMed] [Google Scholar]
- [34].Geretti E, Klagsbrun M. Neuropilins: novel targets for anti-angiogenesis therapies. Cell adhesion & migration. 2007;1:56–61. doi: 10.4161/cam.1.2.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]