Abstract
The ability to silence, in a temporally precise fashion, the electrical activity of specific neurons embedded within intact brain tissue, is important for understanding the role that those neurons play in behaviors, brain disorders, and neural computations. “Optogenetic” silencers, genetically encoded molecules that, when expressed in targeted cells within neural networks, enable their electrical activity to be quieted in response to pulses of light, are enabling these kinds of causal circuit analyses studies. Two major classes of optogenetic silencer are in broad use in species ranging from worm to monkey: light-driven inward chloride pumps, or halorhodopsins, and light-driven outward proton pumps, such as archaerhodopsins and fungal light-driven proton pumps. Both classes of molecule, when expressed in neurons via viral or other transgenic means, enable the targeted neurons to be hyperpolarized by light. We here review the current status of these sets of molecules, and discuss how they are being discovered and engineered. We also discuss their expression properties, ionic properties, spectral characteristics, and kinetics. Such tools may not only find many uses in the quieting of electrical activity for basic science studies, but may also, in the future, find clinical uses for their ability to safely and transiently shut down cellular electrical activity in a precise fashion.
Keywords: optogenetics, opsins, neural silencing, halorhodopsin, archaerhodopsin, channelrhodopsin, control, cell types, neural circuits, causality
Introduction
The ability to silence, in a temporally precise fashion, the electrical activity of specific neurons embedded within intact brain tissue, is important for understanding the role that those neurons play in behaviors, brain disorders, and neural computations. Over the last several years, we and others have discovered that a large number of light-driven ion pumps naturally occurring in archaea, fungi, and other species, can be genetically expressed in neurons, enabling them to be hyperpolarized in response to light. These molecules, microbial (type I) opsins, are seven-transmembrane proteins, which translocate specific ions from one side of the membrane in which they are expressed, to the other, in response to light. For example, halorhodopsins are light-driven inward chloride pumps, which, when expressed in neurons, enable them to be hyperpolarized by orange light (Fig. 1Ai). Archaerhodopsins are light-driven outward proton pumps which, when expressed in neurons, also result in hyperpolarization (Fig. 1Bi). These proteins are monolithic, and encoded for by relatively small genes, under a kilobase long, small enough that genetic delivery to specific cells (e.g., via viruses, transfection, electroporation, transgenesis, or other methods) is achievable using many conventional methods in animals commonly used in neuroscience, from C. elegans to non-human primate (1–21). Furthermore, thanks to the proliferation of optical devices both driven by microscopy innovation as well as from other fields (e.g., telecommunications and computing), many devices exist or are easily engineered for the delivery of light to specific brain regions, even deep within the mammalian brain (conveyed via implanted optical fibers or waveguides; for example, a typical 200 micron diameter fiber with end-of-tip irradiance of 100–200 mW/mm2 can illuminate a volume of a few cubic millimeters), and even in a wirelessly controlled fashion (22–25). In vitro, conventional microscope fluorescence illuminators, such as xenon lamps, mercury lamps, confocal or two-photon lasers, and LEDs, equipped with fast shutters, deflectors, or controllers (3, 26–31), suffice to drive these molecules, the majority of which operate under illumination with incident light in the range 0.1–10 mW/mm2 at typical levels of expression in typical cells.
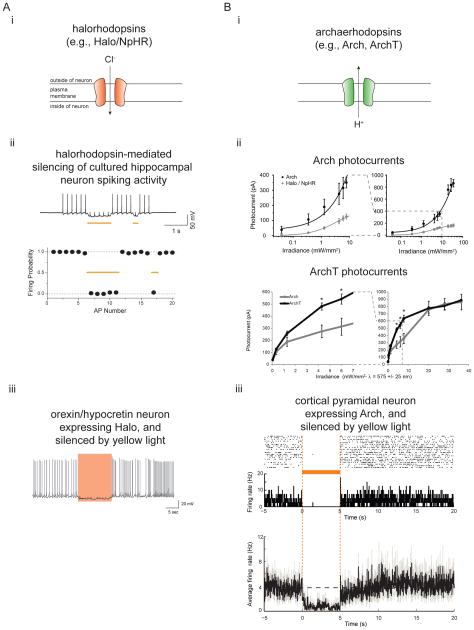
Figure 1. Two major classes of microbial opsin for light-driven hyperpolarization of neurons.
A, Halorhodopsins, light-driven inward chloride pumps. i, diagram of the physiological response of halorhodopsins when expressed in the plasma membranes of neurons and exposed to light. ii, light-driven spike blockade, demonstrated for a representative cultured hippocampal neuron (top), as well as a population of cultured hippocampal neurons (bottom). Neurons expressed Halo/NpHR, and received 20 pulses of somatic current injection (~300 pA, 4 ms, 5 Hz), accompanied by two periods of yellow light delivery (yellow bars). Adapted from ref. (36). iii, a neuron in a transgenic mouse expressing Halo in orexin/hypocretin neurons, being whole-cell patch clamp recorded during yellow light delivery, and exhibiting neural silencing. Adapted from ref. (41). B, Archaerhodopsins, light-driven outward proton pumps. i, diagram of the physiological response of archaerhodopsins when expressed in the plasma membranes of neurons and exposed to light. ii, photocurrents of Arch (top) and ArchT (bottom), measured as a function of 575 ± 25 nm light irradiance, in patch-clamped cultured neurons, for low (left) and high (right) light powers. Adapted from refs. (42, 43). iii, neural activity in a representative neuron recorded in awake mouse brain (top, middle), as well as a population of neurons in awake mouse brain (bottom). Shown is neural activity before, during, and after 5 seconds of yellow light illumination, displayed both as a spike raster plot (top), and as a histogram of instantaneous firing rate averaged across trials (middle, bottom; bin size, 20 ms). Adapted from ref. (42).
The speed of operation of these molecules is high, as they respond to light within milliseconds, and shut off rapidly after cessation of light delivery. For operation, these proteins require the co-factor all-trans-retinal as their essential chromophore, which acts to capture light. This chemical, being a natural beta carotene derivative, occurs in many organisms (such as mice, rats, and primates) at high enough background levels so that external chemical supplementation is not needed; for species such as C. elegans and Drosophila, whose background levels are lower, feeding of the all-trans-retinal to the organism suffices to enable supplementation. Because of this ease of use, adoption of these molecules for use in neuroscience has been rapid.
Overview of light-driven ion pumps
The first fully genetically encoded optogenetic tool to be utilized in neuroscience was the light-gated cation channel channelrhodopsin-2 (ChR2) from the green alga Chlamydomonas reinhardtii. This light-gated nonspecific cation channel, when expressed in neurons, enable the neurons to be depolarized with blue light (31, 32). The channelrhodopsins, and the origins of optogenetic tools, are reviewed elsewhere in this volume (CITE LIN CHAPTER HERE), as well as in other earlier references (33, 34). Like halorhodopsins and light-driven proton pumps, channelrhodopsins are all-trans-retinal binding seven-transmembrane proteins, and so there are some similarities in structure and function across these different opsin classes. Unlike channelrhodopsins, which open up a channel pore when gated by light, halorhodopsins and light-driven proton pumps translocate one ion per photon absorbed, which may in principle limit light sensitivity, but also means that they can pump ions against a concentration gradient, causing significant voltage swings less limited by the reversal potential of specific ions in neurons.
Halorhodopsins, as exemplified by the light-driven inward chloride pump halorhodopsin from the species Natronomonas pharaonis (Halo/NpHR (35)), were found in 2007 to function in mammalian neurons, where they mediate hyperpolarizing currents of 40–100 pA (36, 37), which is sufficient to support modest neural silencing (Fig. 1Aii). Because of the low currents that the original Halo could produce in mammalian neurons, due to limited protein trafficking in mammalian neurons that hampers expression at high levels (38–40), usage of the original Halo has largely been confined to use in invertebrates such as C. elegans, where it appears to function sufficiently in neurons and other excitable cells, to modulate behavior (37). In 2011, however, a group reported for the first time the use of the original Halo to mediate behavioral changes in living mammals (41), expressing Halo in the orexin/hypocretin neurons of the mouse brain via the generation of a transgenic mouse line. The orexin/hypocretin neurons of this mouse, when illuminated, undergo reductions in spiking (Fig. 1Aiii), and when such illumination is performed in the awake mouse, the animals enter slow-wave sleep, thus showing that brief silencing of this pathway is sufficient to induce sleep. Light-driven proton pumps, as exemplified by the archaeal protein archaerhodopsin-3 (Arch/aR-3) from Halorubrum sodomense (Fig. 1Bi), were found in 2010 to function well in mammalian neurons in the service of light-driven neural silencing (42). Arch, and its close relative ArchT, from Halorubrum sp. TP009 (43), can mediate photocurrents of 900 picoamps in cultured neurons (Fig. 1Bii), and can mediate complete light-driven neural silencing of neural activity in mice and primates (42, 43), in response to yellow or green light (Fig. 1Biii).
Light-driven inward chloride pumps and light-driven outward proton pumps are found in species of archaea (44–48), bacteria (49–55), fungi (56, 57) and algae (58). Because halorhodopsins, bacteriorhodopsins, and archaerhodopsins have been crystallized, and characterized by spectroscopy, mutagenesis, and physiology, much is known about their structure-function relationships (59–63). To date, these light-driven ion pumps have been found chiefly by analyzing genomes for sequences that resemble those of light-activated ion pumps and channels, followed by assessment of these gene products in heterologous expression systems (chiefly cell lines, neurons, and intact mouse brain) (64). Interestingly, a great many light-activated ion pumps from species all over the tree of life – even the very first one ever characterized, the Halobium salinarum bacteriorhodopsin – were able to mediate neural hyperpolarizations when assessed in cultured mouse neurons (42).
Properties of opsins, and how they affect performance
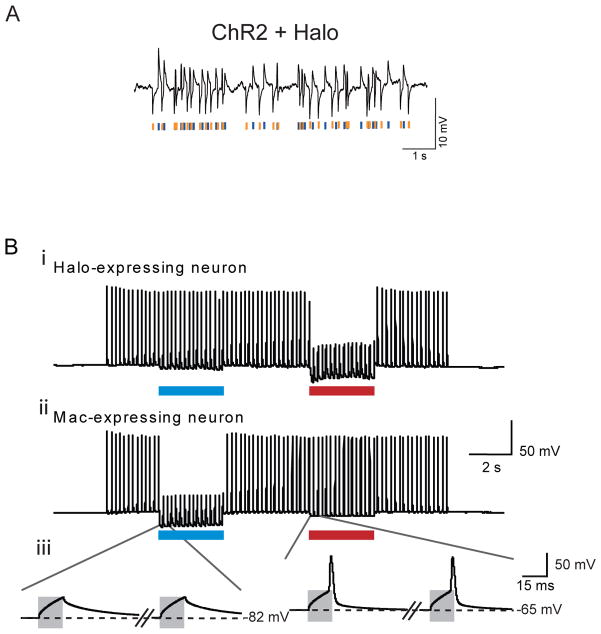
Each of these opsins has a different characteristic action spectrum, which describes the set of colors that optimally drive the opsin to function. Although chloride pumps are chiefly driven by yellow light, and possess largely stereotyped action spectra, proton pumps exist in many different kingdoms of life that are driven by many different colors of light. For example, the fungal light-driven proton pump from Leptosphaeria maculans (Mac) is drivable by blue-green light, and thus, alongside the earlier molecule Halo, which can be driven by yellow-red light, enables two-color silencing of different sets of neurons (Fig. 2B) (38). That is, a Mac-expressing neuron will be quieted by blue light, but not red light; in contrast, a Halo-expressing neuron will be quieted by red light, and not blue light. ChR2, which is blue light driven, can also be used alongside silencers such as Halo which are yellow or red light driven, even in the same cell, enabling depolarization and hyperpolarization of the doubly targeted cell by blue and yellow light respectively (Fig. 2A). The delivery of multiple opsins to the same cell can be facilitate by the use of 2A linker peptides to combine the genes for multiple opsins into a single open reading frame (14, 36), useful for bi-directionally assessing the causal role that a given set of neurons plays in a single animal, or for perturbing complex properties of neural dynamics such as neural synchrony (36).
Figure 2. Silencing of neurons with specific colors of light.
A, Hyperpolarization and depolarization events induced in a representative cultured hippocampal neuron expressing both ChR2 and Halo, by a Poisson train (mean inter-pulse interval λ = 100 ms) of alternating pulses of yellow and blue light (10 ms duration), denoted by yellow and blue bars, respectively. Adapted from ref. (36). B, multicolor silencing of two neural populations, enabled by blue- and red-light drivable ion pumps of different genomic classes. Action potentials evoked by current injection into patch clamped cultured neurons transfected with the archaeal opsin Halo (i) were selectively silenced by the red light but not by blue light, and vice-versa in neurons expressing the fungal opsin Mac (ii). Gray boxes in the inset (iii) indicate periods of patch clamp current injection. Adapted from ref. (42).
In this review, we will summarize the properties of these opsins, surveying how they are expressed in cells, how they conduct ions across membranes, how they cycle in response to light, and how they respond to light of different colors and powers. The optogenetic molecules here described were isolated from archaea and fungi, species whose lipid and ionic compositions, and external environment, differ greatly from those of neurons. Accordingly, the heterologous expression of these molecules in neurons may result in performance characteristics that are very different from what would be expected from their expression in their native cells (64). Indeed, this means that the performance of opsins must be assessed directly in the cell types of interest, as it can be difficult to predict how these high-speed molecules will function in a specific milieu, from their properties as characterized in another one.
Some general guidelines and considerations apply, however. First, to insure efficient translation, it is useful to codon-optimize molecules for the target species. Second, to boost membrane expression, it is often helpful to append extra protein sequences to opsins to improve their protein folding, membrane trafficking and localization, and even subcellular compartmentalization (38–40, 42, 65). Third, although many groups use these molecules to change the voltage of neurons, it is also useful to explicitly consider the ions translocated by these pumps; for example, they can be used to change the levels of specific ions in cells or subcellular compartments, in a temporally-precise way. Fourth, the photocycle kinetics can be used to determine which molecule might be best for a given scientific application; for example, various light-driven ion pumps may enter different states that may render them nonfunctional for periods of time (Fig. 4). It is critical to remember that these light-driven ion pumps are not simple on-off switches; they undergo a series of structural rearrangements, with many intermediate states between light reception and final restoration of the molecule to the initial state. Over the last several decades, a myriad of structure-function studies have been performed on these molecules (35, 47, 48, 59–61, 66–84), linking specific amino acids with specific kinetic and spectral properties of the molecules. Fifth, the light-sensitivity of a molecule can determine how well it will function in the brain; improving light sensitivity can enable larger volumes of brain to be controlled than possible with less light-sensitive molecules (e.g., Fig. 1Bii, bottom). In the following sections, we explore these properties, for light-driven chloride pumps and light-driven proton pumps.
Figure 4. Photocycle models for the two major optical neural silencer classes.
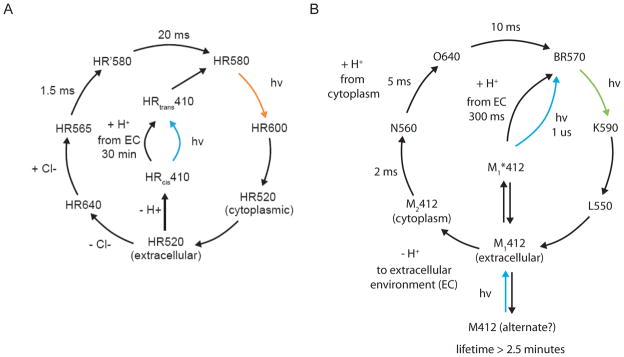
A, The halorhodopsin photocycle at high-power continuous illumination on the timescale of a typical photocycle (i.e. conditions used for neural silencing, > few ms). The HR410 intermediate is the origin of long-lived inactivation in neural silencing (e.g., (36, 75)). B, The photocycle of the H. salinarum bacteriorhodopsin. The photocycle has been simplified to reflect the dominant photocycle at large continuous illumination on the timescale of a typical photocycle (i.e. conditions used for neural silencing, > few ms). The M412 alternate intermediate is the origin of long-lived inactivation with bacteriorhodopsin. In contrast, Arch spontaneously quickly recovers from this state in the dark.
Protein expression and photocurrent magnitude, of light-driven ion pumps
The Natronomonas pharaonis halorhodopsin Halo/NpHR, the first microbial opsin to be used for optical neural silencing (Fig. 1A), has a reversal potential of approximately –400 mV (85), and although it comes from an archaeon that lives in very high salinity environments, the optimal chloride concentration for its operation is actually very close to that found in neurons (in contrast to that of other halorhodopsins such as the H. salinarum halorhodopsin, which demands far higher chloride concentrations) (86). This may be one of the reasons why it works in some neurons in some species (Fig. 1Aii, 1Aiii). However, at high expression levels, Halo forms intracellular aggregates (39, 40), an issue that can be ameliorated by the appending of trafficking sequences from the Kir2.1 channel (39, 40, 87), which reduces aggregation and increases photocurrent manyfold over that of the baseline molecule. Other signal sequences, such as a prolactin localization sequence, also increase halorhodopsin photocurrent (33); it is important to note, however, that different sequences may do different things in different cell types in different species.
The Halorubrum sodomense archaerhodopsin Arch (Fig. 1B), possesses strong photocurrents that exceed the original N. pharaonis halorhodopsin currents by an order of magnitude, both at low and at high light power (Fig. 1Bii, top) (42). Arch can mediate nearly complete silencing of neural activity in awake behaving mice (Fig. 1Biii). Arch does not require trafficking sequences to achieve these high levels of performance, although they can may help boost expression and/or current yet further (42). Other opsins that are similar in amino acid sequence to Arch have been described in genomic and gene sequence databases; all of these Arch relatives, found in other species of the Halorubrum genus, also express well in neurons (43). One of these Arch relatives, the archaerhodopsin from H. sp. TP009, presents photocurrents that are about 3.5x more light sensitive than those of Arch (Fig. 1Bii, bottom), supporting the silencing of broad brain regions, and neurons in the cortex of the awake behaving macaque (43). Light-driven proton pumps are powerful at neural silencing, a perhaps surprising fact given the relative scarcity of protons relative to other major charge carriers in the brain (such as Na+, K+, or Cl−); the fast kinetics of archaerhodopsins (88, 89), and their good trafficking in mammalian cells, amongst other attributes, may support the high performance of archaerhodopsins. These light-driven outward proton pumps, interestingly, do not change cellular pH to a greater extent than do opsins such as ChR2 (whose proton conductance is 106 times its sodium conductance). Specifically, illuminating Arch with relatively bright light when expressed in cultured neurons resulted in alkalinizations of neurons by 0.1–0.15 pH units, a change in pH of smaller magnitude than that seen with ChR2, perhaps due to the high proton conductance of channelrhodopsins (32, 90, 91). From an end-user standpoint, illumination of Arch- and ArchT-expressing neurons, in awake mice and macaques, for periods of many minutes, did not alter spike waveform or spike frequency when comparing before vs. after silencing.
Kinetics of light-driven ion pumps
From a kinetic standpoint, all of the halorhodopsins we have examined to date inactivate by roughly 30% every 15 seconds under typical illumination conditions (1–10 mW/mm2, of yellow, e.g. 593 nm, light; see Fig. 3A, 3B), consistent with the photocycle topology shown in Fig. 4A (36, 42). Furthermore, recovery in the dark of halorhodopsins from this inactivated state is slow, taking tens of minutes for full recovery (36, 75, 77). Although this rundown may somewhat compromise performance over very long illumination periods, it is possible to drive the molecule out of this inactive state by using UV or blue light (36, 75) (Fig. 3Aii, 3B). The entire photocycle is summarized in Fig. 4A, with the limiting time constants noted in the diagram, and with the states denoted by the light absorption properties of the state, as is conventional. The outer circle of the photocycle describes the dominant mode of operation, in which, after photon absorption, a chloride ion is released into the cytoplasm, and then a second chloride ion is taken up from the extracellular space, thus fulfilling the chloride pumping action. In the center of the photocycle diagram, however, is a proton-pumping series of states that involve the long-lasting inactive state described above, here denoted HR410. This HR410 state is the inactivated state that can be reprimed back to the initial state using blue light.
Figure 3. Kinetic properties of the two major optical neural silencer classes.
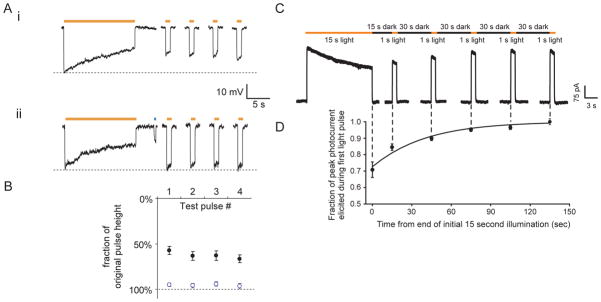
A, (i) Halo-mediated hyperpolarizations in a representative current-clamped hippocampal neuron during 15 seconds of continuous yellow light, followed by four 1-second test pulses of yellow light (one every 30 seconds, starting 10 seconds after the end of the first 15-second period of yellow light). (ii) Halo-mediated hyperpolarizations for the same cell exhibited in (i), but when Halo function is facilitated by a 400-ms pulse of blue light in between the 15-second period of yellow light and the first 1-second test pulse. Adapted from ref. (36). B, Population data for blue-light facilitation of Halo recovery. Plotted are the hyperpolarizations elicited by the four 1-second test pulses of yellow light, normalized to the peak hyperpolarization induced by the original 15-second yellow light pulse (mean + std. err.). Black dots represent experiments when no blue light pulse was delivered (as in Fig. 3Ai). Open blue dots represent experiments when 400 ms of blue light was delivered to facilitate recovery (as in Fig. 3Aii). Adapted from ref. (36). C, Raw current trace of a neuron lentivirally-infected with Arch, illuminated by a 15 s light pulse (575 ± 25 nm, irradiance 7.8 mW/mm2), followed by 1 s test pulses delivered starting 15, 45, 75, 105, and 135 seconds after the end of the 15 s light pulse. Adapted from ref. (42). D, Population data of averaged Arch photocurrents sampled at the times indicated by the vertical dotted lines that extend into Fig. 3C. Adapted from ref. (42).
In contrast to the kinetic properties of halorhodopsins, archaerhodopsins such as Arch and ArchT spontaneously recover in the dark, even after extensive illumination (Fig. 3C, 3D). The time constant of recovery is in the range of tens of seconds, in contrast to that of halorhodopsins, which is in the range of tens of minutes. Although archaerhodopsin photocycles are still relatively uncharacterized compared to those of earlier-discovered opsins, some insight into proton pump operation can be derived from consideration of the H. salinarum bacteriorhodopsin photocycle (Fig. 4B). The topology of the photocycle shares some similarities with that of halorhodopsins, although many of the timescales are significantly different.
Towards the future
Improvements in optogenetic silencers are rapidly transpiring, both driven by the burgeoning amount of microbial genome being sequenced, as well as the influx of bioengineering approaches such as mutagenesis and protein engineering. It is likely that the kinetic and amplitude properties of these molecules will continue to improve, given that only a small part of genomic space has been explored, and only a small number of mutations and protein changes (e.g., the appending of Kir2.1 trafficking sequences) have been explored. From a scientific standpoint, the optical silencers have been broadly applied in the service of assessing the causal role that specific neurons play in the behaviors and neural computations implemented by brain circuits. Now that these molecules have been safely and efficaciously used to modulate neural activity in awake macaque (7, 92), it is also intriguing to ponder whether new neural control prosthetics will arise, capable of silencing overactive neurons or deleting pathological neural activity patterns, in human patients, in the future.
Acknowledgments
E.S.B. acknowledges funding by the NIH Director’s New Innovator Award (DP2OD002002) as well as NIH Grants 1R01NS075421, 1R01DA029639, 1RC1MH088182, 1RC2DE020919, 1R01NS067199 and 1R43NS070453; the NSF CAREER award as well as NSF Grants EFRI 0835878, DMS 0848804, and DMS 1042134; Benesse Foundation, Jerry and Marge Burnett, Department of Defense CDMRP Post-Traumatic Stress Disorder Program, Google, Harvard/MIT Joint Grants Program in Basic Neuroscience, Human Frontiers Science Program, MIT Alumni Class Funds, MIT Intelligence Initiative, MIT McGovern Institute and the McGovern Institute Neurotechnology Award Program, MIT Media Lab, MIT Mind-Machine Project, MIT Neurotechnology Fund, NARSAD, Paul Allen Distinguished Investigator Award, Alfred P. Sloan Foundation, SFN Research Award for Innovation in Neuroscience, and the Wallace H. Coulter Foundation.
Footnotes
The authors declare no competing financial interests.
References
- 1.Wang H, Peca J, Matsuzaki M, Matsuzaki K, Noguchi J, Qiu L, Wang D, Zhang F, Boyden E, Deisseroth K, Kasai H, Hall WC, Feng G, Augustine GJ. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proc Natl Acad Sci U S A. 2007;104(19):8143–8. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huber D, Petreanu L, Ghitani N, Ranade S, Hromadka T, Mainen Z, Svoboda K. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451(7174):61–4. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10(5):663–8. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, Chiel H, Hegemann P, Landmesser LT, Herlitze S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci U S A. 2005;102(49):17816–21. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishizuka T, Kakuda M, Araki R, Yawo H. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci Res. 2006;54(2):85–94. doi: 10.1016/j.neures.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, Wang LP, Boyden ES, Deisseroth K. Channelrhodopsin-2 and optical control of excitable cells. Nat Methods. 2006;3(10):785–92. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- 7.Han X, Qian X, Bernstein JG, Zhou HH, Franzesi GT, Stern P, Bronson RT, Graybiel AM, Desimone R, Boyden ES. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62(2):191–8. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28(28):7025–30. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhlman SJ, Huang ZJ. High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS ONE. 2008;3(4):e2005. doi: 10.1371/journal.pone.0002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglass AD, Kraves S, Deisseroth K, Schier AF, Engert F. Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons. Curr Biol. 2008;18(15):1133–7. doi: 10.1016/j.cub.2008.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15(24):2279–84. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 12.Schroll C, Riemensperger T, Bucher D, Ehmer J, Voller T, Erbguth K, Gerber B, Hendel T, Nagel G, Buchner E, Fiala A. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16(17):1741–7. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 13.Dittgen T, Nimmerjahn A, Komai S, Licznerski P, Waters J, Margrie TW, Helmchen F, Denk W, Brecht M, Osten P. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc Natl Acad Sci U S A. 2004;101(52):18206–11. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X, Qian X, Stern P, Chuong AS, Boyden ES. Informational Lesions: Optical Perturbation of Spike Timing and Neural Synchrony Via Microbial Opsin Gene Fusions. Front Mol Neurosci. 2009 doi: 10.3389/neuro.02.012.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chhatwal JP, Hammack SE, Jasnow AM, Rainnie DG, Ressler KJ. Identification of cell-type-specific promoters within the brain using lentiviral vectors. Gene Ther. 2007;14(7):575–83. doi: 10.1038/sj.gt.3302898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450(7168):420–4. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan W, Janczewski WA, Yang P, Shao XM, Callaway EM, Feldman JL. Silencing preBotzinger complex somatostatin-expressing neurons induces persistent apnea in awake rat. Nat Neurosci. 2008;11(5):538–40. doi: 10.1038/nn.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toni N, Laplagne DA, Zhao C, Lombardi G, Ribak CE, Gage FH, Schinder AF. Neurons born in the adult dentate gyrus form functional synapses with target cells. Nat Neurosci. 2008;11(8):901–7. doi: 10.1038/nn.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan SC, Bernstein JG, Boyden ES. Scalable Fluidic Injector Arrays for Viral Targeting of Intact 3-D Brain Circuits. Journal of Visualized Experiments. 2009 doi: 10.3791/1489. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi A, Cui J, Ma YP, Olshevskaya E, Pu M, Dizhoor AM, Pan ZH. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron. 2006;50(1):23–33. doi: 10.1016/j.neuron.2006.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagali PS, Balya D, Awatramani GB, Munch TA, Kim DS, Busskamp V, Cepko CL, Roska B. Light-activated channels targeted to ON bipolar cells restore visual function in retinal degeneration. Nat Neurosci. 2008;11(6):667–75. doi: 10.1038/nn.2117. [DOI] [PubMed] [Google Scholar]
- 22.Gradinaru V, Thompson KR, Zhang F, Mogri M, Kay K, Schneider MB, Deisseroth K. Targeting and readout strategies for fast optical neural control in vitro and in vivo. J Neurosci. 2007;27(52):14231–8. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravanis AM, Wang LP, Zhang F, Meltzer LA, Mogri MZ, Schneider MB, Deisseroth K. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4(3):S143–56. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- 24.Wentz CT, Bernstein JG, Monahan P, Guerra A, Rodriguez A, Boyden ES. A Wirelessly Powered and Controlled Device for Optical Neural Control of Freely-Behaving Animals. Journal of Neural Engineering. 2011;8(4):046021. doi: 10.1088/1741-2560/8/4/046021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zorzos AN, Boyden ES, Fonstad CG. Multiwaveguide implantable probe for light delivery to sets of distributed brain targets. Opt Lett. 2010;35(24):4133–5. doi: 10.1364/OL.35.004133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farah N, Reutsky I, Shoham S. Patterned optical activation of retinal ganglion cells. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:6369–71. doi: 10.1109/IEMBS.2007.4353812. [DOI] [PubMed] [Google Scholar]
- 27.Guo ZV, Hart AC, Ramanathan S. Optical interrogation of neural circuits in Caenorhabditis elegans. Nat Methods. 2009;6(12):891–6. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campagnola L, Wang H, Zylka MJ. Fiber-coupled light-emitting diode for localized photostimulation of neurons expressing channelrhodopsin-2. J Neurosci Methods. 2008;169(1):27–33. doi: 10.1016/j.jneumeth.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Rickgauer JP, Tank DW, editors. Neuroscience. Washington D.C: Society for Neuroscience; Nov 17, 2008. Optimizing two-photon activation of channelrhodopsin-2 for stimulation at cellular resolution. [Google Scholar]
- 30.Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457(7233):1142–5. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8(9):1263–8. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 32.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100(24):13940–5. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chow BY, Han X, Bernstein JG, Monahan PE, Boyden ES. Light-Activated Ion Pumps and Channels for Temporally Precise Optical Control of Activity in Genetically Targeted Neurons. In: Kramer JJCaRH., editor. Photosensitive Molecules for Controlling Biological Function. Humana Press; 2011. pp. 99–132. [Google Scholar]
- 34.Boyden ES. A history of optogenetics: the development of tools for controlling brain circuits with light. F1000 Biology Reports. 2011;3(11) doi: 10.3410/B3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanyi JK, Duschl A, Hatfield GW, May K, Oesterhelt D. The primary structure of a halorhodopsin from Natronobacterium pharaonis. Structural, functional and evolutionary implications for bacterial rhodopsins and halorhodopsins. J Biol Chem. 1990;265(3):1253–60. [PubMed] [Google Scholar]
- 36.Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS ONE. 2007;2(3):e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446(7136):633–9. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 38.Chow B, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, Boyden ES. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2009 doi: 10.1038/nature08652. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36(1–4):129–39. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao S, Cunha C, Zhang F, Liu Q, Gloss B, Deisseroth K, Augustine GJ, Feng G. Improved expression of halorhodopsin for light-induced silencing of neuronal activity. Brain Cell Biol. 2008;36(1–4):141–54. doi: 10.1007/s11068-008-9034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsunematsu T, Kilduff TS, Boyden ES, Takahashi S, Tominaga M, Yamanaka A. Acute Optogenetic Silencing of Orexin/Hypocretin Neurons Induces Slow-Wave Sleep in Mice. J Neurosci. 2011;31(29):10529–39. doi: 10.1523/JNEUROSCI.0784-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, Boyden ES. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463(7277):98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han X, Chow BY, Zhou H, Klapoetke N, Chuong A, Rajimehr R, Yang A, Baratta MV, Winkle J, Desimone R, Boyden ES. A high-light sensitivity optical neural silencer: development and application to optogenetic control of non-human primate cortex. Frontiers in Systems Neuroscience. 2011;5:18. doi: 10.3389/fnsys.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ihara K, Umemura T, Katagiri I, Kitajima-Ihara T, Sugiyama Y, Kimura Y, Mukohata Y. Evolution of the archaeal rhodopsins: evolution rate changes by gene duplication and functional differentiation. J Mol Biol. 1999;285(1):163–74. doi: 10.1006/jmbi.1998.2286. [DOI] [PubMed] [Google Scholar]
- 45.Mukohata Y, Ihara K, Tamura T, Sugiyama Y. Halobacterial rhodopsins. J Biochem. 1999;125(4):649–57. doi: 10.1093/oxfordjournals.jbchem.a022332. [DOI] [PubMed] [Google Scholar]
- 46.Klare JP, Chizhov I, Engelhard M. Microbial rhodopsins: scaffolds for ion pumps, channels, and sensors. Results Probl Cell Differ. 2008;45:73–122. doi: 10.1007/400_2007_041. [DOI] [PubMed] [Google Scholar]
- 47.Lanyi JK. Bacteriorhodopsin. Annual Review of Physiology. 2004;66(1):665–88. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 48.Lanyi JK. Halorhodopsin: A Light-Driven Chloride Ion Pump. Annual Review of Biophysics and Biophysical Chemistry. 1986;15(1):11–28. doi: 10.1146/annurev.bb.15.060186.000303. [DOI] [PubMed] [Google Scholar]
- 49.Antón J, Peña A, Valens M, Santos F, Glöckner FO, Bauer M, Dopazo J, Herrero J, Rosselló-Mora R, Amann R. Salinibacter ruber: genomics and biogeography. In: Gunde-Cimerman NAP, Oren A, editors. Adaptation to Life in High Salt Concentrations in Archaea, Bacteria and Eukarya. Dordrecht: Kluwer Academic Publishers; 2005. pp. 257–66. [Google Scholar]
- 50.Balashov SP, Imasheva ES, Boichenko VA, Anton J, Wang JM, Lanyi JK. Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science. 2005;309(5743):2061–4. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beja O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411(6839):786–9. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 52.Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289(5486):1902–6. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 53.Friedrich T, Geibel S, Kalmbach R, Chizhov I, Ataka K, Heberle J, Engelhard M, Bamberg E. Proteorhodopsin is a light-driven proton pump with variable vectoriality. J Mol Biol. 2002;321(5):821–38. doi: 10.1016/s0022-2836(02)00696-4. [DOI] [PubMed] [Google Scholar]
- 54.Kelemen BR, Du M, Jensen RB. Proteorhodopsin in living color: diversity of spectral properties within living bacterial cells. Biochim Biophys Acta. 2003;1618(1):25–32. doi: 10.1016/j.bbamem.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Kim SY, Waschuk SA, Brown LS, Jung KH. Screening and characterization of proteorhodopsin color-tuning mutations in Escherichia coli with endogenous retinal synthesis. Biochim Biophys Acta. 2008;1777(6):504–13. doi: 10.1016/j.bbabio.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 56.Brown LS. Fungal rhodopsins and opsin-related proteins: eukaryotic homologues of bacteriorhodopsin with unknown functions. Photochem Photobiol Sci. 2004;3(6):555–65. doi: 10.1039/b315527g. [DOI] [PubMed] [Google Scholar]
- 57.Waschuk SA, Bezerra AG, Shi L, Brown LS. Leptosphaeria rhodopsin: Bacteriorhodopsin-like proton pump from a eukaryote. Proc Natl Acad Sci U S A. 2005;102(19):6879–83. doi: 10.1073/pnas.0409659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsunoda SP, Ewers D, Gazzarrini S, Moroni A, Gradmann D, Hegemann P. H+-Pumping Rhodopsin from the Marine Alga Acetabularia. 2006;91(4):1471–9. doi: 10.1529/biophysj.106.086421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Essen LO. Halorhodopsin: light-driven ion pumping made simple? Curr Opin Struct Biol. 2002;12(4):516–22. doi: 10.1016/s0959-440x(02)00356-1. [DOI] [PubMed] [Google Scholar]
- 60.Kolbe M, Besir H, Essen LO, Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 A resolution. Science. 2000;288(5470):1390–6. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- 61.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 A resolution. J Mol Biol. 1999;291(4):899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 62.Yoshimura K, Kouyama T. Structural role of bacterioruberin in the trimeric structure of archaerhodopsin-2. J Mol Biol. 2008;375(5):1267–81. doi: 10.1016/j.jmb.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 63.Enami N, Yoshimura K, Murakami M, Okumura H, Ihara K, Kouyama T. Crystal structures of archaerhodopsin-1 and -2: Common structural motif in archaeal light-driven proton pumps. J Mol Biol. 2006;358(3):675–85. doi: 10.1016/j.jmb.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 64.Chow BY, Chuong AS, Klapoetke NC, Boyden ES. Synthetic physiology strategies for adapting tools from nature for genetically targeted control of fast biological processes. Methods Enzymol. 2011;497:425–43. doi: 10.1016/B978-0-12-385075-1.00018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greenberg KP, Pham A, Werblin FS. Differential targeting of optical neuromodulators to ganglion cell soma and dendrites allows dynamic control of center-surround antagonism. Neuron. 2011;69(4):713–20. doi: 10.1016/j.neuron.2011.01.024. [DOI] [PubMed] [Google Scholar]
- 66.Braiman MS, Stern LJ, Chao BH, Khorana HG. Structure-function studies on bacteriorhodopsin. IV. Purification and renaturation of bacterio-opsin polypeptide expressed in Escherichia coli. J Biol Chem. 1987;262(19):9271–6. [PubMed] [Google Scholar]
- 67.Gilles-Gonzalez MA, Engelman DM, Khorana HG. Structure-function studies of bacteriorhodopsin XV. Effects of deletions in loops B-C and E-F on bacteriorhodopsin chromophore and structure. J Biol Chem. 1991;266(13):8545–50. [PubMed] [Google Scholar]
- 68.Mogi T, Stern LJ, Chao BH, Khorana HG. Structure-function studies on bacteriorhodopsin. VIII. Substitutions of the membrane-embedded prolines 50, 91, and 186: the effects are determined by the substituting amino acids. J Biol Chem. 1989;264(24):14192–6. [PubMed] [Google Scholar]
- 69.Mogi T, Marti T, Khorana HG. Structure-function studies on bacteriorhodopsin. IX. Substitutions of tryptophan residues affect protein-retinal interactions in bacteriorhodopsin. J Biol Chem. 1989;264(24):14197–201. [PubMed] [Google Scholar]
- 70.Mogi T, Stern LJ, Hackett NR, Khorana HG. Bacteriorhodopsin mutants containing single tyrosine to phenylalanine substitutions are all active in proton translocation. Proc Natl Acad Sci U S A. 1987;84(16):5595–9. doi: 10.1073/pnas.84.16.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marti T, Otto H, Mogi T, Rosselet SJ, Heyn MP, Khorana HG. Bacteriorhodopsin mutants containing single substitutions of serine or threonine residues are all active in proton translocation. J Biol Chem. 1991;266(11):6919–27. [PubMed] [Google Scholar]
- 72.Marinetti T, Subramaniam S, Mogi T, Marti T, Khorana HG. Replacement of aspartic residues 85, 96, 115, or 212 affects the quantum yield and kinetics of proton release and uptake by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1989;86(2):529–33. doi: 10.1073/pnas.86.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mogi T, Stern LJ, Marti T, Chao BH, Khorana HG. Aspartic acid substitutions affect proton translocation by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1988;85(12):4148–52. doi: 10.1073/pnas.85.12.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Subramaniam S, Greenhalgh DA, Khorana HG. Aspartic acid 85 in bacteriorhodopsin functions both as proton acceptor and negative counterion to the Schiff base. J Biol Chem. 1992;267(36):25730–3. [PubMed] [Google Scholar]
- 75.Bamberg E, Tittor J, Oesterhelt D. Light-driven proton or chloride pumping by halorhodopsin. Proc Natl Acad Sci U S A. 1993;90(2):639–43. doi: 10.1073/pnas.90.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown LS, Needleman R, Lanyi JK. Interaction of proton and chloride transfer pathways in recombinant bacteriorhodopsin with chloride transport activity: implications for the chloride translocation mechanism. Biochemistry. 1996;35(50):16048–54. doi: 10.1021/bi9622938. [DOI] [PubMed] [Google Scholar]
- 77.Hegemann P, Oesterhelt D, Steiner M. The photocycle of the chloride pump halorhodopsin. I: Azide-catalyzed deprotonation of the chromophore is a side reaction of photocycle intermediates inactivating the pump. Embo J. 1985;4(9):2347–50. doi: 10.1002/j.1460-2075.1985.tb03937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blanck A, Oesterhelt D. The halo-opsin gene. II. Sequence, primary structure of halorhodopsin and comparison with bacteriorhodopsin. Embo J. 1987;6(1):265–73. doi: 10.1002/j.1460-2075.1987.tb04749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Henderson R, Schertler GF. The structure of bacteriorhodopsin and its relevance to the visual opsins and other seven-helix G-protein coupled receptors. Philos Trans R Soc Lond B Biol Sci. 1990;326(1236):379–89. doi: 10.1098/rstb.1990.0019. [DOI] [PubMed] [Google Scholar]
- 80.Rudiger M, Oesterhelt D. Specific arginine and threonine residues control anion binding and transport in the light-driven chloride pump halorhodopsin. Embo J. 1997;16(13):3813–21. doi: 10.1093/emboj/16.13.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Varo G, Brown LS, Sasaki J, Kandori H, Maeda A, Needleman R, Lanyi JK. Light-driven chloride ion transport by halorhodopsin from Natronobacterium pharaonis. 1. The photochemical cycle. Biochemistry. 1995;34(44):14490–9. doi: 10.1021/bi00044a027. [DOI] [PubMed] [Google Scholar]
- 82.Tittor J, Haupts U, Haupts C, Oesterhelt D, Becker A, Bamberg E. Chloride and proton transport in bacteriorhodopsin mutant D85T: different modes of ion translocation in a retinal protein. J Mol Biol. 1997;271(3):405–16. doi: 10.1006/jmbi.1997.1204. [DOI] [PubMed] [Google Scholar]
- 83.Tittor J, Oesterhelt D, Bamberg E. Bacteriorhodopsin mutants D85N, D85T and D85,96N as proton pumps. Biophys Chem. 1995;56(1–2):153–7. doi: 10.1016/0301-4622(95)00027-u. [DOI] [PubMed] [Google Scholar]
- 84.Varo G, Needleman R, Lanyi JK. Light-driven chloride ion transport by halorhodopsin from Natronobacterium pharaonis. 2. Chloride release and uptake, protein conformation change, and thermodynamics. Biochemistry. 1995;34(44):14500–7. doi: 10.1021/bi00044a028. [DOI] [PubMed] [Google Scholar]
- 85.Seki A, Miyauchi S, Hayashi S, Kikukawa T, Kubo M, Demura M, Ganapathy V, Kamo N. Heterologous expression of Pharaonis halorhodopsin in Xenopus laevis oocytes and electrophysiological characterization of its light-driven Cl- pump activity. Biophys J. 2007;92(7):2559–69. doi: 10.1529/biophysj.106.093153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okuno D, Asaumi M, Muneyuki E. Chloride concentration dependency of the electrogenic activity of halorhodopsin. Biochemistry. 1999;38(17):5422–9. doi: 10.1021/bi9826456. [DOI] [PubMed] [Google Scholar]
- 87.Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141(1):154–65. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ming M, Lu M, Balashov SP, Ebrey TG, Li Q, Ding J. pH dependence of light-driven proton pumping by an archaerhodopsin from Tibet: comparison with bacteriorhodopsin. Biophys J. 2006;90(9):3322–32. doi: 10.1529/biophysj.105.076547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lukashev EP, Govindjee R, Kono M, Ebrey TG, Sugiyama Y, Mukohata Y. pH dependence of the absorption spectra and photochemical transformations of the archaerhodopsins. Photochem Photobiol. 1994;60(1):69–75. doi: 10.1111/j.1751-1097.1994.tb03945.x. [DOI] [PubMed] [Google Scholar]
- 90.Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96(5):1803–14. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Berthold P, Tsunoda SP, Ernst OP, Mages W, Gradmann D, Hegemann P. Channelrhodopsin-1 Initiates Phototaxis and Photophobic Responses in Chlamydomonas by Immediate Light-Induced Depolarization. Plant Cell. 2008;20(6):1665–77. doi: 10.1105/tpc.108.057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Diester I, Kaufman MT, Mogri M, Pashaie R, Goo W, Yizhar O, Ramakrishnan C, Deisseroth K, Shenoy KV. An optogenetic toolbox designed for primates. Nat Neurosci. 2011;14(3):387–97. doi: 10.1038/nn.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]