Abstract
While the use of detergents is necessary for a variety of protein isolation preparation protocols, they are not compatible with mass spectral analysis due to ion suppression and adduct formation. This manuscript describes optimization of detergent removal, using commercially available SDS depletion spin columns containing an affinity resin, providing for both increased protein recovery and thorough SDS removal. Ion mobility spectrometry coupled with mass spectrometry (IMS-MS) allowed for a concurrent analysis of both analyte and detergent. In the case of both proteins and peptides, higher detergent concentrations than previously reported provided an increase of sample recovery; however there was a limit as SDS was detected by IMS-MS at higher levels of SDS indicating incomplete detergent depletion. The results also suggest optimal conditions for SDS removal are dependent on the sample concentration. Overall, this study provides a useful guide for proteomic studies where SDS is required for efficient sample preparation.
Keywords: Ion mobility, SDS depletion, sample preparation, detergents
Technical Brief
Sodium dodecyl sulfate (SDS) is a common detergent used for processing protein samples to aid with solubilization, reduction of protein aggregation, isoelectric focusing, and separation/purification [1]. While SDS has many great properties, which make it a popular reagent among laboratories, one major caveat is the incompatibility with mass spectral analyses as it leads to decreased signal to noise ratio and resolution of the ions of interest [1, 2]. The introduction of new techniques that rely on SDS such as Gel-Eluted Liquid Fraction Entrapment Electrophoresis (GELFrEE) [3] have increased the need to effectively remove SDS prior to both top-down (intact proteins) and bottom-up (proteolytic peptides) mass spectrometry (MS)studies.
Standard sample clean up with various solid-phase extraction (SPE) or desalting techniques have not been effective in SDS removal to date, prompting investigation of a variety of removal methods, which have resulted in several commercial products. Filter-aided sample preparation (FASP) [4], high salt precipitation kits [5], and SDS specific binding spin cartridges are all popular methods of detergent depletion for proteomic sample preparation [6, 7]. However, a majority of these methods are hindered by low protein recovery, labor intensiveness, irreproducibility, incompatibility with high throughput/online LCMS methods, or incomplete SDS removal. To address these shortcomings, both increasing protein recovery and effectiveness of SDS removal were evaluated in this study. Detergent removal using the spin column format (Pierce, Rockford, IL) [6] was chosen for optimization because the cartridges are inexpensive, easy to use, and have potential for incorporation into an online LCMS system. Additionally, an increase in peptide identification using spin columns over the FASP method was recently demonstrated by Bereman et al. [7].
In order to accurately assess the successful optimization of SDS removal, two variables need to be defined: percent protein recovery after SDS depletion, and extent of SDS removal. Detection of SDS, or ion suppression from SDS, is difficult to analyze using traditional MS analyses because there is little to no separation of the detergent and analyte of interest. Previously, SDS was quantified using colorimetric methods, or GC analysis of the SDS derivative 1-dodecanol [8, 9]. While these methods accurately determine the concentration of SDS, a separate analysis is needed for detection of proteins or peptides. By coupling ion mobility spectrometry (IMS) to MS, compounds can be separated based on structural attributes allowing proteins and peptides to be separated from SDS with no further sample preparation or manipulation [10]. Previously, Bagag et al. demonstrated separation of detergents and peptides using high-field asymmetric waveform ion mobility spectrometry (FAIMS) for simple peptide mixtures [11]. However IMS-MS analyses of detergents combined with intact proteins or complex mixtures have yet to be evaluated using IMS-MS. Ultimately, gas phase separations need to be performed on the LC timescale with complex mixtures, a feature allowed by the instrumentation described in this study. Simultaneous detection of separated SDS and proteins/peptides makes this an ideal system for the evaluation of SDS depletion.
Intact ubiquitin (Sigma-Aldrich, St Louis, MO) or a tryptic digest of Shewanella oneidensis whole cell lysate were used as analytes. Detergent removal spin columns (Pierce, Rockford, IL) were used according to manufactures instructions. Briefly, spin columns were stored at 4 °C, equilibrated to room temperature and the storage buffer was removed by centrifugation at 1,500 rcf for 1 minute. The resin was then washed twice with 400 μL of 50 mM ammonium bicarbonate pH 7 and removed by centrifugation at 1,500 rcf for 1 minute. 150 μL of 0.1–1.0 mg/mL peptides/proteins containing 0–10 % SDS were applied to the spin column resin and incubated for 2 minutes. A final centrifugation at 1,500 rcf for 2 minutes was performed to elute the SDS free sample. Protein concentration and percent recovery were determined using a standard bicinchoninic acid (BCA) assay. Both depletion and BCA assays were performed in technical replicate. Values reported for percent recovery using data from the BCA assay are an average with a mean standard deviation of 4%, with 1% and 10% as minimum and maximum values respectively. BCA standards with and without SDS present were used to assess signal interference. Prior to IMS-MS analysis samples were diluted 1:1 with acetonitrile containing 0.1% formic acid.
Sample mixtures were analyzed by direct infusion or reverse phase liquid chromatography (RPLC) on a NanoAcquity (Waters Corp, Milford, MA) system using a custom packed C18 column (75-μm i.d., 360-μm o.d., 40-cm capillary, Polymicro Technologies Inc., Phoenix, AZ). A 50-minute linear gradient of nanopure water of 0.1% formic acid (Sigma-Aldrich, St Louis, MO) to acetonitrile (Fisher Scientific, Waltham, MA) containing 0.1% formic acid was applied. The eluting peptides/proteins were analyzed with positive ESI using a custom built IMS-TOF MS instrument as previously described [10]. A detailed description of the instrument control software and data acquisition scheme is reported elsewhere [14].
A range of protein/peptide concentrations with varying levels of detergent were depleted of SDS using SDS-specific binding resin in a spin column format. Sample recovery after SDS depletion was first determined using a standard BCA assay (Table 1), where no interfering signal was observed from stock SDS solutions. Specifically, at the higher concentration of 1 mg/mL ubiquitin, there is a noticeable increase in percent protein recovery between 0 and 4% SDS (from 56% to 83% recovery). However when the concentration of ubiquitin was decreased to 0.1 mg/mL, this increase in protein recovery (40% to 70%) is not observed until 7% SDS prior to depletion. A similar trend is observed for peptide samples with an increase in recovery as the concentration of SDS prior to depletion was increased. For both peptide and protein samples, a noticeable increase in protein recovery was observed when > 3% of SDS was present prior to depletion. Notably, percent protein recovery at the same SDS concentration was consistently higher for samples at 1 mg/ml compared to 0.1 mg/mL ubiquitin. Presumably this is due to non-specific interactions between the analyte and the column. Because the same resin volume was used for both concentrations of ubiquitin, the fraction lost due to non-specific binding would be higher for the lower concentration. Presumably recovery would improve for samples of lower concentration if a smaller column was prepared/used. The affinity resin used in this study is commercially available, making optimization for lower sample quantities with less resin feasible.
Table 1.
| % SDS | % Recovery 1 mg/mL Ubiquitin | % Recovery 0.1 mg/mL Ubiquitin | % Recovery 0.1 mg/mL Peptides |
|---|---|---|---|
|
| |||
| 0 | 56 | 40 | 25 |
| 1 | 72 | 32 | 33 |
| 2 | 74 | 32 | 43 |
| 3 | 75 | 37 | 58 |
| 4 | 83 | 42 | 58 |
| 5 | 93 | 43 | 80 |
| 6 | - | 57 | - |
| 7 | - | 70 | - |
| 8 | - | 78 | - |
| 9 | - | 84 | - |
| 10 | - | 94 | - |
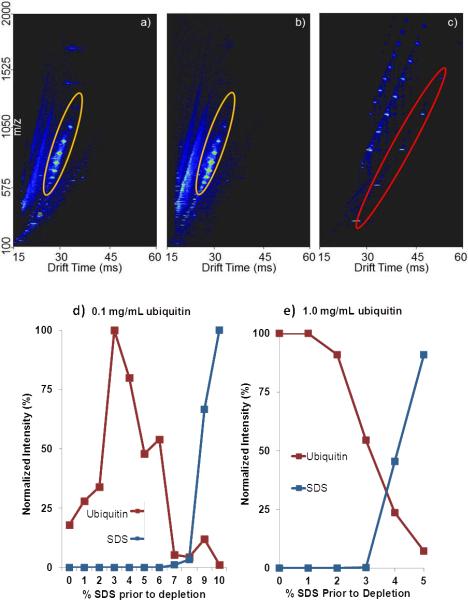
Despite increasing protein recovery with increasing percentage of SDS, presumably there is a concentration of SDS where the resin is saturated and the eluate contains both protein and SDS. To test this, samples processed with detergent removal spin columns were infused directly into an IMS-TOF MS system. Ubiquitin and SDS were both easily visualized in the IMS-MS plots (Figure 1a–c) with signature patterns of SDS starting at a drift time of 26 to 60 msec, and ubiquitin ranging from 26 to 30 msec. Ubiquitin signal increases due to increased recovery until SDS dominates the IMS-MS spectrum. For protein samples at 1.0 mg/ml ubiquitin, SDS was detected in the IMS-MS spectra at ≥ 3% SDS prior to depletion (Figure 1e). In comparison, for samples containing 0.1 mg/mL ubiquitin, SDS was only detected with IMS-MS when the SDS concentration was ≥ 7% prior to depletion (Figure 1d). The observed increase in binding capacity for SDS, at the lower analyte concentration, could be due to a reduced binding site competition between the analyte and detergent. Therefore a smaller analyte to SDS ratio provides more binding sites available to SDS, allowing for complete depletion at higher SDS concentrations because of less non-specific binding by the analyte. As expected, ubiquitin specific ion current decreased as the presence and intensity of SDS increased, despite higher percent protein recovery observed in the BCA assay results.
Figure 1.
IMS-MS spectra for 0% (a), 6% (b), and 10% (c) SDS (red oval) prior to depletion for 0.1 mg/mL ubiquitin (orange oval). Ubiquitin signal is clearly observed for 0 and 6% SDS prior to depletion, at 10% SDS prior to depletion no ubiquitin signal is detected. (d) and (e) Plots of ubiquitin signal intensity over a range of SDS concentrations prior to depletion (0.1 mg/ml ubiquitin and 1.0 mg/ml ubiquitin).
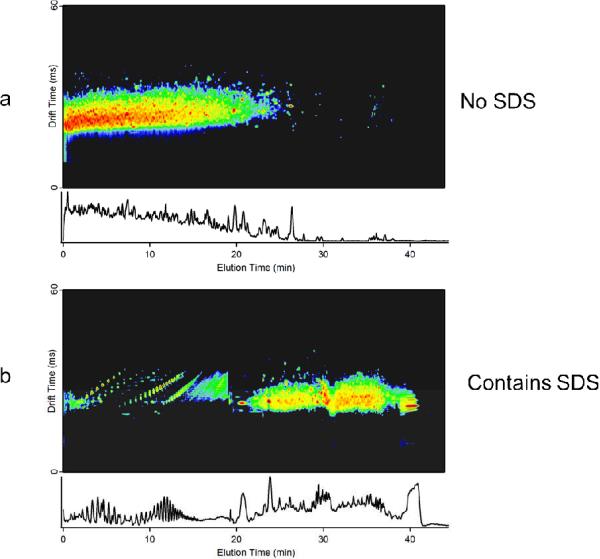
The limit of detection for SDS in the IMS-MS system was determined using SDS alone and SDS in solution with ubiquitin. Starting with a known concentration of SDS, or SDS and ubiquitin, serial dilutions with water or ubiquitin were performed until the SDS, or ubiquitin, signal was no longer detectable. For SDS alone, signal was visible at 0.001 % but was not detected at 0.0001 %. In the presence of 0.1 mg/mL ubiquitin, SDS was also visible at 0.001 % however the signal was less intense than in the SDS alone solution, indicating some suppression from the presence of ubiquitin. Ubiquitin signal was detected when the concentration of SDS was as high as 0.1%; however no protein signal was observed when the concentration of SDS was increased to 0.25%. Therefore, the concurrent SDS-ubiquitin detection range for this IMS-MS study was from 0.1% to 0.001% SDS, well below concentrations used in this study and traditional sample preparation methods, making IMS-MS a suitable analytical technique to evaluate the effectiveness of SDS removal methods.” To understand the effect of this technique on a more complicated sample, S. oneidensis whole cell lysate digest was processed for SDS removal, and separated online with RPLC prior to IMS-MS analysis. Interference with expected chromatography, i.e. “chromatogram smearing” and retention time delay (Figure 2), was observed slightly at 4% SDS prior to depletion with very pronounced distortion at 5% SDS, indicating the presence of SDS in these samples. To confirm interference from SDS, the number of LCMS features [15] were determined for each RPLC separation; each feature represents a potential peptide in the LC IMS-MS dataset. For duplicate LC IMS-MS standard acquisitions at 0% SDS prior to depletion, the average number of features detected was 24,500 with a range of 23,000 to 26,000. For samples processed with SDS depletion cartridges, the highest number of features identified was approximately 24,000 at both 2% and 3% SDS concentrations prior to detection. These results are in the same range as the standard acquisitions above that did not contain any SDS, indicating that SDS has been depleted below the limit of interference and limit of detection. At 4% SDS prior to depletion the number of features was reduced to 15,050 and at 5% the number dropped below 11,000. This confirms that beginning at 4% SDS the detergent removal cartridge fails. Despite previous reports indicating that SDS and peptides can be analyzed using FAIMS [11], because of the observed effect of SDS on the chromatography and potential damage to analytical columns, it is recommended that detergent depletion is performed on samples for LCMS analyses. For peptides at or below 0.1 mg/mL, 2–3 % SDS provided the best protein recovery and the most efficient SDS removal.
Figure 2.
IMS-MS 2D display of S. oneidensis tryptic digest RPLC analysis a) without SDS and b) containing SDS with chromatography interference. When SDS is present peptide retention time is shifted to later in the elution profile and SDS is visible from 0 to 20 min.
In this study, we observed a change in sample recovery dependent upon the concentration of SDS prior to depletion. One possible explanation for the increased recovery observed for higher concentrations of SDS is that as the concentration of SDS increases and binds to the affinity resin, the available non-specific binding sites for peptides/proteins decrease. As SDS begins to saturate the resin, less analyte is lost due to non-specific binding. Similarly, when comparing samples at different concentrations, at a static concentration of SDS, reduced protein recovery was observed for samples at a lower concentration due to the competition of binding sites between SDS and protein/analyte. The number of binding sites available for nonspecific binding at each SDS concentration is constant; therefore total sample loss is similar, resulting in 10–20% loss of analyte using a higher concentration, and 40–50% loss using samples of a lower concentration. These results are consistent with a previous study noting that protein recovery improved significantly above 0.1 mg/ml BSA [6]. Therefore, as determined by IMS-MS analysis, to avoid sample loss when removing detergent from samples using affinity resins, we suggest increasing the SDS concentration coordinately with the sample concentration to maximize protein recovery. Additionally, when employing SDS depletion in a routine fashion, it is advised to perform a similar optimization for sample recovery as it may vary based on analyte, sample matrix, and concentration.
Acknowledgments
This research was supported by the U. S. Department of Energy (DOE) Office of Biological and Environmental Research and by the NIH National Center for Research Resources (RR018522), DOE/BER Genomics: Genome Sciences program and NIGMS (P41 GM103493). Work was performed in the Environmental Molecular Science Laboratory (EMSL), a DOE national scientific user facility located on the campus of Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RLO 1830. The authors also wish to thank Carrie Nicora and Heather Brewer for insightful discussion.
Abbreviations
- BCA
bicinchoninic acid
- FASP
filter-aided sample preparation
- GELFrEE
Gel-Eluted Liquid Fraction Entrapment Electrophoresis
- IMS-MS
Ion mobility spectrometry coupled with mass spectrometry
- SPE
solid-phase extraction
- FAIMS
high-field asymmetric waveform ion mobility spectrometry
Footnotes
The authors have declared no conflict of interest.
References
- [1].Botelho D, Wall MJ, Vieira DB, Fitzsimmons S, et al. Top-down and bottom-up proteomics of SDS-containing solutions following mass-based separation. J Proteome Res. 2010;9:2863–2870. doi: 10.1021/pr900949p. [DOI] [PubMed] [Google Scholar]
- [2].Staudenmann W, Hatt PD, Hoving S, Lehmann A, et al. Sample handling for proteome analysis. Electrophoresis. 1998;19:901–908. doi: 10.1002/elps.1150190605. [DOI] [PubMed] [Google Scholar]
- [3].Tran JC, Doucette AA. Gel-eluted liquid fraction entrapment electrophoresis: an electrophoretic method for broad molecular weight range proteome separation. Anal Chem. 2008;80:1568–1573. doi: 10.1021/ac702197w. [DOI] [PubMed] [Google Scholar]
- [4].Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- [5].Zhou JY, Dann GP, Shi T, Wang L, et al. Simple Sodium Dodecyl Sulfate-Assisted Sample Preparation Method for LC-MS-Based Proteomics Applications. Anal Chem. 2012;84:2862–2867. doi: 10.1021/ac203394r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Antharavally BS, Mallia KA, Rosenblatt MM, Salunkhe AM, et al. Efficient removal of detergents from proteins and peptides in a spin column format. Anal Biochem. 2011;416:39–44. doi: 10.1016/j.ab.2011.05.013. [DOI] [PubMed] [Google Scholar]
- [7].Bereman MS, Egertson JD, Maccoss MJ. Comparison between procedures using SDS for shotgun proteomic analyses of complex samples. Proteomics. 2011;11:2931–2935. doi: 10.1002/pmic.201100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Campbell R, Winkler MA, Wu H. Quantification of sodium dodecyl sulfate in microliter biochemical samples by gas chromatography. Anal Biochem. 2004;335:98–102. doi: 10.1016/j.ab.2004.08.026. [DOI] [PubMed] [Google Scholar]
- [9].Rusconi F, Valton E, Nguyen R, Dufourc E. Quantification of sodium dodecyl sulfate in microliter-volume biochemical samples by visible light spectroscopy. Anal Biochem. 2001;295:31–37. doi: 10.1006/abio.2001.5164. [DOI] [PubMed] [Google Scholar]
- [10].Baker ES, Clowers BH, Li F, Tang K, et al. Ion mobility spectrometry-mass spectrometry performance using electrodynamic ion funnels and elevated drift gas pressures. J Am Soc Mass Spectrom. 2007;18:1176–1187. doi: 10.1016/j.jasms.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bagag A, Giuliani A, Canon F, Refregiers M, Le Naour F. Separation of peptides from detergents using ion mobility spectrometry. Rapid Commun Mass Spectrom. 2011;25:3436–3440. doi: 10.1002/rcm.5242. [DOI] [PubMed] [Google Scholar]
- [14].Ibrahim Y, Belov ME, Tolmachev AV, Prior DC, Smith RD. Ion funnel trap interface for orthogonal time-of-flight mass spectrometry. Anal Chem. 2007;79:7845–7852. doi: 10.1021/ac071091m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Monroe ME, Tolic N, Jaitly N, Shaw JL, et al. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics. 2007;23:2021–2023. doi: 10.1093/bioinformatics/btm281. [DOI] [PubMed] [Google Scholar]