Abstract
Resolution of systematic relationships among members of the Culex pipiens (L.) complex has important implications for public health as well as for studies on the evolution of sibling species. Currently held views contend that in California considerable genetic introgression occurs between Cx. pipiens and Cx. quinquefasciatus Say, and as such, these taxa behave as if they are a single species. Development of high throughput SNP genotyping tools for the analysis of Cx. pipiens complex population structure is therefore desirable. As a first step toward this goal, we sequenced 12 gene fragments from specimens collected in Marin and Fresno counties. On average, we found a higher single nucleotide polymorphism (SNP) density than any other mosquito species reported thus far. Coding regions contained significantly higher GC content (median 54.7%) than noncoding regions (42.4%; Wilcoxon rank sum test, P = 5.29 × 10−5). Differences in SNP allele frequencies observed between mosquitoes from Marin and Fresno counties indicated significant genetic divergence and suggest that SNP markers will be useful for future detailed population genetic studies of this group. The high density of SNPs highlights the difficulty in identifying species within the complex and may be associated with the large degree of phenotypic variation observed in this group of mosquitoes.
Keywords: Culex pipiens, Culex quinquefasciatus, single nucleotide polymorphism, California
Defining species, subspecies, and forms of members of the Culex pipiens (L.) complex has been a subject of much debate despite comprehensive studies using comparative morphology (Dobrotworsky 1967, Jupp 1978, Miles and Paterson 1979), behavior (Urbanelli et al. 1985, 1997; Byrne and Nichols 1998; Chevillon et al. 1998; Spielman 2001; Cornel et al. 2003; Gomes et al. 2009; Reusken et al. 2010), and population genetics using isozymes (Tabachnick and Powell 1983, Weitzel et al. 2009) and microsatellite DNA polymorphism (Fonseca et al. 2004, Keyghobadi et al. 2006, Edillo et al. 2007, Huang et al. 2008, Bataille et al. 2009, Gomes et al. 2009).
Within California, members of the complex are dispersed across various ecoregions: Cx. quinquefasciatus Say in the warmer south, Cx. pipiens in the cooler north, and hybrids in central California (Barr 1982, Tabachnik and Powell 1983, Urbanelli et al. 1995, Cornel et al. 2003). Highly autogenous and stenogamous mosquitoes resembling Cx. pipiens collected as larvae under an apartment complex in the city of San Rafael (Marin County) suggests that Cx. p. molestus may also occur in California (McAbee et al. 2003). Autogenous mosquitoes do not require blood feeding to develop eggs. If these mosquitoes are stenogamous and mate only in a restricted underground spaces (e.g., storm sewers) they are likely genetically isolated.
Cx. pipiens s.l. are confirmed vectors of West Nile virus in California (Goddard et al. 2003, McAbee et al. 2008) and consequently targets of intense control efforts. We were motivated to isolate and characterize single nucleotide polymorphisms (SNPs) to further population genomics studies of this important group of mosquito vectors.
In 2007, the Cx. quinquefasciatus genome project released the latest sequence assembly data of 3,171 scaffolds (http://metazoa.ensembl.org/Culex_quinquefasciatus), and formally published in 2010 (Arensburger et al. 2010). Unfortunately, the genome sequence has yet to be assembled onto chromosomes. We sequenced 12 gene fragments from specimens collected in Marin and Fresno counties in Central California. We chose these two central California counties for our initial SNP characterization because they are likely to include the range of Cx. pipiens s.l. members currently identified in California (Cornel et al. 2003) and therefore are likely to represent the full range of genetic diversity in this region. Cold tolerant Cx. pipens (Cpp) and autogenous and stenogamous “molestus (Cpm)” are sympatric in Marin County (McAbee et al. 2003), whereas Fresno County provides habitat for Cpp, Cx. quinquefasciatus (Cpq) and their hybrids (McAbee et al. 2008). In this article, we summarize the results of a preliminary study aimed at describing SNPs and other types of mutations observed in functional genes of Culex pipiens s.l. mosquitoes from Central California and that may prove useful for downstream population genetics studies of this group of mosquitoes.
Results
The number of single nucleotide polymorphisms varied from 8 to 77 per gene fragment. The mean number of mutations was 44.3 (±24.2) and the median 43; this is equivalent to a SNP every 13 nucleotides on average. GenBank accession numbers of sequences analyzed for this study are provided in Table 1. The most polymorphic regions were found in ESTB1 and ODR (see total number of mutations, μt, in Table 2). The median of %GC in introns was 42.4% while the median in coding regions was 54.7%. GC content was significantly higher in coding regions than introns (Wilcoxon rank sum test, P = 5.29 × 10−5). Sequences were designated “noncoding” and/or “coding” based on exons reported for the Cpq genome on Ensembl (Arensburger et al. 2010).
Table 1.
Primers and sequence information
| Gene | Abbr. | Primer sequences (5′-3′) | Super config | Coordinates | Ensembl Gene ID | Genbank IDb |
|---|---|---|---|---|---|---|
| Acetylcholinesterase 2 | ACE2 | F: AGATGTGGAATCCCAACACG | 3.6 | 256902–257310 | Q1JS07_CULPI | HQ881614 - HQ881646 |
| (Exon 2–3) | R: TCGAGGCCACGATTACGTT | (CPIJ000662) | ||||
| Beta tubulin | bTUB | F: CCCCGCGCCGTCCTGGTC | 3.41 | 905442–905933 | CPIJ0003260 | HQ881598HQ881613 |
| (Exon 2) | R: ATCGCCGTACGTGGGTGTGGTGAG | |||||
| Esterase B1 | ESTB1 | F: GACGGAACCGTTGGACTGTA | 3.512 | 34578–35143 | Q8WQ89_CULPI | HQ881715–HQ881780 |
| (Esterase 3a, exon 2–4) | R: CATGTGGTAGTGCACGGAAC | (CPIJ013918) | ||||
| Fork-head transcription factor | FOXO | F: CCGCTCAAGACCAACTTTTC | 3.2176 | 8094–8803 | CPIJ020030 | HQ881781–HQ881814 |
| (Exon 1) | R: GAATACCGCGAGTACATCTGG | |||||
| Heat shock protein 70 | HSP70 | F: AGCACATCGCATGGAACATT | 3.316 | 28588–29319 | Q52QQ9_CULPI | HQ881815–HQ881866 |
| (Upstream + exon1) | R: TCGTTGAAGTAAGCCGGAAC | 39468–40439 | (CPIJ011081 or CPIJ011082) | |||
| CPIJ011083 | ||||||
| Myosin light chain 2 | MyoLC2 | F: GAGAAGAAGGAAAAGAAGACCAA | 3.963 | 119311–119895 | CPIJ017123 | HQ881867–HQ881890 |
| (Exon 2–3) | R: CCAGTGAGTGAGGGCATAACG | |||||
| Odorant receptor Or2 | OR2 | F: GATTTCTTGCAACGCATCG | 3.32 | 1491759–1492455 | CPIJ002479 | HQ881891–HQ881954 |
| (Exon 2–4) | R: TGTACGCCACCACGATGATA | |||||
| Thiamin pyrophosphokinase 1 | TPPK1 | F: CCGACTTCACCAAATCCCTCAA | 3.639 | 42781–43221 | CPIJ015475 | HQ881955–HQ881978 |
| (Exon 4) | R: ACATGTCCTTCGGCGTCGTG | |||||
| Trypsin 5G1 | TRYP | F: GCTCTTGATACGACACGCTC | 3.103 | 418493–419184 | CPIJ006019 | HQ881979–HQ882010 |
| (Upstream + exon 1) | R: CGATCGGTAATCTGGTTGGT | 415462–416153 | CPIJ006018 | |||
| V type ATPase B subunit | VATPSB | F: TCGATTGCCCGTGGACAGAAGATT | 3.585 | 131911–132352 | CPIJ014699 | HQ881647–HQ881668 |
| (Exon 2) | R: ACATGTAACCGGGGAAACCACGAC | |||||
| Vitellogenin | VIT | F: CCGTGAAGACCACCAAAACT | 3.656 | 52338–52918 | CPIJ015387 | HQ882011–HQ882076 |
| (Exon 1–3) | R: CTCCACCGGAAGGTACTTGA | 3.656 | 49310–49890 | CPIJ015386 | ||
| 3.802 | 156909–157398 | CPIJ016051 | ||||
| 5.8 S RNA coding | 5.8S | F: AGGACACATGAACACCGACA | 3.1464 | 56477–56919 | CPIJ039552 | HQ881669–HQ881714 |
| (Exon 1+ downstream) | R: AATTCAGGGGGTAGTCACACA | 47767–48208 | CPIJ039622 |
The only closest match with Culex genome sequence is esterase (EST) B1, while similar sequences in NCBI nucleotide database (identity >95%) are annotated as EST-3, EST-A, EST-A1, or EST-A2.
GenBank accession no. range for isolate sequences.
Table 2.
Summary of nucleotide polymorphism in single copy genes
| Gene | la | Groupb | N c | FST | P d | ndiffe | Dxy (%)f | μ fix g | μ share h | μ pMmF i | μ pFmM j | μ t k | π (%)l | Hdm | %GCncn | %GCco | TDp |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACE2 | 638 | Marin | 32 | 0.696 | 0.0006 | 28.801 | 4.56% | 0 | 3 | 0 | 42 | 45 | 0.18% | 0.778 | 39.0% | 53.0% | 1.186 |
| Fresno | 16 | 2.59% | 0.892 | 0.873 | |||||||||||||
| bTUB | 492 | Marin | 20 | 0.118 | 0.1089 | 1.492 | 0.30% | 0 | 1 | 4 | 3 | 8 | 0.23% | 0.742 | — | 61.0% | –0.603 |
| Fresno | 12 | 0.31% | 0.848 | 0.466 | |||||||||||||
| ESTB1 | 568 | Marin | 34 | 0.463 | 0 | 26.178 | 4.73% | 0 | 58 | 10 | 3 | 71 | 2.53% | 0.620 | 33.6% | 51.8% | –0.582 |
| Fresno | 32 | 2.69% | 0.429 | –0.054 | |||||||||||||
| FOXO | 710 | Marin | 36 | 0.168 | 0.0001 | 5.233 | 0.74% | 0 | 11 | 6 | 16 | 33 | 0.39% | 0.586 | — | 56.0% | –1.062 |
| Fresno | 32 | 0.893 | –0.412 | ||||||||||||||
| MyoLC2 | 585 | Marin | 38 | 0.576 | 0.0001 | 9.379 | 1.60% | 0 | 9 | 5 | 9 | 23 | 0.37% | 0.768 | 51.1% | 59.1% | –1.120 |
| Fresno | 10 | 0.99% | 0.889 | –0.413 | |||||||||||||
| Or2 | 709 | Marin | 34 | 0.254 | 0.0113 | 22.549 | 3.19% | 0 | 52 | 13 | 3 | 68 | 3.07% | 0.817 | 35.9% | 50.4% | 1.293 |
| Fresno | 32 | 1.69% | 0.659 | –0.466 | |||||||||||||
| TPPK | 441 | Marin | 36 | 0.278 | 0.0159 | 11.755 | 2.67% | 0 | 14 | 4 | 23 | 41 | 1.02% | 0.744 | — | 52.1% | 0.120 |
| Fresno | 12 | 2.83% | 0.955 | 0.086 | |||||||||||||
| VATPS | 442 | Marin | 32 | 0.542 | 0.0041 | 7.193 | 1.63% | 0 | 2 | 3 | 10 | 15 | 0.65% | 0.772 | — | 55.4% | –0.115 |
| Fresno | 12 | 0.84% | 0.939 | –0.583 |
Amplicon length;
sequence group by county;
no. total alleles;
P value from Genetic Differentiation Estimates using χ2 table in DnaSP;
avg no. nucleotide differences between populations;
avg no. nucleotide substitutions per site between populations;
no. fixed differences between populations;
shared mutations between populations;
mutations polymorphic in Marin county but monomorphic in Fresno county;
mutations polymorphic in Fresno county but monomorphic in Marin county;
total no. mutations;
nucleotide diversity within pop;
haplotype diversity within pop;
GC content in percent in noncoding regions;
GC content in coding regions;
Tajima SD FST values are highlighted in bold if gene flow estimation was significant after significance threshold was adjusted for multiple comparison (P < 0.00427).
SNPs that cause frame shifts were lacking, except for one specimen in the TRYP fragment. The 25–29th nucleotide (reference sequence: GCCA) of TRYP exon one encodes the ninth (alanine) and part of the 10th amino acids (threonine). These four nucleotides were replaced by single nucleotide T for all specimens from Marin County, resulting in a deletion of one amino acid and nonsynonymous mutation from threonine to serine. A single specimen from Fresno County possessed an additional insertion of C between GCC and A that may result in a frame shift in the transcript. Another notable mutation was an insertion/deletion of an entire intron in VIT. This intron, between exon two and exon three of VIT, is common (31/34) in Marin County, but only 47% (=15/32) of the samples from Fresno County contained this intron.
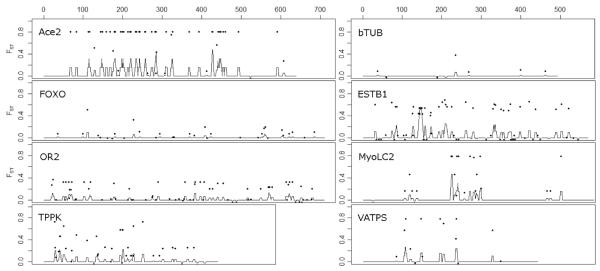
The gene flow estimation method implemented in DnaSP detected significantly limited gene flow between Marin and Fresno Counties for five gene fragments AceII, ESTB1, FOXO, MyoLC, and VATPS. The mean FST between the two counties for these five genes was 0.489 (±0.198), and the average number of nucleotide differences was 15.4 (±11.2) (Table 2). The FST between Fresno and Marin for each locus is illustrated in Fig. 1. We conducted Fisher exact tests to examine the relationship between allele abundance and county for each mutation and found significant divergence in all genes except bTUB. A list of selected significantly diverged mutations is presented in Table 3, and a full list of mutations and calculated Fisher exact test P values are provided in Supplemental Table S1 (available online only).
Fig. 1.
Distribution of SNPs within single copy gene fragments studied (in solid dot ●). Y axis represent FST comparing Fresno and Marin county populations for each SNP. Lines indicate running mean of FST over 5 bp window.
Table 3.
Selected SNPs significantly diverged between Central and Marin county
| Gene | Mutation IDa | Descriptionb | Allele | Fresnoc | Marind | P valuee |
|---|---|---|---|---|---|---|
| ACE2 | Ace2-068 | SNP at 68th nucleotide | A/G | 4/12 | 32/0 | 2.61E-08 |
| ESTB1 | ESTB1-1-502 | SNP at 502nd nucleotide | A/G | 4/28 | 33/7 | 2.51E-09 |
| FOXO | FOXO-111 | SNP at 111th nucleotide | A/T | 31/1 | 18/30 | 7.38E-08 |
| HSP70 | HSP70-3-438 | SNP at 438th nucleotide | C/T | 4/12 | 16/0 | 1.61E-05 |
| MyoLC | MyoLC-280 | SNP at 280th nucleotide | C/T | 10/0 | 6/32 | 1.22E-06 |
| Or2 | ODO-1-050 | SNP at 50th nucleotide | A/T | 26/6 | 12/36 | 8.86E-07 |
| TPPK | TPPK-051 | SNP at 51st nucleotide | G/A | 4/8 | 34/2 | 4.89E-05 |
| TRYP | TRYP-085 | [A/-] indel at 85th nucleotide | –/A | 17/13 | 0/22 | 8.94E-06 |
| VATPS | VATPS-108 | SNP at 108th nucleotide | A/C | 10/2 | 2/30 | 1.57E-06 |
| VIT | VIT-2-193 | SNP at 193th nucleotide | T/C | 24/8 | 3/45 | 1.21E-10 |
| 5.8S | Q-226 | GAC repeat starting at nucleotide 226 | 3 GAC/4 GAC | 4/12 | 32/0 | 2.61E-08 |
Mutation ID corresponding to a locus,
description of a mutation,
no. of observed alleles in Fresno county,
no. of observed alleles in Marin county,
Fisher’s exact test P value.
Full list of mutations and corresponding P values are provided in the supplement material, Table S2.
Mosquitoes collected from Fresno County were more similar to the published Johannesburg strain (JNB) of Cpq genome sequence than those from Marin County (Table 4). Only fragments of TRYP amplified from three specimens from Fresno County were identical to the JNB reference sequence; none of the remaining 13 gene fragments was the same. The TRYP fragment was monomorphic in specimens from Marin County, but was only 95% identical to the published JNB sequence.
Table 4.
Sequence comparison with reference sequences
| Gene | Fresnoa | %iden 2b | Marinc | %iden 2b |
|---|---|---|---|---|
| ACE2 | RL1 | 95.27% | A1–4 | 89.47% |
| bTUB | 7CI-5 | 99.80% | B3–2 | 99.59% |
| ESTB1 | UL1 | 91.73% | B2–5 | 91.44% |
| FOXO | RL1 | 99.86% | A1–4 | 99.15% |
| HSP70 | UD3 | 96.32% | A1–4 | 94.41% |
| MyoLC2 | 7CI-6 | 99.32% | B3–1 | 98.12% |
| Or2 | UD1 | 99.72% | B2–4 | 94.93% |
| TRYP | UD3 | 100.00% | A1–4 | 94.81% |
| TPPK | 7CI-6 | 96.15% | A1–1 | 95.69% |
| VATPS | 7CI-5 | 99.77% | B3–1 | 97.96% |
| VIT | RL2 | 83.84% | B7–4 | 73.07% |
| 5.8S | UD3 | 98.88% | A2–5 | 97.97% |
| Mean | 96.72% | 93.88% |
Fresno county sample ID,
percent identity between Cx. quinquefasciatus genome sequence and a sequence from a field specimen,
Marin county sample ID.
Despite significant divergence between counties (Tables 2 and 3), no fixed SNP differences were observed. Between the two counties, 41% of mutations were shared, 28% of mutations were polymorphic in Marin County and monomorphic in Fresno County, and the remaining 31% of mutations were polymorphic in Fresno Country and monomorphic in Marin County. Tajima SD test indicates all polymorphisms are neutral (P > 0.1); although future studies with a larger sample size are necessary to confirm this.
In addition to SNPs, we observed microsatellites and long insertion/delitions (indels). Multiple micro-satelllites were observed in the 5.8S rRNA sequences that included a single GC repeat in Marin County compared with one or two repeats in Fresno County, three GAC repeats in Marin County compared with three or four repeats in Fresno County, and one or two GTTC repeats in both counties. In an intron between exon two and three of Esterase B1, a GGT motif occurred with 0–2 repeats in both counties. Mutations within exon one of HSP70 were observed that included a 5bp-long (AGTTA) indel and a 11–13bp (TTCACATA[-/C][-/A]AAGT) indel. Because these indels were observed within a coding region, they may affect transcription of HSP70. Some specimens from Fresno County had a TT insertion in TRYP whereas this insertion was absent in mosquitoes from Marin County. In addition, some sequences for the TRYP gene from Fresno County have a GCC insertion that can lead to the insertion of an amino acid. The Vitellogenin gene fragment contained an indel of up to 111 bp in some individuals from both counties, but the insertion of this long fragment was more common in Marin County specimens.
Discussion
Extensive polymorphism was observed in the 12 genes analyzed (total 7,094 bp) in Cx. pipiens s.l. from Marin and Fresno counties. On average, a SNP occurred every 13 bp. This SNP frequency is much greater than the SNP frequency in Anopheles gambiae (Giles), which has a SNP approximately every 250 bp (Holt et al. 2002). Previous An. gambiae research reported one SNP every 125 coding base pairs in nuclear genomic sequences obtained from laboratory strains of An. gambiae (Morlais et al. 2004). The Cx. pipiens SNP density is also higher than in Aedes aegypti (L.), which is reported to have one SNPs every 83 bp (Morlais and Severson 2003), although the genes interrogated are not the same, so this is not a direct comparison. Nonetheless, it is clear that on average the SNP density is very high in the Cx. pipiens genome. The high SNP frequency among California Cx. pipiens s.l. reported here creates significant challenges for genome-wide high-throughput genotyping because primer design is constrained by polymorphism in sequence flanking the target SNP.
Sequence fragments for HSP70, TRYP, VIT, and 5.8S rRNA matched multiple genes in the JNB Cpq genome with a sequence identity >95%. This indicates that some of the observed polymorphism may be because of gene duplications rather than point mutations within a single gene. Specimens presumed to be heterozygous for alleles (e.g., within the same gene fragment) differed by as much as 5% (Table 4). Although this high amount of heterogeneity may be because of allelic variation in a single copy gene it is equally plausible that it represents nonallelic variation in multi-copy genes. This challenges attempts to predict genes in the Cpq and related genomes because single copy genes with high variation can be annotated as a multi-copy gene with high sequence similarity. The issue of gene duplication and the occurrence of multi-copy gene families should be resolved as assembly and annotation of the Cpq genome improves.
Of interest, we reported private microsatellite alleles and SNPs associated with Marin or Fresno County. These county specific SNPs may be useful for future studies aimed at describing introgression of Cpp and Cpq in California. Recently, Huang and colleagues (2011) reported two SNPs within the 28S rDNA sequence and applied them to study introgression between Cpp and Cpq (Huang et al. 2011). One of the many SNPs we reported was a private microsatellite allele in 5.8S rDNA, downstream of the 28S fragment, present in Fresno county but absent in Marin (Table 3). Whether this private polymorphism originated from introgression with Cpq and its utility as a diagnostic marker differentiating Cpq from Cpp remains to be seen.
The observation that in the Cx. pipiens s.l. genome coding regions have higher GC content than noncoding regions is consistent with other organisms (Burge and Karlin 1997, Wuitschick and Karrer 1999, Pozzoli et al. 2008). In addition, a positive correlation between GC content and recombination has been reported in insects (Marais et al. 2001, Takano-Shimizu 2001), humans (Ikemura and Wada 1991, Galtier et al. 2001), and other animals (Hurst et al. 1999, Galtier et al. 2001, Williams and Hurst 2000). This is important because variation in GC content can influence the accuracy in gene predictions (Burge and Karlin 1997). GC content is also reported to be associated with staining intensities of human chromsomes (Furey and Haussler 2003). Whether banding patterns in Cx. pipiens polytene chromosomes are associated with congregates of coding genes remains to be investigated.
This study demonstrates that significant polymorphism and between-population divergence in the genomes of California Cx. pipiens complex members exists and that SNPs can be useful markers for the study of the population genetics of this group. SNPs have significant advantages over other markers (e.g., microsatellites) for population genetic studies. Although microsatellite markers have been developed for Cx. pipiens s.l. (Fonseca et al. 2004, Edillo et al. 2007, Kilpatrick et al., 2007) in our experience the protocols applied for assaying California populations (unpublished) are prone to numerous polymerase chain reaction (PCR) failures (=null alleles). This is not surprising given the very high frequency of SNPs in the genome of this group as observed in this study. It should be expected that with a large number of polymorphisms, amplification of gene fragments, or microsatellites would be difficult because of polymorphisms in polymerase chain reaction (PCR) primer annealing sites. In addition, whereas a typical microsatellite-based population genetics study uses 20–25 markers, a SNP-based study can easily include several hundred markers from various positions across the genome. This greatly improves genome coverage, facilitating analyses of ecological, and/or phenotypic association studies. Association mapping studies using SNPs can be used to identify causative loci responsible for phenotypes of interest, such as space requirement for mating (eurygamy requiring a large space for mating vs. stenogamy that can mate in a narrow space like small cage or tube), host preference (bird vs. mammalian), oviposition site preferences (above ground vs. below and clean vs. eutrophic water bodies), insecticide resistance, dispersal, and other behaviors. Such association studies should contribute to the resolution of some of the systematic mysteries in the Cx. pipiens complex and a better understanding of the genetics of insecticide resistance, which is increasingly being recognized as a threat to controlling this group of mosquitoes (Hemingway and Ranson 2000, McAbee et al. 2003).
Methods
Mosquito Samples
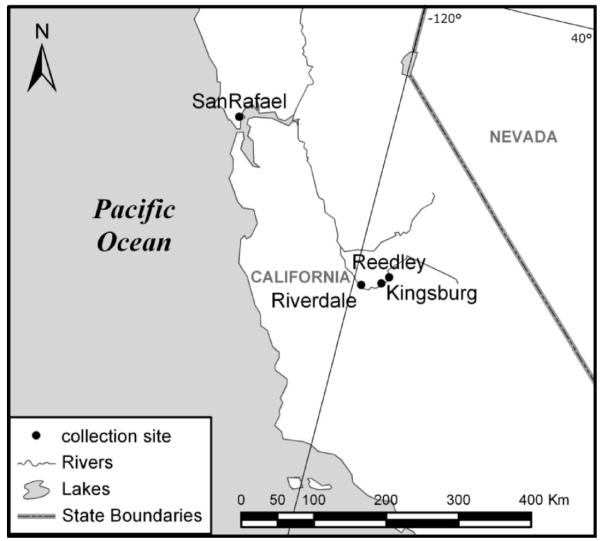
Mosquitoes were collected as larvae in an apartment basement and as adults aboveground in areas surrounding the apartment complex using CO2 baited CDC traps (Sudia and Chamberlain 1962) (in San Rafael, Marin County (37°58′24.73″N, 122°31′51.91″W). Mosquitoes were also collected in Fresno County from within the towns of Riverdale (36°25′51.82″N, 119°51′34.50″W), Kingsburg (36°30′ 49.82″N,119°33′14.46″W), and Reedley (36°35′46.82″N,119°27′1.45″W) between May, 2007 and October, 2009 in CO2 baited CDC traps. Mosquitoes were morphologically identified as members of the Cx. pipiens complex (Bohart and Washino 1978). Location of collection sites are indicated in Fig. 2.
Fig. 2.
Map of collection sites.
DNA Extraction and Sequencing
In total, 35 individual whole mosquitoes, 19 from Marin County and 16 from Fresno County, were lysed using a Qiagen Tissulyser and genomic DNA extracted using a Bio-Sprint 96 DNA Blood Kit (Qiagen, Chatsworth, CA). Published genomic DNA or mRNA sequences derived from multiple isolates of known genes for Cx. pipiens compex (Arensburger et al. 2010, Hasan et al. 2009) were blasted against the Cpq supercontig sequences using the Ensembl Blast tool (http://metazoa.ensembl.org/Culex_quinquefasciatus/blastview). The supercontig sequences with the highest identity score to the query gene sequences were selected for sequencing. Primers were designed using Primer3 (http://frodo.wi.mit.edu/primer3/), and primer sequences used are provided in Table 1. For optimal sequencing results, we limited GC content of each primer to be between 45 ≈ 60%, and primer melting temperature to be between 57 and 63°C.
In total, 12 gene fragments were sequenced including: acetylcholinase two (ACE2), beta tubulin (bTUB), esterase B1 (ESTB1), forkhead transcription factor (FOXO), heat shock protein 70 (HSP70), myosin light chain two (MyoLC2), odorant receptor Or2 (Or2), thiamine pyrophosphokinase one (TPPK), trypsin 5G1 (TRYP), v-type ATP synthase B (VATPS), vitellogenin (VIT), and 5.8S rRNA (5.8S). Each gene was located on a different supercontig (Table 1). ESTB1 is annotated with different names in other literature such as EST-3, EST-A, EST-A1, or EST-A2 (Arensburger et al. 2010, Rooker et al. 1996, Ben Cheikh et al. 2009). Each 50 μl PCR reaction contained 0.5 μM of forward and reverse primers, 1X PCR reaction buffer (Applied Biosystems, Carlsbad, CA), 1.5 mM MgCl2, 200 μM dNTP mix, 1.25U Ampli-TaqDNA polymerase (Applied Biosystems, Carlsbad, CA) and 2 μl of DNA template. The thermocycler was programmed for all PCR reactions to denature for 5 min at 95°C followed by 35 cycles of 95°C for 30 s, annealing temperatures ranging from 48 to 54°C for 30 s, 72°C for 30 s and then a final 5 min at 72°C. For each gene fragment, the PCR reaction was adjusted by either modifying the PCR mix and/or thermal cycling annealing conditions for optimal amplification. Amplicons were sequenced at the UCDNA Sequencing Facility (College of Biological Sciences, UC Davis) using an ABI 3730 Genetic Analyzer (Applied Biosystems). Gene fragments were also sequenced in both directions (forward/reverse) and SNPs were identified only if the SNP was found in both directions. ChromasLite ver. 2.01 was used to view chromatograms and convert chromatograms to text sequences. BioEdit (Ibis Therapeutics, Carlsbad, CA) and/or Geneious (Drummond et al. 2010) software were used for sequence alignment. Certain individuals were heterozygous for indel mutations in some genes causing mixed base pair nucleotide alignments after the indel mutation. To resolve these mixed sequences, we used in-house haplotype finder software, which extracts two haplotype sequences from an entangled chromatogram caused by two haploids having indel mutations.
Genetic Data and Statistics
DnaSP ver 5.10 software was used for analyzing DNA polymorphisms among mosquitoes (Librado and Rozas 2009) and R software (http://www.r-project.org) was used for Fisher exact tests. Means and standard deviations are noted as M ± SD.
Supplementary Material
Acknowledgment
We thank Jim Wanderscheid and his team at the Marin/Sonoma Mosquito and Vector Control District and Steve Mulligan and his team at the Consolidated Mosquito Abatement District for their support. This research was supported by National Institutes of Health grant T32 AI074550. Rebecca Trout Fryxell is supported by the National Institute of Health T32 AI074550 Training Grant.
References Cited
- Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr AR. The Culex pipiens complex. In: Steiner WWM, Tabachnik WJ, Rai KS, Narang S, editors. Recent developments in the genetics of insect disease vectors. Stipes, Urbana, IL: 1982. pp. 551–572. [Google Scholar]
- Bataille A, Cunningham A, Cedeno V, Cruz M, Eastwood G, Fonseca DM, Causton CE, Azuero R, Loayza J, Martinez J. D. Cruz, Goodman SJ. Evidence for regular ongoing introductions of mosquito disease vectors into the Galapagos Islands. Proc. of Biol. Sci. 2009;276:3769–3775. doi: 10.1098/rspb.2009.0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Cheikh R, Berticat C, Berthomieu A, Pasteur N, Cheikh H. Ben, Weill M. Genes conferring re-sistance to organophosphorus insecticides in Culex pipiens (Diptera: Culicidae) from Tunisia. J. Med. Entomol. 2009;46:523–530. doi: 10.1603/033.046.0317. [DOI] [PubMed] [Google Scholar]
- Bohart RM, Washino RK. Mosquitoes of California. University of California Division of Agricultural Sciences; Berkeley, CA: 1978. [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Byrne K, Nichols RA. Culex pipiens in London underground tunnels: differentiation between surface and subterranean populations. Heredity. 1998;82:7–15. doi: 10.1038/sj.hdy.6884120. [DOI] [PubMed] [Google Scholar]
- Chevillon C, Rivet Y, Raymond M, Rousset F, Smouse PE, Pasteur N. Migration/selection balance and ecotypic differentiation in the mosquito Culex pipiens. Mol. Ecol. 1998;7:197–208. [Google Scholar]
- Cornel AJ, McAbee RD, Rasgon J, Stanich MA, Scott TW, Coetzee M. Differences in extent of genetic introgression between sympatric Culex pipiens and Culex quinquefasciatus (Diptera: Culicidae) in California and South Africa. J. Med. Entomol. 2003;40:36–51. doi: 10.1603/0022-2585-40.1.36. [DOI] [PubMed] [Google Scholar]
- Dobrotworsky NV. The problem of the Culex pipiens complex in the South Pacific (including Australia) Bull. World Health Organ. 1967;37:251–255. [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. Geneious v5.1. 2010 http://www.geneious.com.
- Edillo FE, Tripet F, McAbee RD, Foppa IM, Lanzaro GC, Cornel AJ, Spielman AA. A set of broadly applicable microsatellite markers for analyzing the structure of Culex pipiens s.l. (Diptera: Culicidae) populations. J. Med. Entomol. 2007;44:145–149. doi: 10.1603/0022-2585(2007)44[145:asobam]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Fonseca DM, Keyghobadi N, Malcolm CA, Mehmet C, Schaffner F, Motoyoshi M, Fleischer RC, Wilkerson RC. Emerging vectors in the Culex pipiens complex. Science. 2004;303:1535–1538. doi: 10.1126/science.1094247. [DOI] [PubMed] [Google Scholar]
- Furey TS, Haussler D, D. Integration of the cytogenetic map with the draft human genome sequence. Hum. Mol. Genet. 2003;12:1037–104. doi: 10.1093/hmg/ddg113. [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 2001;159:907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard LB, Roth AE, Reisen WK, Scott TW. Vertical transmission of West Nile virus by three California Culex (Diptera: Culicidae) J. Med. Entomol. 2003;40:743–746. doi: 10.1603/0022-2585-40.6.743. [DOI] [PubMed] [Google Scholar]
- Gomes B, Sousa CA, Novo MT, Freitas FB, Alves R, Corte-Real AR, Salgueiro P, Donnelly MJ, Almeida APG, Pinto J. Asymmetric introgression between sympatric molestus and pipiens forms of Culex pipiens (Diptera: Culicidae) in the Comporta region, Portugal. BMC Evolut. Biol. 2009;9:262. doi: 10.1186/1471-2148-9-262. doi:10.1186/147-2148-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan AU, Suguri S, Ahmed SM, Fujimoto C, Harada M, Rahman SM, Zaman RU, Kakehi Y. Molecular phylogeography of Culex quinquefasciatus mosquitoes in central Bangladesh. Acta Trop. 2009;112:106–114. doi: 10.1016/j.actatropica.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Hemingway J, Ranson H. Insecticide resistance in insect vectors of human disease. Annu. Rev. Entomol. 2000;45:371–391. doi: 10.1146/annurev.ento.45.1.371. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramania GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Huang S, Molaei G, Andreadis TG, G. T. Genetic insights into population structure of Culex pipiens (Diptera:Culicidae) in Northeastern Unites States by using microsatellites analysis. Am. J. Trop. Med. Hyg. 2008;79:518–527. [PubMed] [Google Scholar]
- Huang S, Molaei G, Andreadis TG. Reexamination of Culex pipiens hybridization zone in the Eastern United States by ribosomal DNA-based single nucleotide polymorphism markers. Am. J. Trop. Med. Hyg. 2011;85:434–441. doi: 10.4269/ajtmh.2011.10-0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Brunton CF, Smith NG. Small introns tend to occur in GC-rich regions in some but not all vertebrates. Trends Genet. 1999;15:437–439. doi: 10.1016/s0168-9525(99)01832-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T, Wada K. Evident diversity of codon usage patterns of human genes with respect to chromosome banding patterns and chromosome numbers; relation between nucleotide sequence data and cytogenetic data. Nucleic Acids Res. 1991;19:4333–4339. doi: 10.1093/nar/19.16.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupp PG. Culex (Culex) pipiens pipiens Linnaeus and Culex (Culex) pipiens quinquefasciatus Say in South Africa: morphological and reproductive evidence in favour of their status as two species. Mosq. Syst. 1978;10:461–473. [Google Scholar]
- Keyghobadi N, La Pointe MD, Fleischer RC, Fonseca DM. Fine-scale population genetic structure of a wildlife disease vector: the southern house mosquito on the islands of Hawaii. Mol. Ecol. 2006;15:3919–3930. doi: 10.1111/j.1365-294X.2006.03069.x. [DOI] [PubMed] [Google Scholar]
- Kilpatrick AM, Kramer LD, Jones MJ, Marra PP, Daszak P, Fonseca DM. Genetic influences on mosquito feeding behavior and the emergence of zoonotic pathogens. Am. J. Trop. Med. Hyg. 2007;77:667–671. [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. (doi:10.1093/bioinformatics/btp187) [DOI] [PubMed] [Google Scholar]
- Marais G, Mouchiroud D, Duret L. Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5688–5692. doi: 10.1073/pnas.091427698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAbee RD, Kang K-D, Stanich MA, Christiansen JA, Wheelock CE, Inman AD, Hammock BD, Cornel AJ. Pyrethroid tolerance in Culex pipiens pipiens var molestus from Marin County, California. Pest. Manag. Sci. 2003;60:359–368. doi: 10.1002/ps.799. [DOI] [PubMed] [Google Scholar]
- McAbee RD, Green EN, Holeman J, Christiansen JA, Frye N, Dealey K, Mulligan FS, Brault AC, Cornel AJ. Identification of Culex pipiens complex mosquitoes in a hybrid zone of West Nile virus transmission in Fresno County, California. Am. J. Trop. Med. Hyg. 2008;78:303–310. [PubMed] [Google Scholar]
- Miles SJ, Paterson HE. Protein variation and systematics in the Culex pipiens group of species. Mosq. Syst. 1979;11:187–202. [Google Scholar]
- Morlais I, Severson DW. Intraspecific DNA variation in nuclear genes of the mosquito Aedes aegypti. Insect Mol Biol. 2003;12:631–639. doi: 10.1046/j.1365-2583.2003.00449.x. [DOI] [PubMed] [Google Scholar]
- Morlais I, Ponçon N, Simard F, Cohuet A, Fontenille D. Intraspecific nucleotide variation in Anopheles gambiae: new insights into the biology of malaria vectors. Am. J. Trop. Med. Hyg. 2004;71:795–802. [PubMed] [Google Scholar]
- Pozzoli U, Menozzi G, Fumagalli M, Cereda M, Comi GP, Cagliani R, Bresolin N, Sironi M. Both selective and neutral processes drive GC content evolution in the human genome. BMC Evol. Biol. 2008;8:99. doi: 10.1186/1471-2148-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken CBEM, de Vries A, Buijs J, Braks MAH, den Hartog W, Scholte EJ. First evidence for presence of Culex pipiens biotype molestus in The Netherlands, and of hybrid biotype pipiens and molestus in northern Europe. J. Vector Ecol. 2010;35:210–212. doi: 10.1111/j.1948-7134.2010.00050.x. [DOI] [PubMed] [Google Scholar]
- Rooker S, Guillemaud T, Bergé J, Pasteur N, Raymond M. Coamplification of esterase A and B genes as a single unit in Culex pipiens mosquitoes. Heredity. 1996;77(Pt 5):555–561. [PubMed] [Google Scholar]
- Spielman A. Structure and seasonality of Nearctic Culex pipiens populations. Ann. N.Y. Acad. Sci. 2001;951:220–234. doi: 10.1111/j.1749-6632.2001.tb02699.x. [DOI] [PubMed] [Google Scholar]
- Sudia WD, Chamberlain RW. Battery-operated light trap, an improved model. Mosq. News. 1962;22:126–129. [PubMed] [Google Scholar]
- Tabachnick WJ, Powell JR. Genetic analysis of Culex pipiens populations in the central valley of California. Ann. Entomol. Soc. Am. 1983;76:715–720. [Google Scholar]
- Takano-Shimizu T. Local changes in GC/AT substitution biases and in crossover frequencies on Drosophila chromosomes. Mol. Biol. Evol. 2001;18:606–619. doi: 10.1093/oxfordjournals.molbev.a003841. [DOI] [PubMed] [Google Scholar]
- Urbanelli S, Villani F, Bullini L. Electrophoretic studies on Culex quinquefasciatus Say from Africa: genetic variability and divergence from Culex pipiens L. (Diptera, Culicidae) Bull. Entomol. Res. 1985;75:291–304. [Google Scholar]
- Urbanelli S, Silvestrini F, Sabatinelli G, Raveloarifera F, Petrarca V, Bullini L. Characterization of the Culex pipiens complex (Diptera: Culicidae) in Madagascar. J. Med. Entomol. 1995;32:778–786. doi: 10.1093/jmedent/32.6.778. [DOI] [PubMed] [Google Scholar]
- Urbanelli S, Silvestrini F, Reisen WK, De Vito E, Bullini L. California hybrid zone between Culex pipiens and Cx. p. quinquefasciatus revisited (Diptera: Culicidae) J. Med. Entomol. 1997;34:116–127. doi: 10.1093/jmedent/34.2.116. [DOI] [PubMed] [Google Scholar]
- Weitzel T, Collado A, Jost A, Pietsch K, Storch V, Becker N. Genetic differentiation of the populations with the Culex pipiens complex and phylogeny of related species. J. Am. Mosq. Control Assoc. 2009;25:6–17. doi: 10.2987/08-5699.1. [DOI] [PubMed] [Google Scholar]
- Williams EJ, Hurst LD. The proteins of linked genes evolve at similar rates. Nature. 2000;407:900–903. doi: 10.1038/35038066. [DOI] [PubMed] [Google Scholar]
- Wuitschick JD, Karrer KM. Analysis of genomic G + C content, codon usage, initiator codon context and translation termination sites in Tetrahymena thermophila. J. Eukaryot. Microbiol. 1999;46:239–247. doi: 10.1111/j.1550-7408.1999.tb05120.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.