Abstract
Protein methyltransferases (PMTs) orchestrate epigenetic modifications through post-translational methylation of various protein substrates including histones. Since dysregulation of this process is widely implicated in many cancers, it is of pertinent interest to screen inhibitors of PMTs, as they offer novel target-based opportunities to discover small molecules with potential chemotherapeutic use. We have thus developed an enzymatic screening strategy, which can be adapted to scintillation proximity imaging assay (SPIA) format, to identify these inhibitors. We took advantage of S-adenosyl-L-[3H-methyl]-methionine availability and monitored the enzymatically catalyzed [3H]-methyl addition on lysine residues of biotinylated peptide substrates. The radiolabeled peptides were subsequently captured by streptavidin coated SPA imaging PS beads. We applied this strategy to four PMTs: SET7/9, SET8, SETD2, and EuHMTase1, and optimized assay conditions to achieve Z′ values ranging from 0.48 to 0.91. The robust performance of this SPIA for the four PMTs was validated in a pilot screen of approximately 7,000 compounds. We identified 80 cumulative hits across the four targets. NF279, a suramin analogue found to specifically inhibit SET7/9 and SETD2 with IC50 values of 1.9 and 1.1 μM, respectively. Another identified compound, Merbromin, a topical antiseptic, was classified as a pan-active inhibitor of the four PMTs. These findings demonstrate that our proposed SPIA strategy is generic for multiple PMTs and can be successfully implemented to identify novel and specific inhibitors of PMTs. The specific PMT inhibitors may constitute a new class of anti-proliferative agents for potential therapeutic use.
Keywords: protein methyl transferases, drug discovery, inhibit or, SET7/9, SET8, SETD2, EuHMTase1, SPA technology, red shifted imaging beads
INTRODUCTION
The human genome encodes more than 60 PMTs including 9 known protein arginine methyltransferases (PRMTs) and at least 50 protein lysine methyltransferases (PKMTs). The common feature of these enzymes is their ability to transfer a methyl group from the S-adenosyl-L-methionine (SAM) cofactor to lysine or arginine side chains of protein substrates.1 Unlike other histone modifying enzymes such as histone acetyltransferases, PMTs display high substrate specificity, consistent with their roles on target-specific posttranslational regulation.2 Protein lysine methylation has recently caught significant attention because of its involvement in diverse biological processes.2 This methylation can be both an inhibitory mark (e.g. H3K4me3, H3K36me3) and an activating mark (e.g. H3K27me3, H4K20me3) of gene transcription.3 Furthermore, different methylation states on the same amino acid residue have been shown to correlate with distinct genomic location and functions.2 The combination of the modifications on histone tails dictates their interaction with various effector proteins, forming the basis of the “histone code”hypothesis.2
In the course to understanding the biology of PMTs, evidence has also been accumulated to reveal their aberrant roles in the context of diseases including cancer. Target methylation that results in destabilizing or down-regulating tumor suppressors, has been reported (e.g. SMYD2, SUV39H1, PRMT5, EuHMTase1 and EZH2).4 Further evidence correlating PMTs with cancer was obtained from primary tumor tissue samples4 in which overexpression of certain PMTs has frequently been observed (e.g. EuHMTase1, PRDM14, SMYD3, NSD2/3 and SUV39H2). These findings collectively argue the important roles of PMTs in cancer biology and claim the value of discovering small molecule inhibitors of PMTs for potential cancer therapy intervention.
A challenge in studying PMTs as recombinant enzymes is their intrinsic low catalytic turnover rates in vitro. Therefore, it is more desirable to measure the methylation reaction by quantifying appearance of end products rather than depletion of starting substrates. Methylated products and the by-product S-adenosyl-L-homocysteine (SAH) could be quantified either directly via autoradiography5, mass spectrometry6–8, or using specific antibodies; or indirectly after processing them into various derivatives for enzyme-coupled colorimetric assays.9–13
Given the current need of novel and target-specific PMTs for cancer therapy, generating simple PMT-activity assays to screen such small-molecule entities has become increasingly important. For example, Bedford and his colleagues formulated an antibody-based ALISA PMT-activity assay and applied it to identify PRMT inhibitors (e.g. AMI1, 5, 6, 9, 18) from a 9K compound library.14 The Imhof lab used a filter-paper-based radiometric assay to screen a pooled mixture of 2,976 compounds and identified Chaetocin15 as an Su(VAR)3–9 inhibitor. Though the applicability of these assays has been proven by identifying inhibitors from small-to-medium-size libraries, they are not sufficient for handling current high throughput screening (HTS) libraries of >100K entries.
The first HTS PMT-activity assay was reported by the Kubicek lab. In this assay, a biotinylated H3 peptide substrate was dimethylated by EuHMTase1 and then immobilized to neuroavidin-coated 384-well plates. The methylated peptide product was then quantified with specific antibodies (primary rabbit α-H3Kme2, secondary europium labeled goat α-rabbit).16 Hits were identified by loss of signal from the inhibited reactions. With this method, the Kubicek lab was able to screen a 125K-compound library and identify seven EuHMTase2 inhibitorsincluding BIX-01294.16
In order to accommodate HTS, PMT-activity assays in a mix-and-measure format are more desirable. Technologies such as AlphaScreen (PerkinElmer), AlphaLISA (PerkinElmer), LANCE Ultra (PerkinElmer) and LanthaScreen (Invitrogen) have been explored as potential HTS platforms for PMTs because their mix-and-measure readiness.2,17,18 These assays adapt a similar principle of action by pairing PMT substrates and anti-methyllysine antibodies with donor and acceptor dyes.3,19–21 Assays for SET7/9-mediated H3K4 monomethylation (AlphaLISA) and EuHMTase2-mediated H3K9 dimethylation (LanthaScreen TR-FRET) were evaluated for HTS compatibility.22,23 Z′ values (> 0.7) of the two assays demonstrated their amenability for HTS. Klink et. al. also measured the Z′ of a competitive fluorescence polarization immunoassay for SAH-derivatized AMP.24 Although Klink’s assay only has a modest Z′ of 0.59, the assay is expected to enable assessment of multiple PMTs by quantifying their commonly-shared reaction byproduct SAH.
Another consideration for developing reliable HTS assays is to control the rate of false-positives especially for emerging detection technologies such as AlphaScreen. Ferguson and coworkers developed an AlphaScreen HTS assay to screen inhibitors of SMYD2 and reported that AlphaScreen detection technology was amenable to a high rate of false-positives.25 A large number of false positives have to be triaged by an efficient secondary assay. They described a radiometric Scintillation Proximity Assay (SPA) approach as a robust secondary assay to validate the hits of SMYD2 after the AlphaScreen-based primary HTS. To measure the enzymatic activities of SET7/9, SET8, SETD2, or EuHMTase1 in an reliable HTS format, we opted for the Scintillation Proximity Imaging Assay (SPIA) technology as our primary HTS assay as it is homogeneous in nature, easy to miniaturize into 384-well microtiter plates, and has been used successfully by our HTS group for a CDC7 kinase26 and DNA ligase IV27 screening projects. The recent introduction of the PS imaging beads results in “red-shifted” assay readouts that appear to be relatively insensitive to colored compounds, especially those absorbing in the yellow, red, and blue ranges of the light spectrum.28 In terms of reagents, the SPA HTS approach is also more generic in comparison with the antibody-based HTS assays because the later requires the use of high-quality antibodies for each individual PMT assay. Development of a robust, reliable HTS platform is required to address the aforementioned gaps in the known methods.
In this paper, we report on the successful adaptation of an SPIA in a robust HTS format for PMTs, which allows a direct measure of the methylation product. Here scintillation signal arises from the proximity between streptavidin SPA imaging PS beads and [3H-Me]-labeled biotinylated peptide (expected PMT product using [3H-Me]-SAM as cofactor). The suppression of the scintillation signal will lead to the identification of PMT inhibitors. We applied our strategy to four PMTs: SET7/9, SET8, SETD2, and EuHMTase1, and optimized assay conditions achieving Z′ values ranging from 0.48 to 0.91. We present the performance of this SPIA strategy, first tested in a pilot screen of around 7,000 compounds. We were able to identify 80 cumulative hits across the four targets.
MATERIALS AND METHODS
Reagents
S-adenosyl-L-[methyl-3H]methionine (10 Ci/mmol in 9:1 sulfuric acid/ethanol) and streptavidin-conjugated polystyrene SPA imaging beads (binding capacity 160 pmol/mg) were purchased from PerkinElmer. Streptavidin-conjugated Sepharose High Performance beads (binding capacity > 300 nmol/mL) were obtained from GE Healthcare. P-81 phosphocellulose filter paper is a product of Whatman. Other reagents were obtained from available commercial sources unless otherwise mentioned. C-terminus-GGK-biotinylated H3 peptide (1–21-aa, ARTKQTARKSTGGKAPRKQLAGGK-Biotin; 20–50-aa, ATKAARKSAPATGGVKKPHRYRPGTVALREGGK-Biotin) and C-terminus-GGK-biotinylated H4 peptide (10–30 aa, LGKGGAKRHRKVLRDNIQGITGGK-Biotin) were synthesized via standard Fmoc-protected solid-phase peptide synthesis and purified by HPLC with 0~40% acetonitrile gradient in 0.1% trifluoroacetic acid/H2O (the Proteomics Resource Center of the Rockefeller University). Integrity of purified peptides was confirmed by mass spectrometry. 384-well microtiter plates (Cat# 3570) used for HTS were purchased from Corning.
Expression and Purification of SET7/9, SET8, SETD2, and EuHTMase1
Plasmids containing the N-terminal His6-tagged methyltransferase domain of human EuHMTase1 (residues 951–1235) and SETD2 (residues 1347–1711) were generously donated by the Min laboratory at the University of Toronto.29 EuHMTase1 (residues 951–1235) was transformed into the E. coli Rosseta -2(DE3) strain (Novagen), and expression of the SET domain was induced by 0.5 mM IPTG in the presence of 25 μM ZnSO4 at 17°C overnight. Active protein fractions were obtained after a purification gradient of 25 – 400 mM imidazole on Ni- sepharose™ 6 Fast Flow resin (GE Healthcare). The N-terminal His6-tagged plasmids of full-length human SET7/9 (hSET7/9) and human SET8 (residues 191–352) were a gift from the Trievel laboratory at the University of Michigan. The procedures for SET7/9, SET8 (residues 191–352), and SETD2 (residues 1347–1711) expression and Ni-NTA purification are similar to those for EuHMTase1, but without 25 μM ZnSO4,30 and followed by a size exclusion column (GE Healthcare). All enzymes were flash-frozen in liquid nitrogen and stored at −80°C before their use. The four PMTs are stable under these conditions for at least 6 months.
Methylation Reaction
The 20 μL methylation reaction was carried out at ambient temperature using two mixtures: A. 10 μL of enzyme mixture in assay buffer, 50 mM Hepes (pH 8.0), 0.005% Tween-20, 5 μg/mL BSA, and 1 mM TCEP; B. 10 μL of a [3H-Me]-SAM cofactor and biotinylated peptide substrate mixture in this buffer. After A and B were mixed for a designated time period, the resulting product was assayed for PMT-activity via our established assays. Assay development ensued concurrently with establishing three PMT assays for optimization of reaction parameters, as detailed below(a three-step approach for assay optimization).
Filter-paper based Assay
Whatman P-81 filter paper is specific for binding proteins or peptides (e.g. the histone peptide substrates described above), but not SAM.31 PMTs transfer the 3H-methyl group from [3H-Me]-SAM to the peptide substrates and the resultant 3H-methylated, filter-paper-bound peptide is quantified via scintillation counter (“filter-paper assay” hereinafter). Briefly, 6 μL of the methylation reaction was spotted onto Whatman P-81 phosphocellulose filter paper (1.2 × 1.2 cm2) to immobilize the 3H-labeled peptide. After drying in ambient air for 20 min, filter paper was immersed into 20 mL of 50 mM Na2CO3/NaHCO3 buffer (pH 9.2), and washed for 10 minutes, 5 times. The washed paper was transferred to a 20 mL scintillation vial containing 1 mL of distilled water and 10 mL of Ultima Gold scintillation cocktail (PerkinElmer). Radioactivity was quantified by a Beckman LS6000IC liquid scintillation counter.
Sepharose-bead based assay
Biotinylated peptides were methylated with [3H-Me]-SAM cofactor and immobilized onto strepavidin sepharose beads. Beads were then washed to remove unreacted [3H-Me]-SAM. Prior to the assay, streptavidin-conjugated Sepharose High Performance beads were prepared by resuspension in a 15-fold volume of PMT-reaction quenching buffer (100 mM MES, pH 6.5). For each assay, 80 μL of the prepared sepharose beads were incubated with the methylation reaction mixture for 10 min to sequester the [3H-Me]-labeled peptide. This suspension was washed 5 times with 100 μL of the quenching buffer. The sepharose beads were suspended in 0.4 mL and mixed with a 0.6 mL H2O/10 mL Ultima Gold scintillation cocktail. Radioactivity was quantified via the Beckman LS6000IC liquid scintillation counter. This “sepharose-bead assay” was applied to validate assay parameters that were optimized with the filter-paper assay (described above).
SPIA based assay
Streptavidin-conjugated SPA imaging PS beads were used in place of sepharose beads (“SPIA-bead assay” hereinafter). SPIA beads were added to PMT reactions, and the signal was detected by the LEADseeker™ Multimodality Imaging System (GE Healthcare). This assay was also implemented for a final validation of the assay parameters, and was further adapted into a HTS format. Briefly, a volume of 10 μL of the enzyme mixture was added to 384-well microtiter lates containing 2 μL 10% DMSO (v/v). The samples were mixed at 625 rpm for 1 min with an Eppendorf Thermomixer, followed by addition of 10 μL buffer containing PMT substrates and cofactor. After the reaction was completed, 0.2 mg of streptavidin-conjugated SPA imaging beads in 60 μL quenching buffer (100 mM MES, pH 6.5) were added to this reaction, mixed again at 625 rpm for 1 min, and then incubated for 1 h at ambient temperature. The SPIA signal was measured with the LEADseeker™ system after spinning down the beads at 2,000 rpm for 1 min.
Chemical Libraries
The library used in the pilot screen included up to 6,912 chemicals obtained from MicroSource, Prestwick, Tocris, Sigma, and other commercial sources,26,32–34 The MicroSource Library contains 2,000 biologically active and structurally diverse compounds from known drugs, experimental bioactives, and pure natural products; it includes a reference collection of 160 synthetic and natural toxic substances (previously characterized as being inhibitors of DNA/RNA synthesis, protein synthesis, cellular respiration, and membrane integrity), a collection of 80 compounds representing classical and experimental pesticides, herbicides, and endocrine disruptors, and a singular collection of 720 natural products and their derivatives. Rest collection included simple and complex oxygen heterocycles, alkaloids, sequiterpenes, diterpenes, pentercyclic triterpenes, sterols, and many other diverse representatives. The Prestwick Chemical Library is a unique collection of 1,119 high purity chemical compounds, all of which are off patent and selected for structural diversity and broad spectrum activities in neuropsychiatry, cardiology, immunology, anti-inflammatory, and analgesia, with known human safety and bioavailability. Approximately 90% of the library comprises marketed drugs and 10% of bioactive alkaloids or related substances.32–34
Chemical screen of SET7/9, SET8, SETD2, and EuHTMase1 using the developed SPIA assays
2 μL of 100 μM solution of each compound in 10% DMSO (v/v) were transferred to 384-well microtiter plates using the PP-384-M Personal Pipettor (Apricot Designs). For the high control, 2 μL of 10% DMSO (v/v) was added to column 13 and 14 in rows A through H. For the low control, the final concentration of 10 mM of HClO4 was added to column 13 and 14 in rows I through P (here we inactivated PMTs by lowering the pH in each well). To initialize our assay, 10 μL of the enzyme mixture was dispensed using the FlexDrop IV dispenser (PerkinElmer) and incubated for 10 min at ambient temperature. 10 μL of the PMT substrate and cofactor mixture was dispensed using the FlexDrop, followed by incubation at ambient temperature. After the reactions were completed, 60 μL of streptavidin-conjugated SPA imaging beads (0.2 mg) were added using the Multidrop384 (Thermo), followed by incubation for 1h at ambient temperature. Assay plates were centrifuged at 2,000 rpm for 1 min and imaged on the LEADseeker™. Data files from the LEADseeker™ were loaded onto the HTS Core’s Oncology Research Information System (ORIS), a custom-built suite for compound registration, plating, and data management powered by ChemAxon Cheminformatic tools (ChemAxon). Data was analyzed for inhibition, and thepositive hits were exported for further analysis.
Z′ factor
The Z′ factor was used to assess the performance of the optimized SPA based assay as previously described.26 The Z′ factor constitutes a dimensionless parameter that ranges from 1 (infinite separation) to < 0. It is defined as: Z′ = 1 − (3σc+ + 3σc−)/|μc+−μc−| where σc+, σc−, μc+ and μc− are the standard deviations (σ) and averages (μ) of the high (c+) and low (c−) control. Z′ factor between 0.5 and 1 indicates that the assay has an excellent separation from controls; Z′ factor between 0 and 0.5 for a marginal separation; <0 for a poor separation fromcontrols. 26
Dose-Response Study of Primary Positives with Filter-Paper Based Assay
The filter-paper assay (described above) was implemented to assess dose responses of potential PMT inhibitors (triplicate with 0.1 – 400 μM compounds). Briefly, enzyme and positive hits were incubated for 10 min before adding the mixture of [3H-Me]-SAM cofactor and biotinylated peptide substrate. Inhibition was expressed as the percentage between the high control (no inhibition) and low control (no enzyme) as follows: percentage inhibition = [(high control − reading)/(high control − low control)] × 100%. The IC50 values were obtained by fitting the percentages of inhibition versus inhibitor concentration using GraphPad Prism5 software.
RESULTS
Assay Optimization
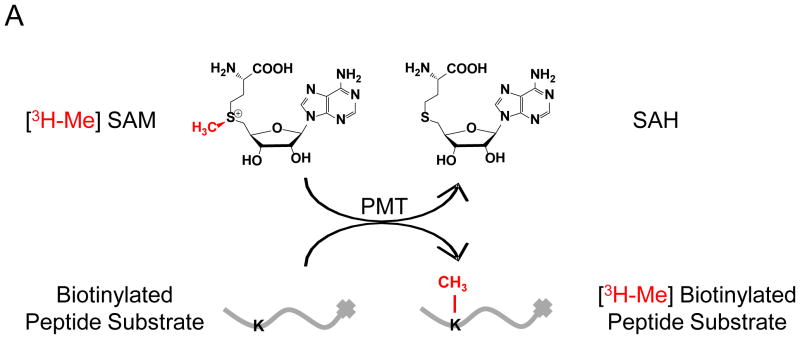
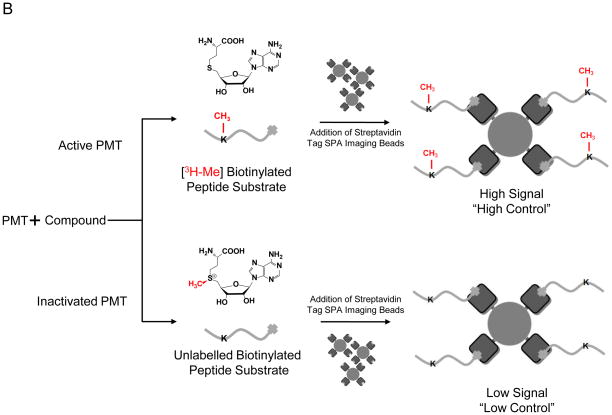
Here we report a direct SPIA-based mix-and-measure HTS assay for PMTs, in which the [3H]-methyl residue of [3H-Me]-SAM (S-adenosyl-L-[3H]-methionine) is enzymatically transferred to biotinylated peptide substrates (Fig 1A). The [3H]-labeled substrates are then captured onto streptavidin-coated SPA imaging PS beads. The resultant proximity between β-particles from the immobilized [3H]-labeled peptide and SPA-bead-coated scintillation fluid leads to fluorescence emission of the scintillation signal, which is suppressed in the presence of PMT inhibitors as schematically depicted in Fig 1B. Assay development began with the optimization of substrates conditions, specifically titration of [3H-Me]-SAM and peptide concentrations together with the amount of SPA imaging PS beads required to capture the biotinylated peptides. Based on parameters of lowering cost and optimizing signal to noise ratio, 0.75 μM (0.15 μCi) [3H-Me]-SAM, 1.5 μM (30 pmol) biotinylated peptide, and 0.2 mg of beads were used (data not shown).
Figure 1. Overview of the SPA-based HTS approach.
(A) The 3H-methyl residue of [3H-Me]-SAM is enzymatically transferred to biotinylated PMT substrates; (B) The methylated reaction can proceed through two pathways: inactivated, in which the compound inhibits the reaction or active, in which the compound leads to no apparent effect. The peptide is then immobilized onto streptavidin-conjugated SPA beads. The proximity between β-particles and bead-coated scintillation fluid generates strong scintillation signal, which is suppressed in the inactive pathway.
We assessed the feasibility of this assay for its adaptation to an HTS format using SET7/9, SET8, SETD2, and EuHMTase1 following our three-step approach (see Materials and Methods). First, we utilized the filter-paper assay to optimize various parameters for each of the PMTs: enzyme concentration, reaction time, DMSO tolerance, and enzyme stability, using biotinylated-H3 (1–21 aa) peptide for SET7/9 and EuHTMase1, biotinylated-H3 (20–50 aa) peptide for SETD2, and biotinylated-H4 (10–30 aa) peptide for SET8. We confirmed results of the filter-paper assay using the sepharose-bead based assay, which resembles SPIA, but is performed with a scintillation counter instead. Finally, sepharose beads were replaced with SPIA beads for the final validation.
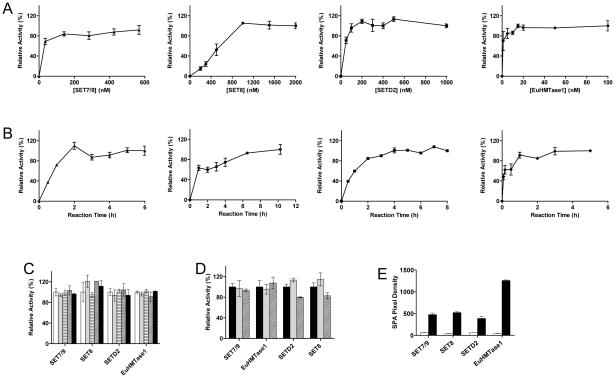
We found that the four PMTs behave differently under our assay conditions, showing maximal conversion at 150 nM for SET7/9, 1.5 μM for SET8, 250 nM for SETD2, and 10 nM for EuHMTase1 (Fig 2A). With these enzyme concentrations, reaction completion was longest for SET8 (8h incubation time), followed by SETD2 (4h), SET7/9 (3h), and EuHMTase1 (1h) (Fig 2B). Using these conditions, we moved ahead to determine enzyme tolerance to DMSO (the common solvent for all compounds). Enzyme activity was shown to tolerate up to 5% DMSO (Fig 2C). Enzymes were shown to be stable in ambient temperature for at least 16 h (Fig 2D).
Figure 2. Optimization of assay parameters.
(A) Enzyme concentration. The assays were carried out for 16 h with the varied concentrations of (▲) SET7/9, (●) SET8, (■) SETD2 and (◆) EuHTMase1.The final concentrations of 150 nM for SET7/9, 1.5 μM for SET8, 250 nM for SETD2, and 10 nM for EuHMTase1 were chosen for HTS; (B) Reaction time. Using these final concentrations, the maximal conversions were reached in 3 h for SET7/9 (▲), 8 h for SET8 (●), 4 h for SETD2 (■), and 1 h for 10 nM EuHMTase1 (◆); (C) DMSO tolerance. The reactions were carried out with 150 nM SET7/9 for 3 h, 1.5 μM SET8 for 8h, 250 nM SETD2 for 4h, and 10 nM EuHMTase1 for 1 h in the presence of a varied amount of DMSO: without (white), 1% (gray), 2% (horizontal lines), 3% (diagonal lines), 5% (black). The four PMTs were tolerant up to 5% DMSO; (D) Enzyme stability. Experiments were performed with 150 nM SET7/9, 1.5 μM SET8, 250 nM SETD2, and 10 nM EuHMTase1. All enzymes were pre-incubated at ambient temperature for 0 (black), 6 (gray), and 16 h (diagonal lines) prior to the assay start. The unchanged reactivity indicated that the four PMTs are stably functional under our assay conditions for at least 16 h; (E) SPA readout under high and low controls. The reactions were carried out under the optimized assay conditions with either active PMTs (black) or HClO4-treated PMTs (white). Upon mixing with 0.2 mg SPA beads, the scintillation signals were recorded with LEADseeker™ Multimodality Imaging System. The ratios of the high-to-low controls of 8 for SET7/9, 3 for SET8, 6 for SETD2, and 30 for EuHTMase1, display an excellent signal-to-noise separation for the present assay format (10 repeats for each data set). Each assay point was performed in triplicate (n=3), and SDs were plotted unless otherwise mentioned.
These optimized assay parameters (reaction time, PMT concentrations, and DMSO tolerance) were further confirmed with the sepharose-bead based assay (see Materials and Methods). Sepharose beads were replaced with SPIA beads for the final validation (0.2 mg SPA beads for 30 pmol of biotinylated peptide, see Materials and Methods). We successfully accomplished the transition from the filter-paper assay to the SPIA assay with high-to-low ratios of 8 for SET7/9, 3 for SET8, 6 for SETD2, and 30 for EuHTMase1 and with < 10% standard deviations for our target (SPIA) assay. This indeed indicated that our SPIA-based approach was suitable for HTS format (Fig 2E).
Chemical Screen for PMT Inhibitors
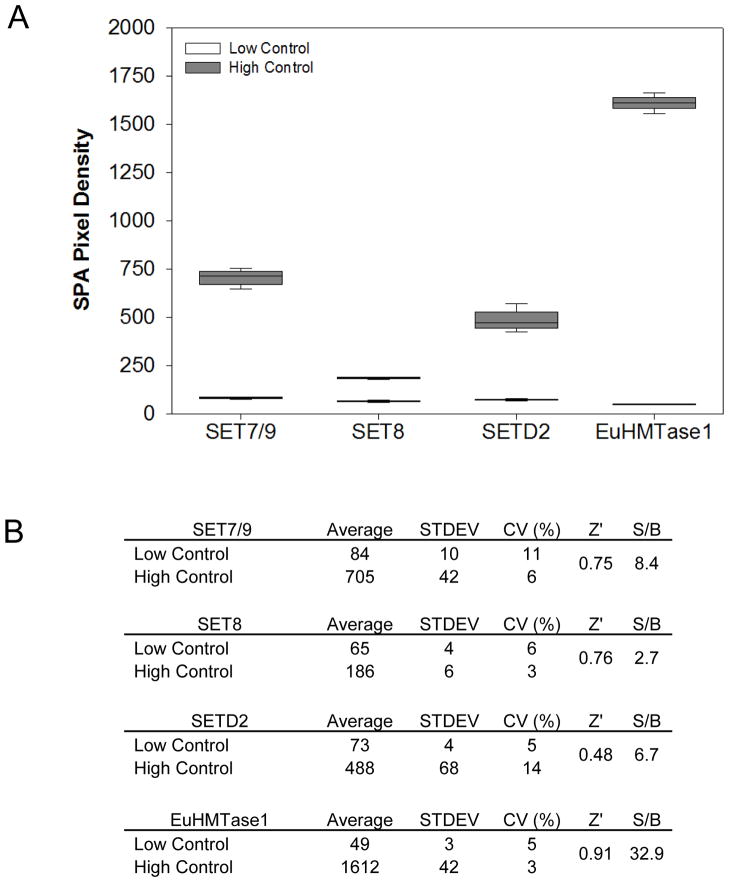
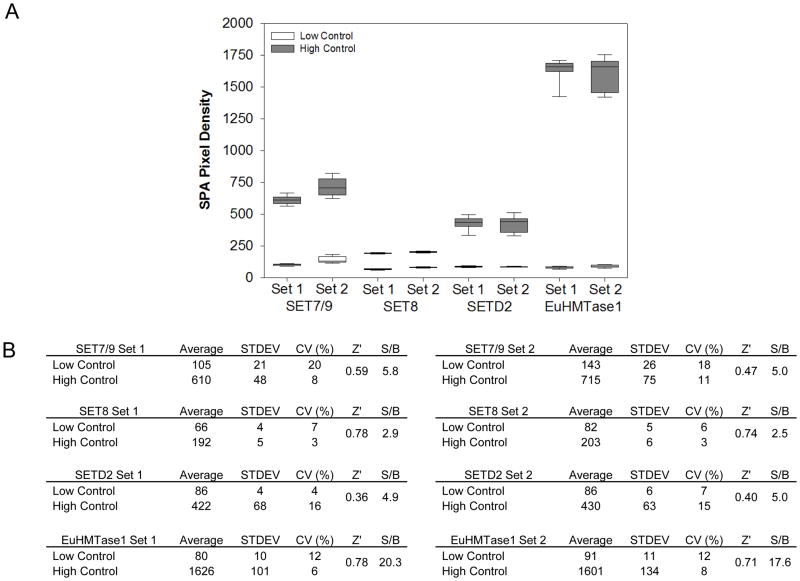
To assess robustness for chemical screening, we performed a control run of three 384- well microtiter plates with 1% DMSO (v/v) for no enzymatic inhibition, or “high control,” and three 384-well microtiter plates with 10 mM of HClO4 for total inhibition, or “low control” (Table 1). Assay evaluation was based on calculating percent coefficients of variation (%CV) and Z′ factors. From the 1,152 data points for each of the controls, CVs for all four tested assays were between 3% and 14%, with the lowest Z′ factor of 0.48. These results indicate adequate robustness and therefore provide enough confidence in moving forward to screening our chemical library (Figs 3A & 3B).
Table 1.
SPIA Assay Protocol for the four PMTs.
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Enzyme Mix | 10 μL | Methyltransferase Mix |
| 2 | Library compounds | 2 μL | 100 μM in 10% DMSO |
| 3 | Low control | 2 μL | 100mM Perchloric Acid |
| 4 | High control | 2 μL | 10% DMSO |
| 5 | Incubation time | 10 min | Ambient Temperature |
| 6 | SAM + Peptide Mix | 10 μL | [3H-Me]-SAM and biotinylated Peptide Mix |
| 7 | Incubation time | 1h to 8h | Ambient Temperature |
| 8 | SPA Bead | 60 μL | SPA bead with Streptavidin Coating |
| 9 | Incubation time | 1h | Ambient Temperature in the Dark |
| 10 | Spin | 1 min | Centrifuge assay plate at 2,000 rpm |
| 11 | SPA Readout | 5 min | LEADSeeker MultiModality Imaging System |
| Step | Notes |
| 1 | Dispensing with the FlexDrop IV. (SET7/9 at 300 nM; SET8 at 3 μM; SETD2 at 500 nM; EuHMTase1 at 20 nM) |
| 2 to 4 | Dispensing on the PP-384-M Personal Pipettor using a custom 384 head. |
| 5 | Plates incubated at room temperature. |
| 6 | Dispensing with the FlexDrop IV. (1.5 μM [3H-Me]-SAM and 3 μM Peptide) |
| 7 | Plates incubated at room temperature. (SET7/9 for 3h; SET8 for 8h; SETD2 for 4h; EuHMTase1 for 1h) |
| 8 | Dispensing with the Multidrop384. |
| 9 | Plates incubated at room temperature. |
| 10 | Plates centrifuged at 2,000 rpm for 1 min. |
| 11 | Radiometric signal quantified with empty epi mirror and empty emission mirror. |
Figure 3. Assay robustness for chemical screening.
(A) Repeated (n=3) 384-well plate high- to-low controls were performed for SET7/9, SET8, SETD2, and EuHTMase1. For the high controls, the wells contained 2 μl of 9:1 H2O/DMSO prior to the addition of PMTs, substrate and cofactor. In comparison, the wells of the low controls contained 2 μl of 100 mM HClO4. Lowering pH inactivated PMTs. (B) Assay for the four enzymes. SET7/9, SET8, SETD2 and EuHMTase1 were evaluated on percent coefficient of variation (%CV) and Z′ factor.
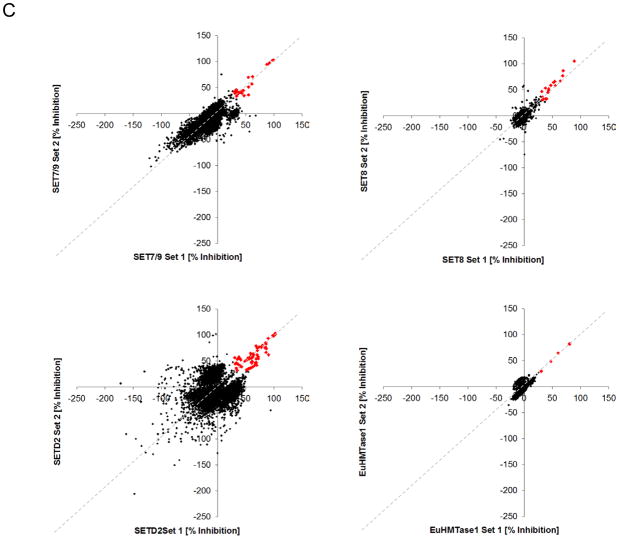
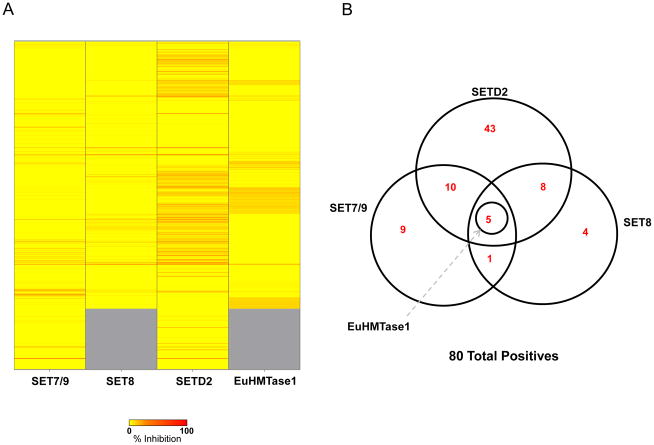
To identify inhibitors of methyltransferases, we screened the four PMTs against our chemical library comprised of compounds gathered from various commercial sources that represent a diverse collection of FDA approved and known bioactive molecules (see Materials and Methods for the compound library). For SET8 and EuHMTase1, the chemical library used for screening contained 5,632 compounds and was tested in duplicate to evaluate assay performance. For SET7/9 and SETD2, the library was expanded to include a new commercial collection, totaling 6,912 compounds, screened in duplicate. “High” and “low” controls served as the quality controls and were included to measure assay robustness. CVs for all assays were low, between 3% and 20%, with the resulting Z′ factors between 0.36 and 0.78 (Figs 4A & 4B). To measure assay reproducibility, data sets were graphed in a correlation plot to show acceptable correlation, as depicted by the pattern slope. The majority of compounds were centrally distributed along the X/Y intercept, indicative of little or no activity (Fig 4C). Using a threshold of 30% inhibition, we identified 18 positives for SET8 and 5 positives for EuHMTase1, with an initial hit rate of 0.32% and 0.09%, respectively, for the 5,632 compound library. Using the same threshold, we identified 25 positives for SET7/9 and 67 positives for SETD2 with initial hit rates of 0.36% and 0.97%, respectively, for the 6,912 compound library. To identify selective inhibitors of tested methyltransferases, we performed an overlap analysis and identified 80 positive hits between the assays of which 69 compounds are unique. Representative hits are shown in Table 2 and positives are comprised of a diverse class of compounds. Selective inhibitors were identified with a majority of compounds hitting SETD2 only. Additionally, approximately 25 compounds hit across multiple enzymes and 5 compounds were active across all four enzymes (Figs 5A & 5B). The 5 compounds included gentian violet and evans blue tetrasodium salt, known non-specific inhibitors of SPIA based assays. Biotin was also identified as a pan-active compound due to its ability to displace the streptavidin labeled SPIA beads. Biotin also functioned as a positive control to further confirm assay reliability.
Figure 4. Scatter plot analysis of the duplicate values of percentage inhibition for each compound in the pilot screening for SET7/9, SET8, SETD2, and EuHMTase1.
(A) Each validation plate contained high and low controls (n=6). The high controls contained 1% DMSO (v/v) and low controls included 10 mM HClO4. (B) The assay for the four enzymes SET7/9, SET8, SETD2, and EuHMTase1 were evaluated by percent coefficient of variation (%CV) and Z′ factor. (C) Libraries of 6,912 compounds for both SET7/9 (upper left) and SETD2 (lower left), and 5,632 for SET8 (upper right), and EuHMTase1 (lower right) were examined (n=2). The scatter plot compares our two data sets in order to examine the correlation between percent inhibitions for each compound. The upper-right region represents potential inhibitors (red) and the compounds that cloud the central region (black) indicate that the majority of the examined compounds are inert against the tested PMTs.
Table 2.
Inhibitors Identified from Screening
| Compound Name | Supplier | Supplier ID | SET7/9 [% IHBN] | SET8 [% IHBN] | SETD2 [% IHBN] | EuHMTase1 [% IHBN] |
|---|---|---|---|---|---|---|
| 2′,2′-Bisepigallocatechin Digallate | MicroSource Discovery Systems | 201507 | 45 | 21 | 66 | −3 |
| 2′,2′-Bisepigallocatechin Monogallate | MicroSource Discovery Systems | 201506 | 53 | 17 | 80 | −3 |
| 6-Hydroxy-DL-DOPA | Sigma-Aldrich | H 2380 | −3 | -- | 62 | -- |
| Acetriazoic Acid | MicroSource Discovery Systems | 1504142 | −9 | −4 | 56 | 12 |
| Acriflavinium Hydrochloride | MicroSource Discovery Systems | 1500618 | −3 | 29 | 42 | −9 |
| Alexidine Hydrochloride | MicroSource Discovery Systems | 1503074 | −110 | 34 | −96 | −22 |
| Anthralin | Prestwick Chemicals | 43 | 8 | 19 | 40 | 10 |
| Aurintricarboxylic Acid | Sigma-Aldrich | A 1895 | 100 | -- | 76 | -- |
| Biotin | Prestwick Chemicals | 418 | 91 | 47 | 74 | 81 |
| Biotin | MicroSource Discovery Systems | 1503009 | 95 | 60 | 81 | 81 |
| Cadmium Acetate | MicroSource Discovery Systems | 330006 | 1 | 65 | 101 | −6 |
| CCG-2046 | Sigma-Aldrich | C 3243 | −7 | -- | 55 | -- |
| Chaulmoogric Acid | MicroSource Discovery Systems | 310016 | −48 | −10 | 48 | −11 |
| Chicago Sky Blue 6B | Tocris Bioscience | 846 | 27 | 24 | 57 | 11 |
| Chicago Sky Blue 6B | Prestwick Chemicals | 425 | 28 | 23 | 48 | 23 |
| Cisplatin | Sigma-Aldrich | P 4394 | −3 | -- | 63 | -- |
| Cisplatin | Tocris Bioscience | 2251 | 13 | 33 | 72 | 7 |
| Cisplatin | MicroSource Discovery Systems | 1502107 | 22 | 39 | 85 | 5 |
| Cyproheptadine Hydrochloride | Prestwick Chemicals | 103 | 36 | 4 | 9 | 4 |
| Cyproheptadine Hydrochloride | Tocris Bioscience | 996 | 42 | −1 | −32 | −2 |
| Dihydrogambogic Acid | MicroSource Discovery Systems | 201524 | 11 | 7 | 58 | −4 |
| Ellagic Acid | MicroSource Discovery Systems | 1502245 | 20 | 2 | 35 | 6 |
| Epigallocatechin 3,5-Digallate | MicroSource Discovery Systems | 201513 | 28 | 6 | 56 | −4 |
| Epigallocatechin-3-Monogallate | MicroSource Discovery Systems | 210239 | 8 | 2 | 42 | 0 |
| Epitheaflavin Monogallate | MicroSource Discovery Systems | 241155 | 17 | 8 | 46 | −3 |
| Evans Blue Tetrasodium Salt | Tocris Bioscience | 845 | 101 | 78 | 103 | 48 |
| Gambogic Acid | MicroSource Discovery Systems | 200007 | −5 | 18 | 80 | −8 |
| Gentian Violet | MicroSource Discovery Systems | 1500315 | 59 | 97 | 76 | 62 |
| Haematoporphyrin Dihydrochloride | MicroSource Discovery Systems | 700024 | −1 | 0 | 54 | −3 |
| Hematein | MicroSource Discovery Systems | 1502253 | 28 | 8 | 58 | 11 |
| Irigenol | MicroSource Discovery Systems | 201182 | 34 | 9 | 61 | 0 |
| Juglone | MicroSource Discovery Systems | 300038 | 13 | 19 | 63 | −1 |
| L-690,330 | Tocris Bioscience | 681 | 31 | 1 | 54 | −4 |
| Me-3,4-Dephostatin | Sigma-Aldrich | M 9440 | −5 | -- | 40 | -- |
| Merbromin | Prestwick Chemicals | 787 | −16 | 15 | 38 | 7 |
| Merbromin | MicroSource Discovery Systems | 1500637 | 27 | 57 | 98 | 7 |
| Methylene Blue | Prestwick Chemicals | 462 | 35 | 43 | 43 | 30 |
| Mitoxanthrone Hydrochloride | Sigma-Aldrich | M 6545 | −47 | -- | 54 | -- |
| Mitoxanthrone Hydrochloride | Prestwick Chemicals | 385 | −47 | 2 | 44 | 15 |
| Mitoxanthrone Hydrochloride | MicroSource Discovery Systems | 1503278 | −9 | 9 | 55 | 10 |
| NF 023 | Sigma-Aldrich | N 8652 | 45 | -- | 26 | -- |
| NF 279 | Tocris Bioscience | 1199 | 62 | −7 | 59 | −9 |
| NF 449 | Tocris Bioscience | 1391 | 67 | 5 | 17 | −10 |
| N-Formylmethionylphenylalanine | MicroSource Discovery Systems | 1502175 | −12 | 31 | −7 | 2 |
| N-Formylmethionylalanine | MicroSource Discovery Systems | 1502172 | 7 | 33 | −13 | 0 |
| Norstictic Acid | MicroSource Discovery Systems | 201716 | 0 | −14 | 50 | −4 |
| Oxaliplatin | Tocris Bioscience | 2623 | −3 | 29 | 49 | 2 |
| Pimethixene Maleate | MicroSource Discovery Systems | 1501114 | 41 | −11 | −22 | −9 |
| Pimethixene Maleate | Prestwick Chemicals | 294 | 41 | 4 | 1 | 2 |
| Primaquine Diphosphate | Prestwick Chemicals | 476 | −19 | 28 | 92 | 7 |
| Propidium Iodide | Prestwick Chemicals | 792 | −34 | −5 | 46 | 6 |
| Protoporphyrin IX | Sigma-Aldrich | P 8293 | 34 | -- | 6 | -- |
| PSB 069 | Tocris Bioscience | 2573 | 35 | 45 | 33 | 26 |
| Purpurogallin | MicroSource Discovery Systems | 210505 | 27 | 7 | 45 | 3 |
| Pyrithione Zinc | MicroSource Discovery Systems | 1500260 | 13 | 26 | 53 | −7 |
| Pyrogallin | MicroSource Discovery Systems | 210515 | 40 | 9 | 57 | −5 |
| Reactive Blue 2 | Sigma-Aldrich | R 115 | 93 | -- | 70 | -- |
| Ruthenium Red | Sigma-Aldrich | R 2751 | -88 | -- | 75 | -- |
| Sennoside A | MicroSource Discovery Systems | 1504078 | 36 | 23 | 60 | 16 |
| Suramin Hexasodium Salt | Tocris Bioscience | 1472 | 39 | 1 | 29 | −6 |
| Suramin Hexasodium Salt | Sigma-Aldrich | S 2671 | 41 | -- | 17 | -- |
| Tannic Acid | MicroSource Discovery Systems | 1504105 | 38 | 35 | 40 | 12 |
| Theaflavanin | MicroSource Discovery Systems | 211709 | 16 | 3 | 65 | 5 |
| Theaflavin Digallate | MicroSource Discovery Systems | 201515 | 38 | 26 | 55 | 0 |
| Tin Protoporphyrin IX Dichloride | Tocris Bioscience | 747 | 0 | 4 | 41 | −2 |
| Zinc Protoporphyrin IX | Tocris Bioscience | 746 | 39 | 26 | 80 | 9 |
Figure 5. Identification of inhibitors (A) Heat map representation of inhibition of our chemical library against SET7/9, SET8, SETD2, and EuHMTase1.
The scale ranges from yellow (no inhibition) to red (100% inhibition). (B) Overlap analysis of SET7/9, SET8, SETD2, and EuHMTase1. With a threshold of 30% inhibition, we identified 80 positives between the four assays: 5 positives for SET7/9, 18 positives for SET8, 67 positives for SETD2, and 5 positives for EuHMTase1. 24 compounds hit across multiple enzymes and 5 of them were active across all four enzymes, including biotin (twice) and SPA-quenching dyes (Gentian violet, Evans blue, and methylene blue). We discerned potential specific inhibitors through pilot screening: 9 compounds for SET7/9, 4 for SET8 and 43 for SETD2.
Hit confirmationwith a secondary assay
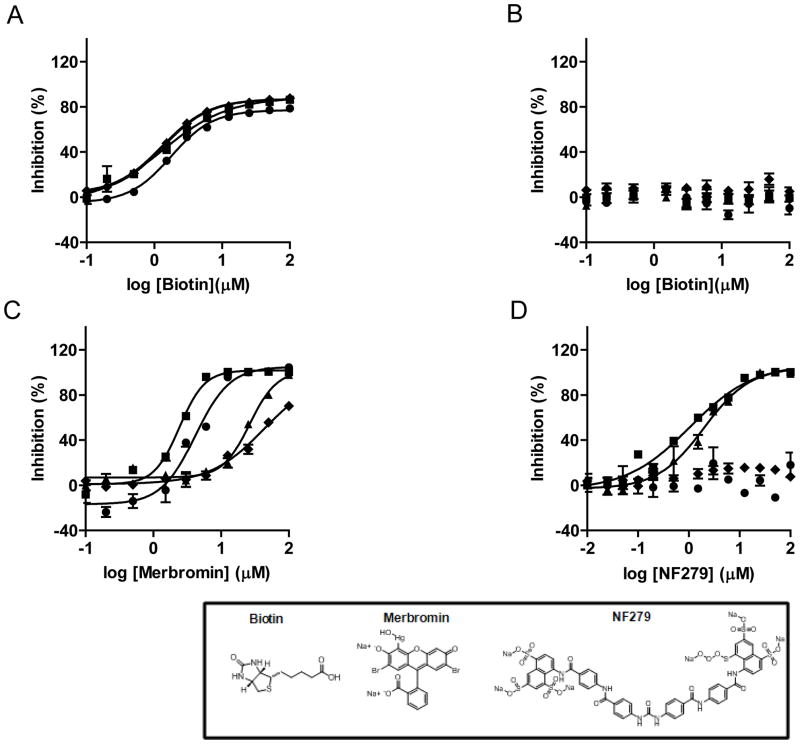
Several compounds may lead to false positives in SPIA format via a nonspecific quenching effect. Independent secondary assays are thus necessary to confirm positive hits after primary HTS. Dose-response curves of three compounds: biotin, merbromin, and NF-279 were further examined with the filter-paper assay (see Materials and Methods), and biotin was also reanalyzed with the SPIA-bead assay. Biotin acts as an artifact that affects the SPA readout by its ability to displace the biotinylated peptide from the streptavidin coated SPA beads, leading to apparent inhibition of SET7/9, SET8, SETD2 and EuHMTase1 by 60–90% at 10 μM. SET7/9, SET8, and SETD2 were inhibited by merbromin at 10 μM (35–98%), and NF-279 showed to be selective towards SET7/9 and SETD2 with 83% and 59% inhibition, respectively, at 10 μM concentration. In our SPIA assay, we observed IC50 values of 1.30 μM, 1.76 μM, 1.33 μM, and 1.33 μM for biotin against SET7/9, SET8, SETD2 and EuHMTase1, respectively (Fig 6A). In the filter-paper assay, no inhibition was observed for up to 100 μM biotin (Fig 6B). Moreover, we were able to confirm inhibition by merbromin within the range observed in the primary HTS with IC50 values of 28 μM for SET7/9, 4.2 μM for SET8, 2.4 μM for SETD2, and 42.9 μM for EuHMtase1 (Fig 6C). Finally, IC50 values of 1.9 μM for SET7/9 and 1.1 μM for SETD2 were obtained for NF-279 (Fig 6D). Such consistency unambiguously argues for the combined capability of the current screen and the secondary confirmation via filter-paper assay to identify PMT inhibitors.
Figure 6. Dose-response curves against SET7/9 (▲), SET8 (●), SETD2 (■), and EuHMTase1 (◆).
(A) The IC50 values for Biotin in the SPIA-bead assay were 1.30 ± 0.04 μM for SET7/9, 1.7 ± 0.1 μM for SET8, 1.3 ± 0.2 μM for SETD2, and 1.33 ± 0.03 μM for EuHMTase1 (B) Using the filter-paper assay to determine IC50 values for Biotin no effect observed, as expected; (C) Merbromin showed IC50 values of 28 ± 1 μM for SET7/9, 4.2 ± 0.4 μM for SET8, 2.4 ± 0.1 μM for SETD2, and 43 ± 15 μM for EuHMTase1 using the filter-paper assay; (D) NF-279, the specific inhibitor against SET7/9 and SETD2, was also tested using the filter-paper assay. It showed IC50 values of 1.9 ± 0.2 μM for SET7/9 and 1.1 ± 0.2 μM for SETD2. Each assay point was performed in triplicate (n=3), and SD is plotted.
DISCUSSION
Continuous efforts have been invested in developing methods to identify inhibitors of protein methyltransferases. Despite the putative significance of these endeavors, few PMT-specific inhibitors have been reported. Most established PMT-activity assays rely on multiple coupling enzymes to process the methylation byproduct SAH into a chromo/fluorogenic derivative.9–13,35,36 Alternatively, the products of PMT reactions can be traced with mass spectrometry or antibodies. Our laboratory has also developed an enzyme-coupled ultrasensitive luciferase assay for PMTs with luminescence readout.12 However, these methods, in particular the enzyme-coupled PMT-activity assays, are prone to false positives and thus generally not amenable to HTS automation.
Here, we describe a robust and direct assay that does not requireseparation step sand uses commercially available streptavidin-coated SPA imaging PS beads and [3H-Me]-SAM. Use of SPIA allows us to diminish signal quenching derived from most dyes, because these beads emit in the red region, which separates away from the absorbance of most dyes in the compound library. Zheng laboratory also reported a scintillation proximity assay for arginine methylation, in which SPA beads that emit in the blue range were chosen.37 We also favor polystyrene as the core material that facilitates dispensing due to slow settling time.
We formulated a three-step strategy that allows efficient transition from assay development to high throughput screening. First with the filter-paper assay, we optimized enzyme concentration, DMSO tolerance, and enzyme stability for our four enzymes of interest. Second, we confirmed the optimized assay parameters using our sepharose-bead assay. As a final step, the sepharose-bead assay was directly translated into SPIA format to enable high-throughput screening of our four PMTs: SET7/9, SET8, SETD2, and EuHMTase1. Of the four enzymes tested, we observed that catalytic efficiency of SET8 was lowest; requiring a concentration of 1.5 μM and an 8 h reaction time with a good signal to noise ratio of 2.7. Also, all four enzymes showed DMSO tolerance of up to 5%, making their testing viable under assay conditions of 1% DMSO. Finally, control testing for the four PMTs yielded Z′ values of 0.48 – 0.91 and thusunequivocally demonstrated the am enability of our assay for HTS.
To validate our approach, we screened the four PMTs in SPIA-bead format against our pilot library in duplicate to assess robustness and reproducibility. We obtained a total of 25 positive hits for SET7/9, 18 for SET8, 67 for SETD2, and 5 for EuHTMase1 (hit rate 0.45–0.97%) in our search for inhibitors. These results were confirmed via our filter-paper assay to determine dose-response and eliminate false positives. To illustrate, the SPIA-bead assay generated IC50 values between 1.3 – 1.7 μM for biotin, which is in parallel with peptide substrate concentration (1.5 μM) due to displacement of biotinylated peptide from SPIA beads. Using the filter-paper assay as the secondary assay, we did not find inhibition of biotin against all enzymes tested. The filter-paper assay was thus demonstrated as a reliable method for hit confirmation. In addition, Merbromin and NF-279 were identified via the SPIA-bead assay and confirmed the hits with the various enzymes. We classified Merbromin as a pan-active inhibitor that shows highest activity towards SETD2 (IC50 of 2.4 μM), followed by SET8 (4.2 μM), SET7/9 (28 μM), and finally EuHMTase1 (42.9 μM). Compound NF-279 is an important confirmation of assay validity, not only because it shows SET7/9 and SETD2 specificity, but also because other suramin analogs have been previously reported to inhibit PMTs activity by interacting with the substrate peptide.38
The SPIA-based mix-and-measure approach for HTS, together with the filter-paper assay for hit confirmation, present a novel and unprecedented assay pair to identify PMT-specific inhibitors. The general applicability of the SPIA-based HTS was exemplified by its capability to screen multiple PMTs that have diverse substrates and exhibit differing levels of methylation. Importantly, our results highlight success of establishing an in vitro screen for PMT inhibitors, which have the potential application of intervening pharmacologically in PMT-involved physiological and pathological processes.
In conclusion, we formulated the three-step strategy to optimize the assay parameters to identify PMT inhibitors in a generic HTS manner. The readiness and robustness of the stepwise HTS approach were demonstrated by their excellent Z′ values and its ability to identify 80 potential PMT inhibitors for four PMTs. We further examined the three inhibitors and showed that the secondary assay is ready to triage false positives or confirm pan- and target-specific PMT inhibitors. As a further step, the SPIA-based HTS can be readily transformed to screen full-size compound libraries. More importantly, the stepwise approach for assay optimization, HTS, hit validation is expected to be transferable for multiple PMTs.
Acknowledgments
We thank the Proteomics Resource Center of the Rockefeller University for preparing biotinylated peptides, Dr. Raymond C. Trievel (University of Michigan) for providing SET7/9 and SET8 plasmids, and Dr. Jinrong Min (University of Toronto) for the SETD2 and EuHMTase1 plasmids. The authors gratefully acknowledge the financial supports from Mr. William H. Goodwin and Mrs. Alice Goodwin Commonwealth Foundation for Cancer Research, The Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center, NIGMS (1R01GM096056), NINDS (R21NS071520), the NIH Director’s New Innovator Award Program (1DP2-OD007335, M.L.), the V Foundation for Cancer Research (2009 V Foundation Scholar Award, M.L.), March of Dimes Foundation (Basil O’connor Starter Scholar Award, M.L.), Starr Cancer Consortium (M.L.), Alfred W. Bressler Scholars Endowment Fund (M.L.). The HTS Core Facility is partially supported by the William Randolph Hearst Fund in Experimental Therapeutics, the Lillian S. Wells Foundation, and IH/NCI Cancer Center Support Grant (5P30 CA008748-44).
ABBREVIATIONS
- EuHMTase1/2
euchromatic histone methyltransferase 1/2
- H3/4K4/9/20
Histone 3/4 lysine 4/9/20
- MTA
methylthio -adenosine
- aa
amino acids
- MTAN
5′-methylthio-adenosine/SAH nucleosidase
- PMT
protein methyltransferase
- PKMT
protein lysine methyltransferase
- PRMT
protein arginine methyltransferase
- SAH
S-adenosyl-L-homocysteine
- SAM
S-adenosyl-L-methionine
- SET
SuVAR, Enhancer of zest, and Trithorax
- SPA
scintillation proximity assay
- SUV3-9
suppressor of var1, 3–9
- TCEP
tris(2-carboxyethyl)phosphine
- NF279
8,8′-[Carbonylbis(imino-4,1-phenylenecarbonylimino-4,1-phenylenecarbonylimino)]bis-1,3,5-naphtlenetrisulfonic acid hexasodium salt
References
- 1.Schapira M. Structural Chemistry of Human SET Domain Protein Methyltransferases. Curr Chem Genomics. 2011;5:85–94. doi: 10.2174/1875397301005010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford MT, Clarke SG. Protein Arginine Methylation in Mammals: Who, What, and Why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 5.Suh-Lailam BB, Hevel JM. A fast and efficient method for quantitative measurement of S-adenosyl-L-methionine-dependent methyltransferase activity with protein substrates. Anal Biochem. 2010;398:218–224. doi: 10.1016/j.ab.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Islam K, Zheng W, Yu H, Deng H, Luo M. Expanding Cofactor Repertoire of Protein Lysine Methyltransferase for Substrate Labeling. ACS Chem Biol. 2011 doi: 10.1021/cb2000567. Epub ahend of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the Mechanism of Multiple Lysine Methylation by the Human Mixed Lineage Leukemia Protein-1 (MLL1) Core Complex. J Biol Chem. 2009;284:24242–24256. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang R, Zheng W, Yu H, Deng H, Luo M. Labeling Substrates of Protein Arginine Methyltransferase with Engineered Enzymes and Matched S-Adenosyl-L-methionine Analogues. J Am Chem Soc. 2011;133:7648–7651. doi: 10.1021/ja2006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases. Anal Biochem. 2005;342(1):86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Dorgan KM, Wooderchak WL, Wynn DP, et al. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem. 2004;326:100–105. doi: 10.1016/j.ab.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Ibáñez G, McBean JL, Astudillo YM, Luo M. An Enzyme-coupled Ultrasensitive LuminescenceAssay for Protein Methyltransferases. Anal Biochem. 2010;401:203–210. doi: 10.1016/j.ab.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Wang CH, Leffler S, Thompson DH, Hrycyna CA. A general fluorescence-based coupled assay for S-adenosylmethionine-dependent methyltransferases. Biochem Biophys Res Comm. 2005;331:351–356. doi: 10.1016/j.bbrc.2005.03.170. [DOI] [PubMed] [Google Scholar]
- 14.Cheng DH, Yadav N, King RW, Swanson MS, Weinstein EJ, Bedford MT. Small molecule regulators of protein arginine methyltransferases. J Biol Chem. 2004;279:23892–23899. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
- 15.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat Chem Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 16.Kubicek S, O’Sullivan RJ, August EM, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25(3):473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Kouzarides T. SnapShot: Histone-modifying enzymes. Cell. 2007;128:802–803. doi: 10.1016/j.cell.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Barski A, Cuddapah S, Cui KR, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Lee YH, Stallcup MR. Minireview: Protein Arginine Methylation of Nonhistone Proteins in Transcriptional Regulation. Mol Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mowen KA, Tang J, Zhu W, et al. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 21.Yamagata K, Daitoku H, Takahashi Y, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Zhao X, Jankovic V, Gural A, et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22:640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu W, Chen HW, Du KY, et al. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 24.Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. Embo J. 2002 Oct 15;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bedford MT, Richard S. Arginine methylation: An emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Takagi T, Shum D, Parisi M, et al. Comparison of Luminescence ADP Production Assay and Radiometric Scintillation Proximity Assay for Cdc7 Kinase. Comb Chem High Throughput Screen. 2011;14:669–687. doi: 10.2174/138620711796504442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng HM, Shum D, Bhinder B, et al. A High Throughput Scintillation Proximity Based Assay for Human DNA Ligase IV. Assay Drug Dev Technol. 2011 doi: 10.1089/adt.2011.0404. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glickman JF, Schmid A, Ferrand S. Scintillation proximity assays in high-throughput screening. Assay Drug Dev Technol. 2008;6:433–455. doi: 10.1089/adt.2008.135. [DOI] [PubMed] [Google Scholar]
- 29.Wu H, Min JR, Lunin VV, et al. Structural Biology of Human H3K9 Methyltransferases. Plos One. 2010;5(1):e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couture JF, Collazo E, Hauk G, Trievel RC. Structural basis for the methylation site specificity of SET7/9. Nat Struct Mol Biol. 2006;13:140–146. doi: 10.1038/nsmb1045. [DOI] [PubMed] [Google Scholar]
- 31.Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci USA. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shelton CC, Tian Y, Shum D, et al. A miniaturized 1536-well format gamma-secretase assay. Assay DrugDev Technol. 2009;7:461–470. doi: 10.1089/adt.2009.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shum D, Smith JL, Hirsch AJ, et al. High-content assay to identify inhibitors of dengue virus infection. Assay Drug Dev Technol. 2010;8:553–570. doi: 10.1089/adt.2010.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seideman JH, Shum D, Djaballah H, Scheinberg DA. A high-throughput screen for alpha particle radiation protectants. Assay Drug Dev Technol. 2010;8:602–614. doi: 10.1089/adt.2010.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakowski TM, Zurita-Lopez C, Clarke SG, Frankel A. Approaches to measuring the activities of protein arginine N-methyltransferases. Anal Biochem. 2010;397:1–11. doi: 10.1016/j.ab.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rathert P, Dhayalan A, Ma HM, Jeltsch A. Specificity of protein lysine methyltransferases and methods for detection of lysine methylation of non-histone proteins. Mol BioSyst. 2008;4:1186–1190. doi: 10.1039/b811673c. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, Xie N, Feng Y, Zheng YG. Scintillation Proximity Assay of Arginine Methylation. J Biomol Screen. 2012 Aug 5;:2011. doi: 10.1177/1087057111414903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Y, Li M, Wang B, Zheng YG. Discovery and Mechanistic Study of a Class of Protein Arginine Methylation Inhibitors. J Med Chem. 2010;53:6028–6039. doi: 10.1021/jm100416n. [DOI] [PubMed] [Google Scholar]