Abstract
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been successfully applied as an identification procedure in clinical microbiology and has been widely used in routine laboratory practice because of its economical and diagnostic benefits. The range of applications of MALDI-TOF MS has been growing constantly, from rapid species identification to labor-intensive proteomic studies of bacterial physiology. The purpose of this review is to summarize the contribution of the studies already performed with MALDI-TOF MS concerning antibiotic resistance and to analyze future perspectives in this field. We believe that current research should continue in four main directions, including the detection of antibiotic modifications by degrading enzymes, the detection of resistance mechanism determinants through proteomic studies of multiresistant bacteria, and the analysis of modifications of target sites, such as ribosomal methylation. The quantification of antibiotics is suggested as a new approach to study influx and efflux in bacterial cells. The results of the presented studies demonstrate that MALDI-TOF MS is a relevant tool for the detection of antibiotic resistance and opens new avenues for both clinical and experimental microbiology.
INTRODUCTION
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) has recently been introduced into many microbiological laboratories for the routine identification of bacteria and fungi. Although the initial costs of a mass spectrometer are relatively high, the cost of identifying one species remains low compared with the cost of standard biochemical or molecular genetic techniques. This use of MALDI-TOF MS is making the diagnostic process approximately 24 h shorter (1, 2).
Recently, other applications focusing on the detection of antibiotic resistance mechanisms have been described. These methodologies may also play an important role in the application of mass spectrometry in diagnostic microbiological laboratories. The aim of this review is to summarize information that has already been published and to analyze future perspectives on MALDI-TOF MS for the detection of antibiotic resistance mechanisms.
REQUIREMENTS FOR RESISTANCE MECHANISM DETECTION
The resistance of bacteria to antibiotics has increased in recent years. Resistant bacteria can significantly complicate the treatment of infections in critically ill patients, especially in surgery, hemato-oncology, and intensive care in general (3, 4). Bacterial isolates that are resistant to all available antibiotics have also been described (5, 6).
Bacteria can resist antibiotic actions by the following mechanisms: the production of enzymes that inactivate antibiotic molecules (e.g., β-lactamases and aminoglycoside-modifying enzymes) (7–9), the hyperproduction or production of novel efflux pumps and other changes in the cell wall (e.g., porin alterations) (10), mutations in target genes (e.g., in ribosomal protein genes or in genes coding for penicillin-binding proteins [PBPs]) (8, 9, 11), the bypass of a metabolic pathway (e.g., the expression of acquired PBPs with a low affinity for antibiotic molecules) (11), and the production of proteins that protect the target site (e.g., quinolone resistance mediated by Qnr) or of target site-modifying enzymes (12).
In some cases, the detection of the resistance mechanism is complementary to standard susceptibility testing procedures, with an emphasis on the categorization of the isolate as resistant, intermediate, or susceptible (13). A quick determination of the resistance mechanism may also play a key role in the initial choice of antimicrobial therapy (14, 15). The detection of methicillin-resistant Staphylococcus aureus (MRSA) strains or carbapenemase-producing enterobacteria (CPE) is necessary for the effective prevention of the transmission of these bacteria (16, 17).
Therefore, routine microbiological laboratories need fast and reliable methods to detect resistance mechanisms in some bacteria that can provide important information for the choice of proper initial infection therapy and data for the experts working in infection control in health care settings.
MALDI-TOF MS analysis can most likely be applied for the analysis of all possible resistance mechanisms. The approaches that have already been reported are based on the following: the analysis of antibiotic molecules and their modified products, the analysis of bacterial cell components, the analysis of ribosomal DNA methylation, and the detection of mutations with minisequencing.
For some of these applications, MALDI-TOF MS provides results that can be used for diagnosis, whereas some of the methods are useful only in reference centers and research laboratories.
DETECTION OF ENZYMATIC ACTIVITY BY MALDI-TOF MS
Direct Detection of β-Lactamase Activity
The most common mechanism of resistance to β-lactams is the hydrolysis of the amide bond of the β-lactam ring (Fig. 1). The hydrolysis degradation product shows a different molecular mass from that of a native molecule. Some β-lactams are decarboxylated after hydrolysis, resulting in another change in molecular weight. According to the active-site structure, β-lactamases are divided to two groups: serine enzymes (molecular classes A, C, and D) and metalloenzymes (class B) (7, 18, 19). For a long time, class A β-lactamases were the most important group (e.g., TEM, SHV, and CTX-M). Recently, class B β-lactamases (metallo-β-lactamases) have become important due to their ability to hydrolyze carbapenems. The genes encoding metallo-β-lactamases were first spread among Pseudomonas spp., but recently, they have spread among members of the Enterobacteriaceae throughout the world. This spread was followed by Enterobacteriaceae producing class A carbapenemases (e.g., KPC and GES) in the present time (20).
Fig 1.
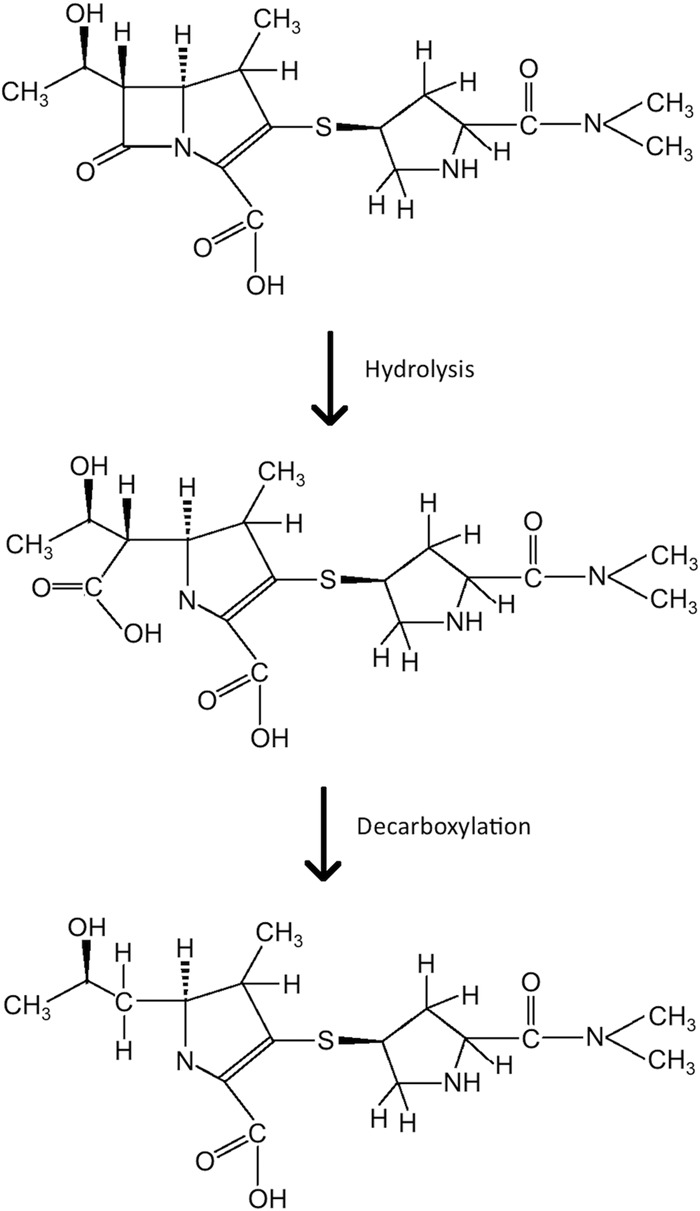
Meropenem and its degradation by β-lactamases.
The rapid detection of clinically important β-lactamases (e.g., extended-spectrum β-lactamases and carbapenemases) in routine diagnostic laboratories may be crucial for initial antibiotic therapy for specific patients as well as for the prevention of the spread of β-lactamase-producing bacteria in health care settings (21). Three main approaches have been introduced into routine laboratories. The most common techniques are based on the ability of some β-lactamases to be inhibited by specific inhibitors (e.g., clavulanic acid for extended-spectrum β-lactamases, amino-phenylboronic acid for KPC-type carbapenemases, and ethylendiamine tetra-acetic acid for metallo-β-lactamases). The inhibition of β-lactamases can be observed synergistically by using the disk diffusion method, the broth dilution method, or a special Etest containing the antibiotic to be tested in combination with an inhibitor (6). Molecular genetics techniques (e.g., PCR and microarrays) are common for the fast detection of some β-lactamases and can detect β-lactamase genes directly from a clinical sample. At this time, many different β-lactamases have been described. Therefore, it is very difficult to propose universal primers for the detection of at least one β-lactamase group. Additionally, a negative result should not be interpreted as the absence of β-lactamase. The direct method used in reference laboratories is based on the spectrophotometric detection of β-lactam hydrolysis using cell extracts containing β-lactamases. This method is labor-intensive and cannot be used routinely in most diagnostic laboratories (22, 23). Another method for the direct detection of carbapenemases in particular is a Hodge test, which can be used routinely. Its interpretation, sensitivity, and specificity, however, may be difficult (24).
In September 2011, the first two studies on direct carbapenemase detection by MALDI-TOF MS were reported (25, 26).
An analysis of antibiotics and their degradation products that are smaller than 1,000 Da by MALDI-TOF MS is possible using a specific sample preparation (27). Because the matrix is also visible in mass spectra, the detected molecule should be of a significantly different mass. For example, the peak representing an intact meropenem molecule can be observed at mass-to-charge ratio (m/z) of 384.5. The dimer of α-cyano-4-hydroxycinnamic acid (CHCA), a common matrix used for bacterial and fungal identification, is observed at m/z 380. The peak of the matrix is usually of a higher intensity than that of meropenem, and thus, the peaks of meropenem and the sodium salt of the meropenem decarboxylated degradation product are not clearly visible in a spectrum. Therefore, another matrix must be chosen.
The detection of β-lactamase activity is performed by using almost the same methodology in all published works. A fresh bacterial culture, usually grown overnight, is suspended in a buffer and centrifuged. The pellet is then resuspended in a reaction buffer containing the β-lactam molecule. After incubation at 35°C for 1 to 3 h, the reaction mixture is centrifuged, and the supernatant is mixed with a proper matrix measured by MALDI-TOF MS (Fig. 2). The spectra containing peaks representing the β-lactam molecule, its salts (usually sodium salts), and/or its degradation products are then analyzed.
Fig 2.
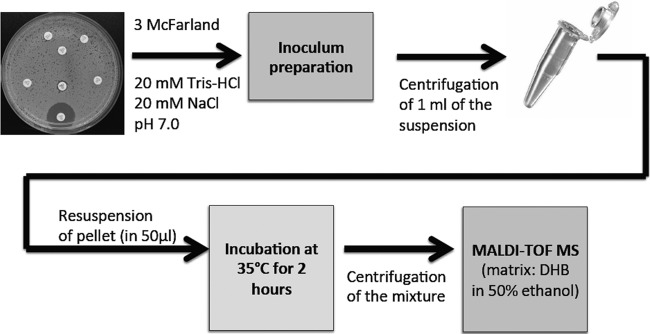
Hydrolysis assay for detection of carbapenemases. The assay is based on a method reported by Hrabák et al. (28). DHB, dihydroxybenzoic acid.
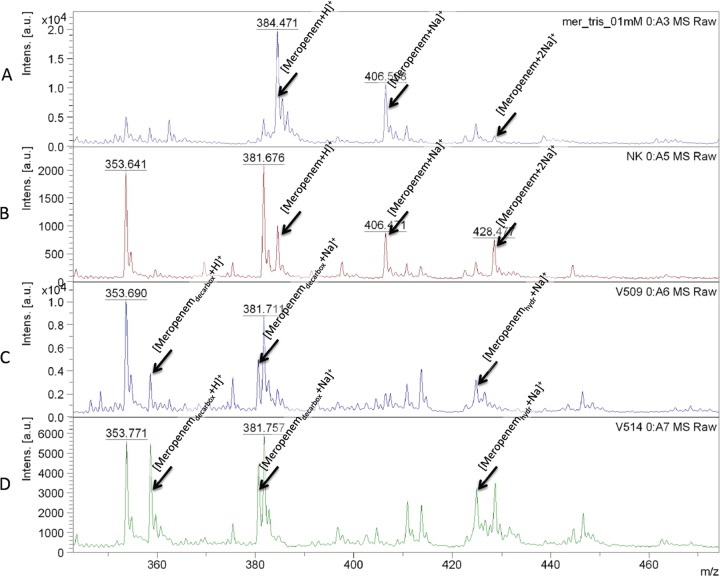
In a study by our group (26), we used the meropenem molecule as an indicator and demonstrated that MALDI-TOF MS is able to detect meropenem and the two types of meropenem sodium salts as well as the relevant degradation products (Fig. 3). The assay was validated using 124 strains, including 30 carbapenemase-producing bacteria representing IMP-7- and VIM-2-producing Pseudomonas aeruginosa strains and VIM-1-, KPC-2-, and NDM-1-producing enterobacteria. The sensitivity and specificity of the assay were both higher than 95%. The specificity of this assay was improved later by a modification of the reaction buffer supplemented with 0.01% sodium dodecyl sulfate. This change allows the concentration of bacteria used to be decreased (28). Comparing these results with those from a previously reported assay (26), the degradation products of meropenem (decarboxylated product at m/z 358.5 and its sodium salt at m/z 380.5) can be observed. For validation, 110 carbapenemase-producing isolates, including NDM-1-, KPC-2-, KPC-3-, VIM-1-, OXA-48-, and OXA-162-producing enterobacteria (e.g., Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae, and Serratia marcescens) and 35 carbapenemase-nonproducing isolates were used. This method was able to detect NDM-1 carbapenemase in Acinetobacter baumannii isolates as well (28). In our experience, OXA-type carbapenemases can also be easily detected in this species (J. Hrabák, unpublished data), similar to findings described by Kempf et al. (29) (see below).
Fig 3.
MALDI-TOF MS spectra of meropenem and its sodium salt variants and degradation products. The reaction was performed according to methods reported previously by Hrabák et al. (28). (A) Spectrum of the meropenem solution; (B) spectrum of the negative control (non-carbapenemase-producing isolate of Klebsiella pneumoniae; (C) spectrum of an NDM-1-producing Acinetobacter baumannii isolate; (D) spectrum of a KPC-2-producing Klebsiella pneumoniae isolate. [Meropenemdecarbox+H]+, decarboxylated degradation product of meropenem after carbapenemase hydrolysis (m/z 358.5); [Meropenemdecarbox+Na]+, decarboxylated sodium salt of the degradation product of meropenem after carbapenemase hydrolysis (m/z 380.5); [Meropenem+H]+, meropenem molecule (m/z 384.5); [Meropenem+Na]+, meropenem sodium salt (m/z 406.5); [Meropenemhydr+Na]+, sodium salt of the meropenem degradation product (m/z 424.5); [Meropenem + 2Na]+, meropenem disodium salt (m/z 428.5); [Meropenemhydr+2Na]+, disodium of the meropenem degradation product (m/z 446.5); [Meropenemhydr+3Na]+, trisodium of the meropenem degradation product (m/z 468.5); a.u., arbitrary units.
Burckhardt and Zimmermann (25) validated a similar method with KPC-2-, NDM-1-, IMP-, and VIM-producing enterobacteria using ertapenem. That group observed the ertapenem molecule (476 Da), its two sodium salt variants (498 and 521 Da), and the decarboxylated degradation product without sodium (450 Da). They also found excellent sensitivity and specificity for the assay.
A study of the detection of different β-lactam molecules was reported by Sparbier et al. (30). That group developed a method for the detection of ampicillin, piperacillin, cefotaxime, ceftazidime, ertapenem, imipenem, and meropenem and analyzed the different degradation products and sodium and/or potassium salt variants. No degradation products of imipenem and meropenem were observed. Some of the molecules (e.g., ampicillin and piperacillin) degraded spontaneously during the incubation time. For the identification of the β-lactamase type, clavulanic acid, tazobactam, and aminophenylboronic acid were introduced as inhibitors in the reaction. This study seems to be of the highest importance for the detection of different β-lactams by MALDI-TOF MS.
Recently, two other studies that validated MALDI-TOF MS for the detection of carbapenemases were reported (29, 31). Kempf et al. (29) used imipenem as an indicator. They tested 3 carbapenemase-producing Klebsiella pneumoniae isolates (KPC and NDM-1), 4 Pseudomonas aeruginosa isolates (VIM and IMP), and 63 OXA-23- and/or OXA-24-producing Acinetobacter baumannii isolates. Imipenem (m/z 300) and its degradation product (m/z 254) were detected by using a CHCA matrix and an AnchorChip target (Bruker Daltonik GmbH, Bremen, Germany). After 2 h of incubation, 67 of 70 carbapenemase-producing isolates were correctly detected, and no false-positive results were found. An extension of the incubation time to 4 h allowed the correct identification of all the carbapenemase producers. The work of that group is of great interest because OXA-type carbapenemases in Acinetobacter baumannii cannot be easily detected by spectrophotometric assays (32).
Another sample preparation for the detection of β-lactamase activity was reported by Hooff et al. (31). That group used a cell-free lysate and a MALDI-TOF and MALDI-triple quadrupole (QqQ) instrument. The data presented in that report clearly demonstrated the ability of the method to detect different β-lactam molecules (penicillin G, ampicillin, cefoxitin, and imipenem) and possible inhibition by clavulanic acid in addition to the reproducibility of the assay.
All of the reported studies describing the direct detection of β-lactamase activity independently presented similar results and allow us to conclude that this methodological approach has great potential to become a routine method for the detection of these enzymes which is comparable to the reference spectrophotometric assay (26). Manual measurements and analysis of raw spectra, however, can be difficult for microbiologists who are not experienced in mass spectrometry. Therefore, special software for the automatic acquisition and interpretation of results should be available to diagnostic laboratories. Another disadvantage of the method is the need for a fresh culture that is used for the analysis. The detection of carbapenemase activity directly from clinical samples without enrichment in specific cultivation media may be difficult and perhaps impossible.
Detection of rRNA Methyltransferase Activity
The methylation of rRNA confers resistance to protein synthesis inhibitors such as aminoglycosides, chloramphenicol, and clindamycin (9). Kirpekar et al. (33) developed a method for the detection of modifications to rRNA by MALDI-TOF MS. This method was later used to analyze the methylation of 23S rRNA by the product of the cfr gene, which is responsible for resistance to chloramphenicol, florfenicol, and clindamycin (34). The detection of methyltransferase activity responsible for resistance to aminoglycosides caused by the methylation of 16S rRNA was described in 2009 by Savic et al. (35). In that analysis, however, it was necessary to use purified ribosomes and purified enzymes. After the reaction in a special buffer, the rRNA was digested with a specific RNase to yield smaller products that were subsequently analyzed by MALDI-TOF MS to detect the methylation of the molecule (which increases the mass by 14 Da). This method is useful for the analysis of methyltransferases, but it should be simplified for use as a routine diagnostic technique or in reference centers.
DIRECT MALDI-TOF MS ANALYSIS OF BACTERIAL EXTRACTS
For the fast detection of resistance determinants, it would be useful to find a procedure that would be comparable to the identification of microbes by MALDI-TOF MS. Until now, however, only a few articles focusing on the development of such techniques have been published. Some researchers used a sample preparation similar to that used for bacterial identification, but most were not successful. We believe that specific sample preparations or measurement conditions must be used (e.g., selective protein extraction) for the detection of resistance markers. In the following sections, previously reported data on methicillin-resistant Staphylococcus aureus (MRSA), β-lactamases, and vancomycin-resistant enterococci are reviewed.
MRSA Detection
The first study on the discrimination between methicillin-susceptible S. aureus (MSSA) and MRSA strains was reported by Edwards-Jones et al. in 2000 (36). The strains were analyzed by MALDI-TOF MS using intact bacterial cells and 5-chloro-2-mercapto-benzothiazole as the matrix. Those authors were able to detect a total of 14 MRSA-specific peaks (e.g., m/z 511, 563, 640, 1,165, 1,229, and 2,127) and 2 MSSA-specific peaks (m/z 2,548 and 2,647) in the m/z ranges of 511 to 2,127 and 2,548 to 2,647, respectively. Seven other S. aureus-specific peaks (e.g., m/z 525, 617, 798, and 826) were also detected. Similar results were reported by Du et al., who were also able to successfully discriminate between MRSA and MSSA (37). In 2002, Bernardo et al. (38) did not find any correlation between MRSA and MSSA profiles, but they demonstrated the reproducibility of the obtained spectra, which allowed this method to be used for the tracking of nosocomial outbreaks. Unlike Edwards-Jones et al. (36), Bernardo et al. (38) used bacterial lysates for analysis.
The selective binding of a protein to a target in MALDI-TOF MS using the ProteinChip array is referred to as surface-enhanced laser desorption ionization–time of flight (SELDI-TOF) MS (39, 40). Shah et al. (40) used this method for the detection of MRSA and successfully identified seven ions that allowed discrimination between MRSA (n = 49) and MSSA (n = 50) strains. The bacteria were diluted in a specific lysis solution, mechanically disrupted, and then frozen and thawed. The supernatant after centrifugation was used for SELDI-TOF detection. Peaks at 3,081, 5,893, and 9,580 Da were detected in MSSA strains, and peaks at 5,709, 7,694, 15,308, and 18,896 Da were detected in MRSA strains. Almost all strains (97 of 99 strains) were correctly assigned as MRSA or MSSA. For routine practice, the preparation of the samples in such a specific way seems to be difficult. However, if we compare this technique with standard PCR detection, the labor intensities are similar.
Majcherczyk et al. (41) described the differentiation between isogenic teicoplanin-susceptible and teicoplanin-resistant strains of methicillin-resistant S. aureus. MALDI-TOF MS was found to have a superior discrimination potential compared to that of pulsed-field gel electrophoresis (PFGE) or peptidoglycan muropeptide digest patterns.
For application in routine diagnosis, the above-mentioned tests need to be further validated.
β-Lactamase Detection
SELDI-TOF MS was also used to discriminate between Escherichia coli strains that harbored different resistance genes (e.g., blaCTX-M-1, blaCTX-M-15, blaSHV-12, blaTEM, cat, strA, sul1, sul2, tetB, dhfr1, and aadA1) (42). With this method, the authors of that study were able to discriminate between bacterial species, but no specific peaks or profiles that allowed the detection of some resistance markers were presented in that article.
Camara and Hays (43) reported a study that focused on the detection of a specific β-lactamase peak for ampicillin-resistant Escherichia coli strains. After the cultivation of resistant strains in Luria-Bertani broth supplemented with ampicillin, proteins were extracted by using a formic acid-isopropyl alcohol-water solution and spotted onto a MALDI-TOF MS target by a sandwich method using sinapinic acid as a matrix. A peak of approximately 29 kDa that represented β-lactamase was successfully detected in the spectra. Those authors noted that this peak was not detectable using whole cells for bacterial identification or using the other matrices (2,5-dihydroxyacetophenone ferulic acid and 2,5-dihydroxybenzoic acid). The 29-kDa protein was then identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and liquid chromatography-MS peptide mass fingerprinting as a β-lactamase. To our knowledge, this is the first and last report that has shown the direct detection of β-lactamases in mass spectra of bacterial lysates.
Recently, Schaumann et al. (44) tried to differentiate between β-lactamase-producing and -nonproducing Enterobacteriaceae and Pseudomonas aeruginosa isolates. The samples were prepared by extraction using trifluoroacetic acid and acetonitrile. Although those researchers obtained clean and reproducible spectra, they were not able to find any difference between β-lactamase producers and nonproducers. No β-lactamase-specific peak was detected. The measurement, however, ranged from m/z 2,000 to 12,000, which seems to be smaller than the expected molecular mass of common β-lactamases (>27 kDa).
Detection of Vancomycin-Resistant Enterococcus spp.
A recently reported study conducted by Griffin and colleagues (45) demonstrated the ability of MALDI-TOF MS to distinguish between vanB-positive Enterococcus faecium and isolates that do not possess this resistance gene, which is responsible for resistance to glycopeptides. In this well-designed study, 67 vanB-positive Enterococcus faecium isolates were used. The sensitivity and specificity of the assay were 96.7% and 98.1%, respectively. It was also shown that this methodology is likely to indicate the relatedness of Enterococcus faecium strains to a degree that is similar to that of the existing gold standard for the analysis of relatedness, PFGE (45). It should be noted that a standard formic acid extraction method was used for MALDI-TOF MS measurements in this application. The authors of that study mentioned that the direct application of some Gram-positive bacteria (including enterococci) to the target without an extraction procedure cannot provide proper results due to the thickness of the peptidoglycan in the cell wall, which does not permit the proper acquisition of the spectra.
PROTEOMIC APPROACHES
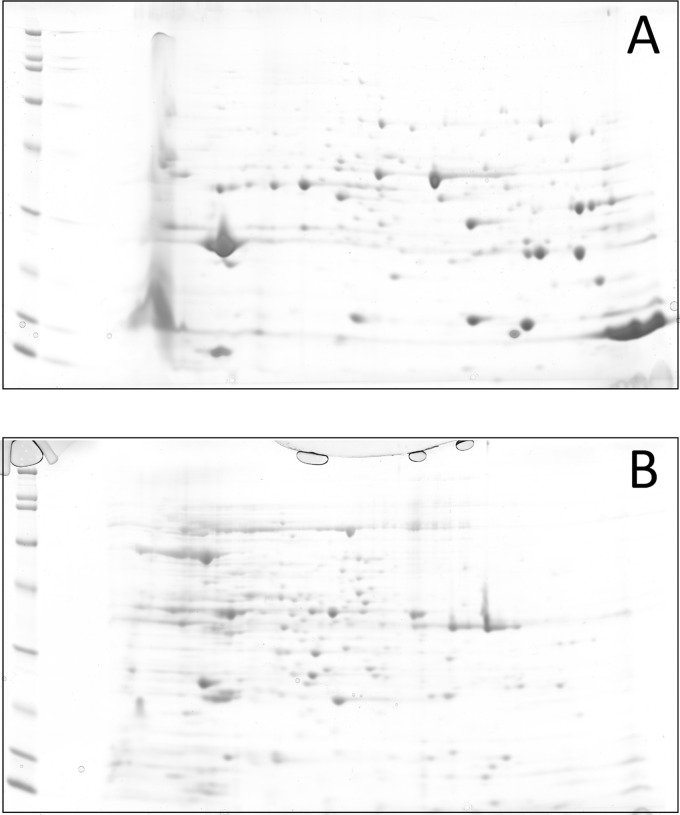
Research on antibiotic resistance is focused mainly on the detection of specific genes and comparison of isolates with molecular genetics techniques. Proteomic studies, however, provide data concerning gene expression and do not focus on only a single gene. As defined in 1995, the proteome is “the entire PROTEin complement expressed by a genOME, or by a cell or tissue type” (46). Proteomics uses primarily electrophoretic techniques for the detection of proteins. The cell extracts obtained by selective lysis are usually separated by two-dimensional (2D) electrophoresis. In the first step, the extract is separated according to the isoelectric point by using isoelectric focusing, and the proteins are then separated by SDS-PAGE (46). An example of the 2D electrophoretic separation of periplasmic and cytoplasmic proteins of Klebsiella pneumoniae is shown in Fig. 4. The protein map is then compared with a database by using specific software. The proteins of interest can be identified by peptide mass fingerprinting and/or amino acid sequencing (39, 46). The identification of the proteins is commonly performed by MALDI-TOF MS or MALDI-tandem TOF (MALDI-TOF/TOF) MS (39). The disadvantages of these techniques are their labor-intensiveness and the fact that the expression of the proteins may depend on the culture conditions (47).
Fig 4.
Example of two-dimensional electrophoretic separation of Klebsiella pneumoniae extracts. (A) Proteins extracted from the periplasmic space by the sucrose method; (B) proteins recovered from the cytoplasm by sonication.
Applications of Proteomic Analysis
A proteomic characterization of vancomycin-resistant Enterococcus sp. isolates recovered from seagulls was reported by Radhouani et al. (47). That group separated protein extracts obtained by the sonication of bacteria by SDS-PAGE as well as 2D electrophoresis with spot identification followed by MALDI-TOF MS identification of the proteins. Those researchers identified nine proteins associated with tetracycline resistance and a protein associated with the ability of the bacterium to form a biofilm. The vancomycin-teicoplanin A-type resistance protein vanA was also detected in Enterococcus durans.
A study focused on a proteome analysis of ampicillin-resistant Fusobacterium nucleatum was reported by Al-Haroni and colleagues (48). The bacteria were resuspended in lysis buffer and mechanically disrupted. This crude extract was used for 2D electrophoretic separation followed by the identification of spots of interest. In resistant isolates, a class D β-lactamase of 29 kDa, a 37-kDa ATP-binding cassette (ABC), a transporter ATP-binding protein, and a 46-kDa enolase were identified.
Three other studies (49–51) used MALDI-TOF MS after SDS-PAGE analysis for protein identification in resistant E. coli, Salmonella enterica, and Mycobacterium tuberculosis strains. All of those groups identified differences in protein expression levels among susceptible and resistant isolates.
A proteomic approach for the analysis of fluconazole-resistant Candida glabrata strains was described by Rogers and colleagues (52). They mechanically disrupted the cells and separated the proteins by 2D electrophoresis. Membrane proteins prepared by a selective procedure were separated by SDS-PAGE only. That group identified several proteins and the differences in their expression levels that may contribute to azole resistance. Further analysis should be performed to demonstrate the real activity of these proteins.
Conclusion
Proteomic analysis is a powerful tool that is used to analyze metabolic pathways in prokaryotic and eukaryotic cells. This process provides more complex data than studies of the expression of single gene or of a cluster of genes. Previous reports demonstrated the usefulness of this technique for determining resistance mechanisms. For new proteins that are identified as potential contributors to antibiotic resistance, their function should be demonstrated genetically (e.g., by gene deletion or cloning).
We do not believe that this technique can be used for the detection of antibiotic resistance mechanisms in routine diagnostic laboratories but can complement molecular genetic techniques in research laboratories until the time when whole-genome sequencing and its interpretation become widely available and fast.
ANALYSIS OF CELL WALL COMPONENTS
The importance of the cell wall in antibiotic resistance is highlighted by the fact that more than half of the targets of current antimicrobials are localized in the cell envelope. Other antibiotics have to overcome the cell wall barrier to reach their targets in the cytosol (10, 53, 54). Proteomic and genomic studies of the outer membrane of Gram-negative bacteria indicated that a complex system of cell components (porins, efflux pumps, and lipopolysaccharides, etc.) is responsible for antibiotic resistance via the regulation of the influx and efflux of different antibiotics (53). MALDI-TOF MS fingerprinting following SDS-PAGE or 2D electrophoresis, as described above, has been used for the identification of outer membrane and periplasmic proteins for which the expression levels differed in resistant isolates relative to the levels in the corresponding sensitive isolates (55–58). Such labor-intensive proteomic studies are useful for building a comprehensive database of bacterial protein fingerprints for comparative studies that provide insight into the roles of various cell components in antibiotic resistance (MASCOT; Matrix Science).
The use of MALDI-TOF MS is not limited to proteomics of the cell wall. Another example of its possible application is the detection of structural changes in lipopolysaccharides. The loss of the negative charge of lipid A due to postsynthetic modifications is especially associated with resistance to cationic antimicrobial peptides such as polymyxins (e.g., colistin) (59). For MALDI-TOF MS analysis, lipopolysaccharide can be extracted from the cell wall by a specific procedure, such as the hot-phenol method (60). In this method, lipopolysaccharide is solubilized by a water-saturated phenol and then precipitated. The samples are then applied with a proper matrix onto an MS target and analyzed. The modification of the structure of lipopolysaccharide (e.g., the loss of the fatty acid chain bound to the nonreducing sugar) causes the molecular weight changes detectable by MALDI-TOF MS. In previously reported works, samples were usually measured in a negative mode. This condition, however, is not usually available in the instruments used in diagnostic laboratories. The changes in lipid A that are caused by 4′-phosphatase in colistin-resistant Porphyromonas gingivalis were observed by Coats et al. (61) as a change of the molecular weight, and a phosphoethanolamide modification of lipid A associated with colistin resistance in Acinetobacter baumannii was reported by Beceiro et al. (62). In both studies, the modified lipid A was identified by MALDI-TOF MS.
Recently, Cai et al. (63) developed a method for the detection of the OmpK36 porin in Klebsiella pneumoniae, which is responsible for the penetration of carbapenems into the periplasm. Those authors used a standard procedure for the isolation of porins by specific extraction with sodium N-lauroyl sarcosinate. The samples were then detected by using MALDI-TOF MS based on a simple determination of the molecular weight. Those authors were able to detect the loss of the main OmpK36 porin, which contributed to resistance to carbapenems. This method appears to make porin detection faster than detection by SDS-PAGE. However, porins are detected according to molecular weight only. A method based on a better MALDI-TOF MS identification of the detected porins (e.g., protein fingerprinting) should be developed. This technique will most likely be used only in research laboratories.
MINISEQUENCING
DNA analysis by MALDI-TOF MS can also be used to detect resistance mechanisms. The principles of MALDI-TOF MS-based sequencing methods were described by Pusch et al. (64). After DNA amplification, the amplicons are purified, and an allele-specific reaction is used to obtain DNA molecules that are measureable by MALDI-TOF MS. For an allele-specific reaction, the most common technique used in microbiological applications was a primer extension assay. In this technique, specific oligonucleotides are used, and the product is extended for only a few bases (Fig. 5). Based on the molecular mass of the product, changes in the DNA structure can be expected. For example, an A→C mutation decreases molecular mass by 24 Da, while A→G mutation increases molecular mass by 16 Da (64). Due to the interactions of DNA with sodium and potassium, which result in salt formation, MALDI-TOF MS is limited to the analysis of molecules smaller than approximately 40 bp; therefore, the direct measurement of PCR products is difficult (64).
Fig 5.
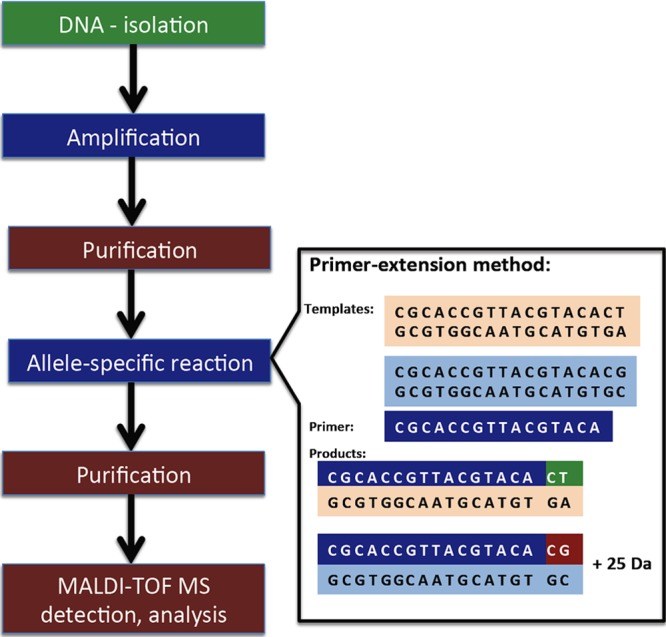
SNP detection by MALDI-TOF MS (according to data reported in reference 64).
This technique was used for the detection of single-nucleotide polymorphisms (SNPs) to identify SHV-type (65) and TEM-type (66) extended-spectrum β-lactamases. Resistance to rifampin and isoniazid in M. tuberculosis (67, 68) was also analyzed by MALDI-TOF MS sequencing.
Mutations in the penA and ponA genes were detected in order to analyze β-lactam resistance in Neisseria gonorrhoeae (69), and subsequent works described the detection of resistance to fluoroquinolones, spectinomycin, and tetracycline (70, 71). That same group reported MALDI-TOF MS minisequencing for the detection of resistance in Streptococcus pneumoniae (72, 73). This approach has also been used to determine point mutations in PBPs in Helicobacter pylori strains resistant to amoxicillin (74).
Despite the number of publications on MALDI-TOF MS-based sequencing for the detection of antibiotic resistance, this approach appears to be labor-intensive and limited by the small size of the DNA fragments that can be sequenced, without any advantage over standard sequencing procedures. Other methods (e.g., PCR-based SNP detection and DNA sequencing) are available and are used by many routine clinical laboratories.
FUTURE PERSPECTIVES
MALDI-TOF MS can be used to detect molecules with a broad range of molecular masses. Antibiotics are usually small molecules (<1,000 Da), which complicates their analysis because of interactions with the matrix and interference with a high level of background (75). Such molecules can be detected by using different matrices and modified approaches for sample preparation (75, 76). Lin et al. (77) reported an interesting methodology based on nanoparticles that were functionalized with a probe protein with an affinity for the detected molecule and coupled to a matrix. They successfully analyzed 6 different drugs: salicylamide, mefenamic acid, ketoprofen, flufenamic acid, sulindac, and prednisolone. This procedure also allows the purification and concentration of the tested molecule from the sample. According to their results, we hypothesize that this method can be used for (i) the detection of resistance mechanism determinants (e.g., modified penicillin-binding proteins and Qnr) and (ii) the direct analysis of antibiotic molecules and their purification from the sample/reaction solution.
This method can most likely be applied not only to the detection of resistance determinants but also to the detection of microbial toxins and drugs in blood samples.
As demonstrated above, MALDI-TOF MS has great potential to become a powerful technique for microbiological detection. Some reported methods need further validation or simplification. We believe that future research on the applications of MALDI-TOF MS for identifying antibiotic resistance mechanisms should focus on four primary topics.
The detection of enzymes that degrade antibiotic molecules through the direct detection of antibiotic modifications. These methods should be validated for routine diagnosis using a simple preparation of the bacterial culture or bacterial extracts. The detection of β-lactamases, especially carbapenemases, has already been validated and is used by some routine as well as reference laboratories. The methods should be developed for the detection of other enzymes (e.g., aminoglycoside-modifying enzymes) applicable in routine and research laboratories. To our knowledge, no such assay has yet been validated.
The detection of resistance mechanism determinants (e.g., Qnr proteins and modified PBPs and β-lactamases) and proteomic studies on multiresistant bacteria that allow the construction of a complete database of protein fingerprints of resistant isolates for the detection of the main proteins responsible for resistance. Recently, a method for the detection of vancomycin-resistant Enterococcus spp. was validated. This method seems to be useful in diagnostic laboratories for the rapid detection of vanB-positive Enterococcus faecium strains. Direct discrimination between MRSA and MSSA strains is a great challenge for MALDI-TOF MS applications in diagnostic laboratories. Despite the reports on this topic, especially work reported recently by Shah et al. (40), these procedures must be further optimized and validated. To the best of our knowledge, no successful method based on the detection of specific peaks in the bacterial lysate has been reported for analyses of other resistant determinants, such as β-lactamases.
The analysis of modification of target sites (e.g., ribosomal methylation). The detection of the methyltransferase activity responsible for resistance to aminoglycosides, chloramphenicol, and clindamycin caused by the methylation of 16S rRNA is still far from application for routine diagnosis. The procedure should be simplified by the use a crude bacterial extract.
The quantification of antibiotics. The development of a method to determine antibiotic concentrations could aid in the analysis of the influx and efflux of an antibiotic.
CONCLUSION
The introduction of MALDI-TOF MS as a routine diagnostic tool could represent a revolution in clinical microbiology. Microbiologists have received a powerful tool with many applications at the molecular level. However, this technique should also be applied with caution. Some microbes cannot be properly identified by MALDI-TOF MS using currently available procedures (78). The advantages and disadvantages of this technique compared with standard microbiological procedures are mentioned in Table 1.
Table 1.
Possible applications of MALDI-TOF MS for clinical microbiology and microbiological research
| MALDI-TOF MS application | Application already developed | Type of laboratories that can use the method | Advantage | Disadvantage | Complementary method | Note |
|---|---|---|---|---|---|---|
| Detection of enzymatic activity | Detection of carbapenemases | Routine laboratories | Direct detection of β-lactamase activity | Needs special skills for interpretation of spectra | Spectrophotometric detection of β-lactam hydrolysis | |
| Detection of other β-lactamases | Reference centers | Comparable with reference spectrophotometric assay | No identification of β-lactamase type (genetic techniques are needed for epidemiological purposes) | Phenotypic, inhibitor-based techniques | ||
| Turnaround time of ca. 3 h if fresh culture is available | Does not provide data about real susceptibility to β-lactams | Molecular genetics methods (e.g., PCR-based methods, sequencing, microarrays) | ||||
| Detection of carbapenemases in Acinetobacter baumannii | Fresh bacterial culture must be available | |||||
| Detection of rRNA methyltransferase activity | Research laboratories | Direct detection of enzymatic modifications | Labor-intensive method | |||
| Needs purification of ribosomes and enzymes | ||||||
| Direct MALDI-TOF MS analysis of bacterial extracts | Detection of vancomycin-resistant enterococci | Routine laboratories | Easy and fast technique for detection of resistance markers | Detection of resistance markers will probably require specific sample prepn | Phenotypic detection of resistance | Validated for detection of vanA in Enterococcus faecium |
| Detection of MRSA | Research laboratories (detection of lipopolysaccharide) | Probably many different resistance markers can be detected | Some proteins in samples with many different molecules may be invisible by MALDI-TOF MS | Molecular genetics techniques | Other techniques need further validation | |
| Detection of β-lactamases | Fresh bacterial culture must be available | |||||
| Detection of OmpK36 porin in Klebsiella pneumoniae | ||||||
| Modification of lipopolysaccharide | ||||||
| Proteomics | Analysis of proteins in resistance isolates | Reference centers | Complex picture of protein expression | Labor-intensive method | Molecular genetics techniques | Not yet fully introduced into research on antibiotic resistance |
| Research laboratories | Easy detection of new resistance mechanisms and their regulation pathways | Expensive technique (e.g., costs of electrophoretic equipment, software for gel analysis) | Methods for selective extraction of cell components should be developed | |||
| Needs specific extraction procedures | ||||||
| Minisequencing | Detection of extended-spectrum β-lactamases | Routine laboratories | Detection of SNP using MALDI-TOF MS | Labor-intensive method | SNP detection by high-resolution melting-curve analysis | Many clinical laboratories are already equipped for and experienced in DNA sequencing or SNP detection by other techniques |
| Detection of mutations responsible for resistance in Neisseria gonorrhoeae, Streptococcus pneumoniae, and Helicobacter pylori, etc. | Time-consuming technique compared with other procedures | Probe-based technique “Classic” sequencing |
Proteomic approaches in studies of resistant strains can complement molecular genetics techniques. Proteome-level studies allow the detection of the behavior of the tested strains, the expression of proteins of interest, and posttranslational modifications. With the exception of whole-genome sequencing, which is not yet available for routine use, all molecular genetics techniques have been restricted to the detection of known resistance determinants. Proteomic analysis may be labor-intensive in routine laboratories, but it should be available in reference centers.
As demonstrated by many studies, MALDI-TOF MS can detect the biological activity of enzymes responsible for the modification of antibiotic molecules, which is not possible with genetic techniques. The results of the studies presented here show that MALDI-TOF MS is a relevant tool for the detection of antibiotic resistance and opens new avenues for both clinical and experimental microbiology.
We do not believe, however, that the application of this technique to the determination of resistance mechanisms can replace standard susceptibility testing, because resistance to antibiotics is a complex process. Therefore, the detection of a limited number of specific determinants cannot provide a complete picture of antibiotic resistance.
ACKNOWLEDGMENTS
We thank C. C. Papagiannitsis for critical reading of the manuscript.
Jaroslav Hrabák and Eva Chudáčková were partially supported by research project grants NT11032-6/2010 from the Ministry of Health of the Czech Republic and MSM0021620819 from the Ministry of Education of the Czech Republic and by the Charles University Research Fund (project number P36).
Biographies

Jaroslav Hrabák, Ph.D., is Assistant Professor of Microbiology at the Faculty of Medicine in Plzeň, Charles University in Prague, Plzeň, Czech Republic, and a collaborator of the National Institute of Public Health, Prague, Czech Republic. His research in the field of antimicrobial resistance is focused mainly on carbapenemase-producing enterobacteria and Pseudomonas spp. and the use of MALDI-TOF MS in microbiology.

Eva Chudáčková, M.D., is a Ph.D. student at the Faculty of Medicine in Plzeň, Charles University in Prague, Plzeň, Czech Republic, where she graduated in general medicine. She works as a clinical microbiologist at the University Hospital Plzeň. Her research interests include carbapenem resistance in enterobacteria, focused on resistant carbapenemase-nonproducing isolates.

Radka Walková, M.D., graduated in general medicine at the Faculty of Medicine in Plzeň and works as a clinical microbiologist at the University Hospital Plzeň. She has been working on research in the field of bacterial resistance to antibiotics, focused on the use of MALDI-TOF MS.
REFERENCES
- 1. Seng P, Rolain J-M, Fournier PE, La Scola B, Drancourt M, Raoult D. 2010. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 5:1733–1754 [DOI] [PubMed] [Google Scholar]
- 2. Wieser A, Schneider L, Jung J, Schubert S. 2012. MALDI-TOF MS in microbiological diagnostics—identification of microorganisms and beyond. Appl. Microbiol. Biotechnol. 93:965–974 [DOI] [PubMed] [Google Scholar]
- 3. Livermore DM. 2012. Fourteen years in resistance. Int. J. Antimicrob. Agents 39:283–294 [DOI] [PubMed] [Google Scholar]
- 4. Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18:263–272 [DOI] [PubMed] [Google Scholar]
- 5. Hrabák J, Niemczyková J, Chudácková E, Fridrichová M, Studentová V, Cervená D, Urbásková P, Zemlicková H. 2011. KPC-2-producing Klebsiella pneumoniae isolated from a Czech patient previously hospitalized in Greece and in vivo selection of colistin resistance. Folia Microbiol. (Praha) 56:361–365 [DOI] [PubMed] [Google Scholar]
- 6. Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bush K, Jacoby GA, Medeiros AA. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shakil S, Khan R, Zarrilli R, Khan AU. 2008. Aminoglycosides versus bacteria—a description of the action, resistance mechanism, and nosocomial battleground. J. Biomed. Sci. 15:5–14 [DOI] [PubMed] [Google Scholar]
- 9. Smith CA, Baker EN. 2002. Aminoglycoside antibiotic resistance by enzymatic deactivation. Curr. Drug Targets Infect. Disord. 2:143–160 [DOI] [PubMed] [Google Scholar]
- 10. Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zapun A, Contreras-Martel C, Vernet T. 2008. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 32:361–385 [DOI] [PubMed] [Google Scholar]
- 12. Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22:664–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leclercq R, Cantón R, Brown DF, Giske CG, Heisig P, Macgowan AP, Mouton JW, Nordmann P, Rodloff AC, Rossolini GM, Soussy CJ, Steinbakk M, Winstanley TG, Kahlmeter G. 21 October 2011. EUCAST expert rules in antimicrobial susceptibility testing. Clin. Microbiol. Infect. [Epub ahead of print.] doi:10.1111/j.1469-0691.2011.03703.x [DOI] [PubMed] [Google Scholar]
- 14. Ledeboer NA, Hodinka RL. 2011. Molecular detection of resistance determinants. J. Clin. Microbiol. 49:S20–S24 doi:10.1128/JCM.00771-11 [Google Scholar]
- 15. Niederman MS. 2009. Treatment options for nosocomial pneumonia due to MRSA. J. Infect. 59(Suppl 1):S25–S31 doi:10.1016/S0163-4453(09)60005-0 [DOI] [PubMed] [Google Scholar]
- 16. Grundmann H, Livermore DM, Giske CG, Canton R, Rossolini GM, Campos J, Vatopoulos A, Gniadkowski M, Toth A, Pfeifer Y, Jarlier V, Carmeli Y, CNSE Working Group 2010. Carbapenem-non-susceptible Enterobacteriaceae in Europe: conclusions from a meeting of national experts. Euro Surveill. 15(46):pii=19711. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19711 [DOI] [PubMed] [Google Scholar]
- 17. Tacconelli E. 2009. Methicillin-resistant Staphylococcus aureus: source control and surveillance organization. Clin. Microbiol. Infect. 15(Suppl 7):31–38 [DOI] [PubMed] [Google Scholar]
- 18. Buynak JD. 2004. The discovery and development of modified penicillin- and cephalosporin-derived beta-lactamase inhibitors. Curr. Med. Chem. 11:1951–1964 [DOI] [PubMed] [Google Scholar]
- 19. Paterson DL, Bonomo RA. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bush K. 2010. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 13:558–564 [DOI] [PubMed] [Google Scholar]
- 21. Nordmann P, Gniadkowski M, Giske CG, Poirel L, Woodford N, Miriagou V, European Network on Carbapenemases 2012. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Infect. 18:432–438 [DOI] [PubMed] [Google Scholar]
- 22. Cornaglia G, Akova M, Amicosante G, Cantón R, Cauda R, Docquier JD, Edelstein M, Frère JM, Fuzi M, Galleni M, Giamarellou H, Gniadkowski M, Koncan R, Libisch B, Luzzaro F, Miriagou V, Navarro F, Nordmann P, Pagani L, Peixe L, Poirel L, Souli M, Tacconelli E, Vatopoulos A, Rossolini GM, ESCMID Study Group for Antimicrobial Resistance Surveillance 2007. Metallo-beta-lactamases as emerging resistance determinants in Gram-negative pathogens: open issues. Int. J. Antimicrob. Agents 29:380–388 [DOI] [PubMed] [Google Scholar]
- 23. Bernabeu S, Poirel L, Nordmann P. 2012. Spectrophotometry-based detection of carbapenemase producers among Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 74:88–90 [DOI] [PubMed] [Google Scholar]
- 24. Girlich D, Poirel L, Nordmann P. 2012. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 50:477–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burckhardt I, Zimmermann S. 2011. Using matrix-assisted laser desorption ionization–time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 49:3321–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hrabák J, Walková R, Studentová V, Chudácková E, Bergerová T. 2011. Carbapenemase activity detection by matrix-assisted laser desorption–ionization time of flight mass spectrometry. J. Clin. Microbiol. 49:3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan C, Xu S, Zhou H, Fu Y, Ye M, Zou H. 2007. Recent developments in methods and technology for analysis of biological samples by MALDI-TOF-MS. Anal. Bioanal. Chem. 387:193–204 [DOI] [PubMed] [Google Scholar]
- 28. Hrabák J, Studentová V, Walková R, Zemlicková H, Jakubu V, Chudácková E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerová T. 2012. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:2441–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel JM, Drissi M, Mesli E, Touati A, Rolain JM. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 7:e31676 doi:10.1371/journal.pone.0031676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. 2012. Matrix-assisted laser desorption ionization–time of flight mass spectrometry-based functional assay for rapid detection of resistance against β-lactam antibiotics. J. Clin. Microbiol. 50:927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hooff GP, van Kampen JJ, Meesters RJ, van Belkum A, Goessens WH, Luider TM. 2012. Characterization of β-lactamase enzyme activity in bacterial lysates using MALDI-mass spectrometry. J. Proteome Res. 11:79–84 [DOI] [PubMed] [Google Scholar]
- 32. Bonnin RA, Naas T, Poirel L, Nordmann P. 2012. Phenotypic, biochemical, and molecular techniques for detection of metallo-β-lactamase NDM in Acinetobacter baumannii. J. Clin. Microbiol. 50:1419–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kirpekar F, Douthwaite S, Roepstorff P. 2000. Mapping posttranscriptional modifications in 5S ribosomal RNA by MALDI mass spectrometry. RNA 6:296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57:1064–1073 [DOI] [PubMed] [Google Scholar]
- 35. Savic M, Lovric J, Tomic TI, Vasiljevic B, Conn GL. 2009. Determination of the target nucleosides for members of two families of 16S rRNA methyltransferases that confer resistance to partially overlapping groups of aminoglycoside antibiotics. Nucleic Acids Res. 37:5420–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Edwards-Jones V, Claydon MA, Evason DJ, Walker J, Fox AJ, Gordon DB. 2000. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J. Med. Microbiol. 49:295–300 [DOI] [PubMed] [Google Scholar]
- 37. Du Z, Yang R, Guo Z, Song Y, Wang J. 2002. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 74:5487–5491 [DOI] [PubMed] [Google Scholar]
- 38. Bernardo K, Pakulat N, Macht M, Krut O, Seifert H, Fleer S, Hünger F, Krönke M. 2002. Identification and discrimination of Staphylococcus aureus strains using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2:747–753 [DOI] [PubMed] [Google Scholar]
- 39. Shah H, Gharbia S. 2010. Mass spectrometry for microbial proteomics. John Wiley & Sons, Inc., Chichester, United Kingdom [Google Scholar]
- 40. Shah HN, Rajakaruna L, Ball G, Misra R, Al-Shahib A, Fang M, Gharbia SE. 2011. Tracing the transition of methicillin resistance in sub-populations of Staphylococcus aureus, using SELDI-TOF mass spectrometry and artificial neural network analysis. Syst. Appl. Microbiol. 34:81–86 [DOI] [PubMed] [Google Scholar]
- 41. Majcherczyk PA, McKenna T, Moreillon P, Vaudaux P. 2006. The discriminatory power of MALDI-TOF mass spectrometry to differentiate between isogenic teicoplanin-susceptible and teicoplanin-resistant strains of methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 255:233–239 [DOI] [PubMed] [Google Scholar]
- 42. Dubska L, Pilatova K, Dolejska M, Bortlicek Z, Frostova T, Literak I, Valik D. 2011. Surface-enhanced laser desorption ionization/time-of-flight (SELDI-TOF) mass spectrometry (MS) as a phenotypic method for rapid identification of antibiotic resistance. Anaerobe 17:444–447 [DOI] [PubMed] [Google Scholar]
- 43. Camara JE, Hays FA. 2007. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Bioanal. Chem. 389:1633–1638 [DOI] [PubMed] [Google Scholar]
- 44. Schaumann R, Knoop N, Genzel GH, Losensky K, Rosenkranz C, Stîngu CS, Schellenberger W, Rodloff AC, Eschrich K. 2012. A step towards the discrimination of beta-lactamase-producing clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa by MALDI-TOF mass spectrometry. Med. Sci. Monit. 18:MT71–MT77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Griffin PM, Price GR, Schooneveldt JM, Schlebusch S, Tilse MH, Urbanski T, Hamilton B, Venter D. 2012. Use of matrix-assisted laser desorption ionization–time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J. Clin. Microbiol. 50:2918–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marshall T, Williams KM. 2002. Proteomics and its impact upon biomedical science. Br. J. Biomed. Sci. 59:47–64 [DOI] [PubMed] [Google Scholar]
- 47. Radhouani H, Poeta P, Pinto L, Miranda J, Coelho C, Carvalho C, Rodrigues J, López M, Torres M, Vitorino R, Domingues P, Igrejas G. 2010. Proteomic characterization of vanA-containing Enterococcus recovered from seagulls at the Berlengas Natural Reserve, W Portugal. Proteome Sci. 8:48 doi:10.1186/1477-5956-8-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Al-Haroni M, Skaug N, Bakken V, Cash P. 2008. Proteomic analysis of ampicillin-resistant oral Fusobacterium nucleatum. Oral Microbiol. Immunol. 23:36–42 [DOI] [PubMed] [Google Scholar]
- 49. Pinto L, Poeta P, Vieira S, Caleja C, Radhouani H, Carvalho C, Vieira-Pinto Themudo P, Torres C, Vitorino R, Domingues P, Igrejas G. 2010. Genomic and proteomic evaluation of antibiotic resistance in Salmonella strains. J. Proteomics 73:1535–1541 [DOI] [PubMed] [Google Scholar]
- 50. dos Santos KV, Diniz CG, de Castro Veloso L, de Andrade HM, da Silva Giusta M, da Fonseca Pires S, Santos AV, Apolonio ACM, de Carvalho MAR, de Macedo Farias L. 2010. Proteomic analysis of Escherichia coli with experimentally induced resistance to piperacillin/tazobactam. Res. Microbiol. 161:268–275 [DOI] [PubMed] [Google Scholar]
- 51. Sharma P, Kumar B, Gupta Y, Singhal N, Katoch VM, Venkatesan K, Bisht D. 2010. Proteomic analysis of streptomycin resistant and sensitive clinical isolates of Mycobacterium tuberculosis. Proteome Sci. 8:59 doi:10.1186/1477-5956-8-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rogers PD, Vermitsky JP, Edlind TD, Hilliard GM. 2006. Proteomic analysis of experimentally induced azole resistance in Candida glabrata. J. Antimicrob. Chemother. 58:434–438 [DOI] [PubMed] [Google Scholar]
- 53. Davin-Regli A, Bolla JM, James CE, Lavigne JP, Chevalier J, Garnotel E, Molitor A, Pages JM. 2008. Membrane permeability and regulation of drug “influx and efflux” in enterobacterial pathogens. Curr. Drug Targets 9:750–759 [DOI] [PubMed] [Google Scholar]
- 54. Drew D, Fröderberg L, Baars L, de Gier JW. 2003. Assembly and overexpression of membrane proteins in Escherichia coli. Biochim. Biophys. Acta 1610:3–10 [DOI] [PubMed] [Google Scholar]
- 55. Gröbner S, Linke D, Schütz W, Fladerer C, Madlung J, Autenrieth IB, Witte W, Pfeifer Y. 2009. Emergence of carbapenem-non-susceptible extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tübingen, Germany. J. Med. Microbiol. 58:912–922 [DOI] [PubMed] [Google Scholar]
- 56. Imperi F, Ciccosanti F, Perdomo A, Tiburzi F, Mancone C, Alonzi T, Ascenzi P, Piacentini M, Visca P, Fimia GM. 2009. Analysis of the periplasmic proteome of Pseudomonas aeruginosa, a metabolically versatile opportunistic pathogen. Proteomics 9:1901–1915 [DOI] [PubMed] [Google Scholar]
- 57. Peng X, Xu C, Ren H, Lin X, Wu L, Wang S. 2005. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Pseudomonas aeruginosa responding to ampicilin, kanamycin, and tetracycline resistance. J. Proteome Res. 4:2257–2265 [DOI] [PubMed] [Google Scholar]
- 58. Xu C, Lin X, Ren H, Zhang Y, Wang S, Peng X. 2006. Analysis of outer membrane proteome of Escherichia coli related to resistance to ampicillin and tetracycline. Proteomics 6:462–473 [DOI] [PubMed] [Google Scholar]
- 59. Tran AX, Lester ME, Stead CM, Raetz CRH, Maskell DJ, McGrath SC, Cotter RJ, Trent MS. 2005. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J. Biol. Chem. 280:28186–28194 [DOI] [PubMed] [Google Scholar]
- 60. Yi EC, Hackett M. 2000. Rapid isolation method for lipopolysaccharide and lipid A from Gram-negative bacteria. Analyst 125:651–656 [DOI] [PubMed] [Google Scholar]
- 61. Coats SR, To TT, Jain S, Braham PH, Darveau RP. 2009. Porphyromonas gingivalis resistance to polymyxin B is determined by the lipid A 4′-phosphatase, PGN_0524. Int. J. Oral Sci. 1:126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55:3370–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cai JC, Hu YY, Zhang R, Zhou HW, Chen G-X. 2012. Detection of Ompk36 porin loss in Klebsiella spp. by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:2179–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pusch W, Wurmbach J-H, Thiele H, Kostrzewa M. 2002. MALDI-TOF mass spectrometry-based SNP genotyping. Pharmacogenomics 3:537–548 [DOI] [PubMed] [Google Scholar]
- 65. Stürenburg E, Storm N, Sobottka I, Horstkotte MA, Scherpe S, Aepfelbacher M, Müller S. 2006. Detection and genotyping of SHV beta-lactamase variants by mass spectrometry after base-specific cleavage of in vitro-generated RNA transcripts. J. Clin. Microbiol. 44:909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ikryannikova LN, Shitikov EA, Zhivankova DG, Il'ina EN, Edelstein MV, Govorun VM. 2008. A MALDI TOF MS-based minisequencing method for rapid detection of TEM-type extended-spectrum beta-lactamases in clinical strains of Enterobacteriaceae. J. Microbiol. Methods 75:385–391 [DOI] [PubMed] [Google Scholar]
- 67. Ikryannikova LN, Afanas'ev MV, Akopian TA, Il'ina EN, Kuz'min AV, Larionova EE, Smirnova TG, Chernousova LN, Govorun VM. 2007. Mass-spectrometry based minisequencing method for the rapid detection of drug resistance in Mycobacterium tuberculosis. J. Microbiol. Methods 70:395–405 [DOI] [PubMed] [Google Scholar]
- 68. Wang Q, Yue J, Zhang L, Xu Y, Chen J, Zhang M, Zhu B, Wang H, Wang H. 2007. A newly identified 191A/C mutation in the Rv2629 gene that was significantly associated with rifampin resistance in Mycobacterium tuberculosis. J. Proteome Res. 6:4564–4571 [DOI] [PubMed] [Google Scholar]
- 69. Malakhova MV, Vereshchagin VA, Il'ina EN, Govorun VM, Zubkov MM, Priputnevich TV, Kisina VI, Kubanova AA. 2006. Analysis of genetic markers of N. gonorrhoeae resistance to beta-lactam antibiotics. Bull. Exp. Biol. Med. 141:610–615 [DOI] [PubMed] [Google Scholar]
- 70. Borovskaya AD, Malakhova MV, Vereshchagin VA, Il'ina EN, Govorun VM, Priputnevich TV, Al-Hafagi N, Kubanova AA. 2007. Analysis of the contribution of molecular mechanisms into formation of gonoccocal resistance to tetracycline. Bull. Exp. Biol. Med. 144:432–437 [DOI] [PubMed] [Google Scholar]
- 71. Il'ina EN, Malakhova MV, Vereshchagin VA, Govorun VM, Priputnevich TV, Kubanova AA. 2007. Direct evaluation of drug resistance parameters in gonococcus. Bull. Exp. Biol. Med. 144:227–230 [DOI] [PubMed] [Google Scholar]
- 72. Malakhova MV, Vereshchagin VA, Il'ina EN, Govorun VM, Filimonova O, Grudinina SA, Sidorenko SV. 2007. MALDI-ToF mass-spectrometry in analysis of genetically determined resistance of Streptococcus pneumoniae to fluoroquinolones. Antibiot. Khimioter. 52:10–17 [PubMed] [Google Scholar]
- 73. Savinova TA, Il'ina EN, Sidorenko SV. 2010. A mass-spectrometric analysis of genetic markers of S. pneumoniae resistance to beta-lactam antibiotics. Mol. Gen. Mikrobiol. Virusol. 3:16–25 [PubMed] [Google Scholar]
- 74. Shen J, Deng DJ, Ke Y, Zhang JZ. 2008. Detection of point mutation in an in vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Zhonghua Liu Xing Bing Xue Za Zhi 29:166–172 [PubMed] [Google Scholar]
- 75. Penno MAS, Ernst M, Hoffmann P. 2009. Optimal preparation methods for automated matrix-assisted laser desorption/ionization time-of-flight mass spectrometry profiling of low molecular weight proteins and peptides. Rapid Commun. Mass Spectrom. 23:2656–2662 [DOI] [PubMed] [Google Scholar]
- 76. Li X, Wu X, Kim JM, Kim SS, Jin J, Li D. 2009. MALDI-TOF-MS analysis of small molecules using modified mesoporous material SBA-15 as assisted matrix. J. Am. Soc. Mass Spectrom. 20:2167–2173 [DOI] [PubMed] [Google Scholar]
- 77. Lin P-C, Tseng M-C, Su A-K, Chen Y-J, Lin C-C. 2007. Functionalized magnetic nanoparticles for small-molecule isolation, identification, and quantification. Anal. Chem. 79:3401–3408 [DOI] [PubMed] [Google Scholar]
- 78. Werno AM, Christner M, Anderson TP, Murdoch DR. 2012. Differentiation of Streptococcus pneumoniae from nonpneumococcal streptococci of the Streptococcus mitis group by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:2863–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]