Abstract
Ribonuclease 7 (RNase 7) is a 14.5-kDa peptide that possesses potent antimicrobial properties against Gram-negative and Gram-positive bacteria and is expressed in a variety of epithelial tissues. Little is known about its mechanisms of action and the determinants of its antimicrobial properties. The objective of this study was to identify the intrinsic functional domains of RNase 7 that influence its activity against uropathogenic bacteria. A series of RNase 7 fragments were generated that contained different components of its secondary motifs starting from both the N terminus and the C terminus of RNase 7. We determined the antimicrobial properties of each fragment against both Gram-positive Staphylococcus saprophyticus and Gram-negative Escherichia coli and Proteus mirabilis. RNase 7 fragments displayed significant differences in their antimicrobial activity profiles. Compared to N-terminal fragments, C-terminal fragments showed uniformly decreased activity against Gram-negative E. coli and P. mirabilis and Gram-positive S. saprophyticus. Fragments that lack β-sheets 1, 3, and 4 also demonstrated significantly decreased activities. We have also identified one fragment with at least 4-fold increased potency against both E. coli and Staphylococcus compared to full-length peptide. We identified distinct regions of the peptide that are independently responsible for Gram-negative and Gram-positive activity. Our results suggest that distinct mechanisms are responsible for RNase 7's antimicrobial activity against various uropathogens.
INTRODUCTION
Antimicrobial peptides (AMPs) are small proteins that possess antimicrobial activity against bacteria, enveloped viruses, fungi, and some protozoa. AMPs are usually cationic secondary to the presence of lysine and/or arginine residues and amphipathic, which allows them to intercalate into hydrophobic bacterial cell membranes and yet remain in solution in aqueous environments. Because pathogenic bacteria are still susceptible to endogenous AMPs, they have been considered a possible treatment against drug-resistant organisms. Unfortunately, our understanding of the fundamental mechanisms of AMP action and the structure-function relationship with bacteria is cursory (1–3). The associations between AMP structure/properties and the antimicrobial activity have been reported and include the (i) net charge, (ii) amphipathicity, (iii) hydrophobicity, and (iv) conformation. The net charge refers to the entire molecular charge. A higher positive net charge increases the electrostatic attraction to negatively charged bacterial cell membranes (4). Amphipathicity refers to the ratio of polar and nonpolar components. Increased amphipathicity as estimated by the hydrophobic moment allows for solubility in pathogen membranes (5). Hydrophobicity quantitates the percentage of hydrophobic residues within a peptide. The ability of an AMP to intercalate into a bacterial membrane increases as hydrophobicity increases (4). Finally, conformation refers to the ways the dimensional topography of a peptide can influence activity. The majority of AMPs have an α-helical and/or β-sheet conformation (4).
Ribonuclease 7 (RNase 7) is a 14.5-kDa AMP, which Harder and Schröder first identified as an abundant protein in the human epidermis while examining protein extracts of normal skin for antimicrobial activity (6). Subsequent studies demonstrated that RNASE7 is expressed in other organs, including the liver, gastrointestinal tract, heart, skeletal muscle, and respiratory tract. This antimicrobial peptide has potent activity against Gram-negative bacteria, Gram-positive positive bacteria, and yeast (6, 7). The mature peptide is arranged as three α-helices and two triple-stranded, antiparallel β-sheets (6, 8). Although the mechanisms for RNase 7's antimicrobial properties are not completely understood, its bactericidal activity has been linked to its capacity to permeate and disrupt the bacterial membrane, independent of its RNase activity (8). Of note, the flexible coil at the N terminus that contains two lysine residues have been shown to be critical for membrane disruption (8).
We have recently demonstrated the expression and relevance of RNase 7 in the human kidney and urinary tract (9). Antibacterial neutralization assays showed that urinary RNase 7 has potent antimicrobial properties against Gram-negative and Gram-positive uropathogenic bacteria. Generation of AMP fragments that contain distinct secondary structures such as α-helices has been used to gain insight into the structure-function activity relationship of other AMPs (10, 11). The objective of the present study was to generate RNase 7 fragments to identify the intrinsic functional domains of RNase 7 that influence its activity on uropathogenic Escherichia coli, Staphylococcus saprophyticus, and Proteus mirabilis.
MATERIALS AND METHODS
Bacterial strains and antibody.
Uropathogenic E. coli (UTI89) was grown in Luria broth (LB) medium and plated on LB agar. Uropathogenic S. saprophyticus (ATCC 15305) was grown in LB and plated on Trypticase soy agar with 5% sheep blood (Fisher Scientific, Pittsburgh, PA). Uropathogenic P. mirabilis (ATCC 7002) was grown in LB and plated on MacConkey II agar (Fisher Scientific). These organisms were chosen as the primary organisms that account for a majority of Gram-negative and Gram-positive urinary tract infections (UTIs) (12). Anti-RNase 7 antibody was generated in rabbits using recombinant human RNase 7 (Biomatik, Wilmington, DE).
Plasmid DNA construction.
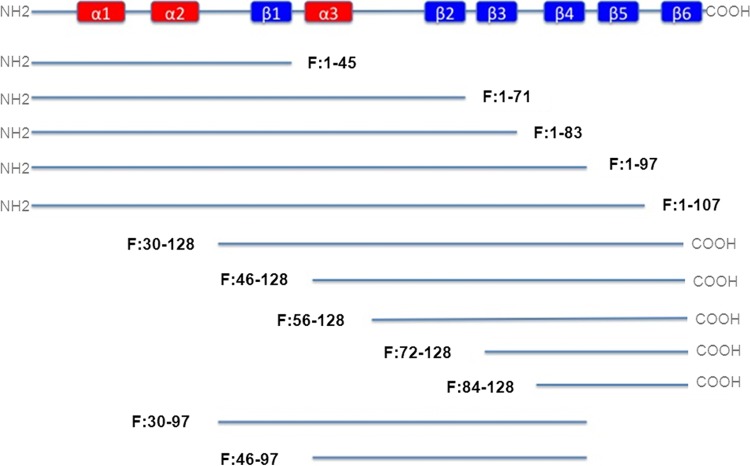
The full-length human RNASE7 sequence was generated from kidney cDNA library by PCR and cloned into E. coli expression vector pDEST17 (Invitrogen, Carlsbad, CA), which adds a six-residue histidine tag at the N terminus. Ten fragmentary constructs of RNase 7 were generated by PCR from full-length RNase 7 template and cloned into vector pDEST17. Each contained different numbers of RNase 7 secondary structures starting from the N or C terminus of RNase 7. Two expression vectors that contained middle fragments (neither the N terminus nor the C terminus) were constructed as well (Fig. 1). The sequences of each construct were confirmed by DNA sequencing.
Fig 1.
Representation of structural elements with N-terminal, C-terminal, and middle fragments. Fragment nomenclature example: RNase 7 fragment amino acids 1 to 45 = F:1-45.
Recombinant peptide expression and purification.
Recombinant peptides were expressed and purified as described previously (13). Briefly, each construct was transformed into E. coli BL21 AI (Invitrogen) to allow for l-arabinose-inducible expression. Bacteria were grown in 2-liter cultures to mid-log phase, and peptide expression was induced by l-arabinose for 3 h. The cells were harvested, and the pellets were resuspended in 30 ml of start buffer (20 mM sodium phosphate [pH 7.4], 500 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail [Sigma, St. Louis, MO]) and lysed by sonication. The cell lysate was pelleted at 13,000 rpm for 20 min. The supernatant was applied to a Ni2+ charged Hi-Trap Chelating HP column (General Electric, Piscataway, NJ). After being washed with wash buffer (20 mM NaPO4 [pH 7.4], 500 mM NaCl) containing 20 mM imidazole and then 50 mM imidazole, the recombinant peptide was eluted with 500 mM imidazole (13). Purified peptides were dialyzed against DN(0) buffer (25 mM Tris [pH 7.0], 0.1 mM EDTA, 10% glycerol), and the peptide concentrations were determined with a Bradford protein assay (Bio-Rad, Hercules, CA) and confirmed with a bicinchoninic acid assay (Pierce, Rockford, IL). Each fragment was subjected to SDS-PAGE and identified by immunoblotting with an anti-RNase7 antibody at a titer of 1:2,000.
Antimicrobial kill assay.
Antimicrobial activities of the recombinant peptides were estimated with a microdilution assay. Briefly, the test bacteria (104 CFU) were incubated with various concentrations of RNase 7, and each fragment was incubated in 50 μl of 0.1× phosphate-buffered saline (PBS) for 3 h at 37°C. After incubation on agar plates overnight at 37°C, the number of CFU at each peptide concentration was determined. Using untreated bacterial aliquots as a baseline, the MIC was defined as the lowest concentration that inhibits the growth of 90% of the inoculation, and the minimum bactericidal concentration (MBC) was defined as the lowest concentration at which <99.99% of the initial inoculum is viable.
Bacterial viability assay.
To confirm the accuracy of our MIC/MBC data, we performed cell death assay using Sytox Green (Invitrogen), which selectively labels nucleic acids of cells with compromised membranes of dead/dying cells. Overnight cultures of bacteria were centrifuged at 5,000 rpm for 5 min. Pellets were then washed with PBS (pH 7.4) and resuspended in PBS to an optical density at 600 nm (OD600) of 0.5 for E. coli (UTI89) and OD600 of 0.8 for S. saprophyticus. According to the manufacturer's instructions, 90 μl of each bacteria strain was incubated with 100 μl of 2 μM Sytox Green for 15 min. Portions (10 μl) of peptides at various concentrations were added for 3 h at 37°C in a 96-well plate. Compromised bacterial membranes of dead/dying cells will be labeled while lives cells are not labeled as noted by Invitrogen. Fluorescence was measured using 485- and 520-nm filters for excitation and emission wavelengths, respectively. Untreated bacteria were used as a 100% live control, and the minimum fluorescence was designated as the Fmin value. To elicit complete killing, bacteria were treated with 70% ethanol for 1 h. This experimental group's fluorescence is referred to as the Fmax value. The percentage of cell death at each concentration for each fragment was calculated as follows: [(Ffragment − Fmin)/(Fmax − Fmin)] × 100.
To obtain images of intact versus disrupted membranes of dying/dead cells, a Live/Dead BacLight bacterial viability kit (Invitrogen) was used. E. coli and S. saprophyticus were grown at 37°C to the mid-log phase, centrifuged at 5,000 rpm for 5 min, and resuspended in water to an OD600 of 0.2. Bacteria were incubated in a 1:1 mixture of Live/Dead BacLight bacterial viability assay. A bacterial aliquot of 50 μl was mixed with recombinant RNase 7 or recombinant fragments to achieve a final concentration of 1 μM. Ten microliters of this mixture was added to poly-l-lysine-coated microscope slides (Polysciences, Inc., Warrington, PA), and images were obtained in triplicate using a Zeiss LSM 710 confocal laser scanning microscope (Carl Zeiss LLC, Thornwood, NY).
Urinary condition functional assays.
Because the urinary environment can undergo significant variations in salt concentration, ionic composition, and pH, an antimicrobial/bacteriostatic assay was performed to evaluate whether pH, calcium, or ionic strength affects the activity of full-length RNase 7 or its fragments. We chose the highest-activity fragment F:1-97 and the lowest-activity fragment F:72-128 for these experiments. The bacteria were inoculated in 1% peptone water and grown to log phase at an OD600 of 0.5. The bacteria were diluted to 1:10 in a 96-well flat-bottom plate (Thermo Scientific/Nunc, Worcester, MA), and 1 μl was added to 100 μl of 1% peptone water with various pH levels or CaCl2 or NaCl concentrations that corresponded to the physiologic range present in urine. To each well, 2 μM RNase 7 or its fragments were added.
Bacterial growth was monitored using an absorbance microplate reader (BioTek Instruments, Winooski, VT) at a final volume of 110 μl. The turbidity of the culture was measured and recorded at t = 0 and every 20 min thereafter for 10 h using the absorbance at 600 nm (OD600). The values of OD600 values were plotted against times to generate growth curves.
CD spectroscopy.
To determine whether recombinant RNase 7 peptides retained secondary structure, circular dichroism (CD) experiments were performed by The Protein Facility at Iowa State University. Briefly, using 0.5-mm path-length cuvettes with 0.5 mg of each peptide/ml in 10 mM Tris-HCl (pH 7.6), the CD spectra for the full-length RNase 7, F:1-97, F:56-128, and F:72-128 were recorded from 190 to 250 nm using a wavelength step of 0.1 nm. The curves were fitted and analyzed using CD-FIT program. All peptide fragments demonstrated retention of some secondary structure with α-helices and β-sheets present in each peptide.
Statistical analyses.
To determine whether the inhibitory (MIC) and killing (MBC) values presented in Table 1 were statistically significantly different compared to full-length peptide, all raw data were subjected to statistical analyses. Briefly, time-to-event analysis was performed for each peptide fragment for each MIC and MBC data point. Chi-square values with Bonferroni adjustments to P values to account for multiple testing for each fragment were determined using SAS software (SAS Institute, Cary, NC). For all fragments, P values of <0.05 were considered statistically significant.
Table 1.
Fragment MICs and MBCsa
| Peptide or fragment |
E. coli |
S. saprophyticus |
P. mirabilis |
|||
|---|---|---|---|---|---|---|
| MIC (μM) | MBC (μM) | MIC (μM) | MBC (μM) | MIC (μM) | MBC (μM) | |
| RNase 7 | 0.26 | 0.53 | 0.22 | 0.44 | 0.88 | 0.88 |
| C-terminal fragments | ||||||
| F:30-128 | 0.52 | 2.07 | 0.88* | 0.88 | >3.5† | >3.5† |
| F:46-128 | 0.34 | 0.68 | 0.22 | 1.75* | 3.5 | >3.5† |
| F:56-128 | 0.83 | 1.65 | 0.22 | 1.75* | >7† | >7† |
| F:72-128 | 3.5† | 7* | 3.5† | >7† | >3.5† | >3.5† |
| F:84-128 | 1.92† | >3.5† | 1.75 | 3.5* | >3.5† | >3.5† |
| N-terminal fragments | ||||||
| F:1-45 | 0.49 | 0.98 | 0.13 | 0.5 | >7† | >7† |
| F:1-71 | 1.93† | 3.86 | 0.23 | >0.92* | 0.44 | 0.88 |
| F:1-83 | 0.43 | 0.87 | 0.34 | 0.69 | 0.44 | 0.88 |
| F:1-97 | 0.07 | 0.14 | 0.04 | 0.08 | 0.44 | 0.88 |
| F:1-107 | 0.57 | 0.57 | 0.49 | >1.96* | 0.44 | 0.44 |
| Middle fragments | ||||||
| F:30-97 | 0.44 | 0.88 | 0.44 | 1.75* | >3.5† | >3.5† |
| F:46-97 | 0.88 | 1.75 | 0.44 | 1.75 | 3.5 | >3.5† |
*, P < 0.05 compared to full length;
, P< 0.001 compared to full length.
Fragment AMP property determination.
To determine whether individual amino acid properties were associated with fragment antimicrobial activity, the net charge and hydrophobicity were calculated with the Collection of Antimicrobial Peptides Comprehensive Antimicrobial Peptide Database AMP tool (14). The mean and maximum hydrophobic moment, a measure of amphipathicity, was calculated with H-moment software (Sanger Centre, Cambridge, United Kingdom) (15).
RESULTS
Recombinant RNase 7 has potent antimicrobial activity.
Antimicrobial activity of recombinant RNase 7 was determined against uropathogenic Gram-negative E. coli and P. mirabilis and uropathogenic Gram-positive S. saprophyticus (Table 1). The MICs of full-length RNase 7 were 0.26 μM for E. coli, 0.88 μM for P. mirabilis, and 0.22 μM for S. saprophyticus. Our results are consistent with previous work demonstrating that RNase 7 possesses potent antimicrobial activity against multiple uropathogenic bacterial species at micromolar concentrations (6, 8).
C-terminal amino acids are not the primary determinant of RNase 7 antimicrobial activity.
The C-terminal fragments generally had activity that was reduced compared to full-length RNase 7 (Table 1). Two fragments, F:46-128 and F:56-128, had MICs similar to that of the full-length protein against S. saprophyticus. Fragments devoid of the initial 71 amino acids (F:72-128 and F:84-128) demonstrated markedly diminished activity. The MIC and MBCs of these fragments were 4- to 16-fold higher than the full-length protein against E. coli and S. saprophyticus (Table 1). None of the C-terminal fragments had activity against P. mirabilis (Table 1). Furthermore, the C-terminal fragments showed decreased E. coli and S. saprophyticus membrane permeabilization and subsequent death compared to full-length peptide (Fig. 2 and see Fig. S1 in the supplemental material).
Fig 2.
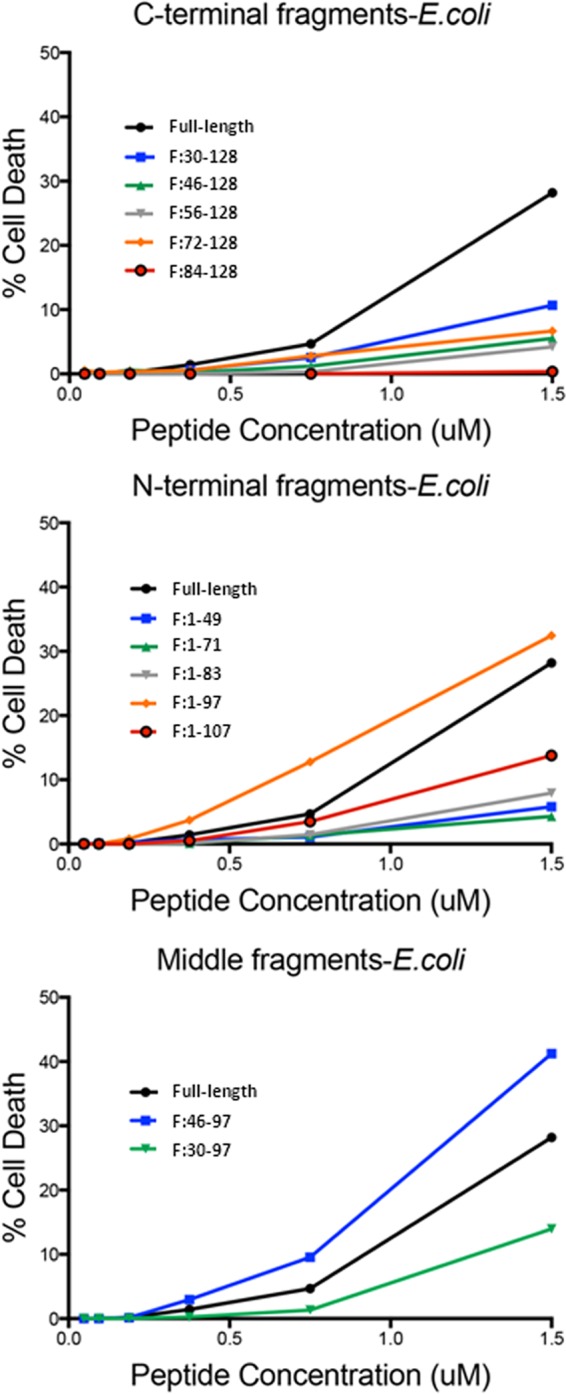
Bacterial cell death assay results for C-terminal (top panel), N-terminal (middle panel), and middle (bottom panel) fragments for E. coli. (Top) C-terminal fragments result in reduced membrane permeabilization and death for E. coli compared to full-length protein. (Middle) The N-terminal fragment F:1-97 has improved activity compared to the full-length protein. (Bottom) The middle fragment F:46-97 demonstrates enhanced permeabilization and death compared to the full-length protein.
N-terminal amino acids are critical for RNase 7 antimicrobial activity.
When compared to the C-terminal fragments, the N-terminal fragments had much greater antimicrobial activity. The N-terminal fragments have a range of activity against E. coli and S. saprophyticus compared to full-length protein (Table 1). Generally, fragments showed variable activity compared to full-length protein with most demonstrating a statistically insignificant altered activity against E. coli and S. saprophyticus (Table 1). Interestingly, F:1-97 had significantly increased activity against E. coli and S. saprophyticus, with MICs and MBCs that were at least 4-fold lower than the full-length protein. Conversely, the F:1-71 fragment had dramatically decreased activity against E. coli, with a 7-fold-higher MIC and MBC compared to the full-length protein. Of note, F:1-71 had the same activity against S. saprophyticus as the full-length protein.
Upon evaluating the antimicrobial activity of the N-terminal fragments against P. mirabilis, all N-terminal fragments except for F:1-45 had similar or increased activity compared to full-length RNase 7. Unlike its activity against the other two uropathogenic bacteria, F:1-45 demonstrated no activity at high concentrations against P. mirabilis.
The differential activities of both F:1-71 and F:1-97 were confirmed with bacterial viability assays (Fig. 2 and see Fig. S1 in the supplemental material). All N-terminal fragments permeabilized and killed E. coli and S. saprophyticus to a similar extent as full-length protein with the exception of F:1-97 (Fig. 2 and see Fig. S1 in the supplemental material). Comparable to its increased antimicrobial activity, fragment F:1-97 demonstrated increased membrane permeabilization and subsequent death against both strains. Finally, fragment F:1-71 demonstrated a permeability similar to that of the full-length protein against S. saprophyticus but not against E. coli, which is consistent with its antimicrobial profile. All permeabilization and antimicrobial assays were confirmed with Live/Dead assays (Fig. 3).
Fig 3.
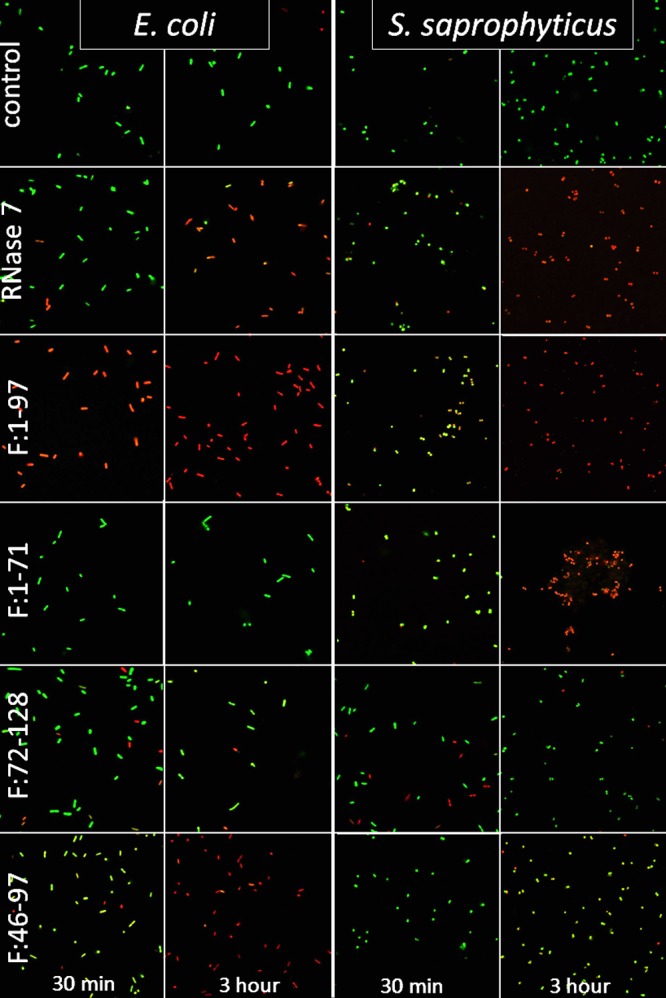
Intact cells are labeled green, and cells with disrupted membranes are labeled red. Membrane viability is demonstrated at 30 min and at 3 h for E. coli (left two columns) and S. saprophyticus (right two columns) after exposure to the negative control, RNase 7 (full-length protein), F:1-97, F:1-71, F:46-97, and F:72-128. All proteins were added at 1 μM. Full-length protein demonstrates membrane disruption against E. coli and S. saprophyticus at 3 h but minimally at 30 min. Fragment F:1-97 lysed most of the E. coli and S. saprophyticus within 30 min and all of it by 3 h. F:1-71 membrane disruption against S. saprophyticus was completed before 3 h; however, the E. coli membranes were largely intact even at 3 h. Fragment F:46-97 demonstrated that marked membrane permeabilization of E. coli was present by 3 h, but the fragment did not appear to be effective against S. saprophyticus. F:72-128 demonstrated negligible membrane disruption against microbes.
S. saprophyticus activity is mediated by a different middle fragment domain than is E. coli activity.
The secondary structures of AMPs have been implicated in determining/influencing antimicrobial activity. RNase 7 is known to have three α-helices and two triple-stranded, antiparallel β-sheets (Fig. 1). Because we identified differential activity between N-terminal fragments and also noted fragment F:1-97's increased activity that could possibly be correlated to secondary structures, we constructed two more fragments—F:30-97 and F:46-97—that were devoid of N-terminal and C-terminal domains (Fig. 1). These fragments were constructed to incorporate the first four β-strands (F:30-97) and β-strands 2 to 4 (F:46-97). The MICs and MBCs of these fragments were 2-fold higher than for the full-length protein. Fragment F:46-97 presents potent membrane penetrating activity against E. coli (Fig. 2C) and negligible activity against S. saprophyticus (Fig. 2F). Fragment F:30-97 penetrates the membrane of both strains with less activity compared to full-length RNase 7. These findings were verified with Live/Dead assays (Fig. 3).
Full-length RNase 7 activity against P. mirabilis seems to be exclusively microbicidal.
The full-length RNase 7 peptide had MIC and MBC values that were identical, indicating only concentrations of peptide that killed were effective against P. mirabilis. Because a lower concentration did not inhibit 90% growth (MIC90) compared to the 99% killing (MBC90), P. mirabilis appears to be resistant to the inhibitory characteristics RNase 7 possesses against the other uropathogens. Furthermore, the N-terminal fragments (other than F:1-45) did demonstrate an MIC at a lower concentration compared to MBC, indicating that the C terminus may play a role in the ability of P. mirabilis to be resistant to the inhibitory effects of RNase 7 (Table 1).
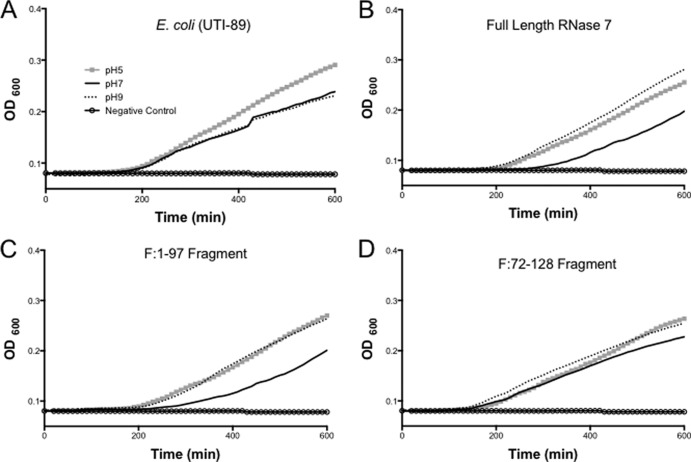
The antimicrobial activities of RNase 7 and fragments are diminished at high pH.
E. coli had the most robust growth in acidic pH of 5, whereas S. saprophyticus and P. mirabilis grew best in alkaline pH 9 media (Fig. 4A and see Fig. S2A and S5A in the supplemental material). Full-length RNase 7 maintained high activity at pH 5 against S. saprophyticus and P. mirabilis, while its activity was optimal at pH 7 against E. coli (Fig. 4B; see also Fig. S2B and S5B in the supplemental material). Fragments F:1-97 (high-activity fragment) and F:72-128 (low-activity fragment) demonstrated similar patterns with a few exceptions. S. saprophyticus actually grew better with the addition of F:72-128 at pH 7 and 9 (Fig. 2D) compared to control conditions (Fig. 2A).
Fig 4.
E. coli growth curves after treatment with different recombinant peptides at various pH levels. (A) E. coli growth is optimum at pH 5 (gray squares) compared to pH 7 and 9. (B) pH 9 (dotted line) decreases full-length RNase 7 activity, as evidenced by enhanced growth compared to that seen at pH 7. (C) F:1-97 has an activity pattern comparable to full-length peptide at various pHs. (D) F:72-128 demonstrates similar activity against E. coli at all pHs.
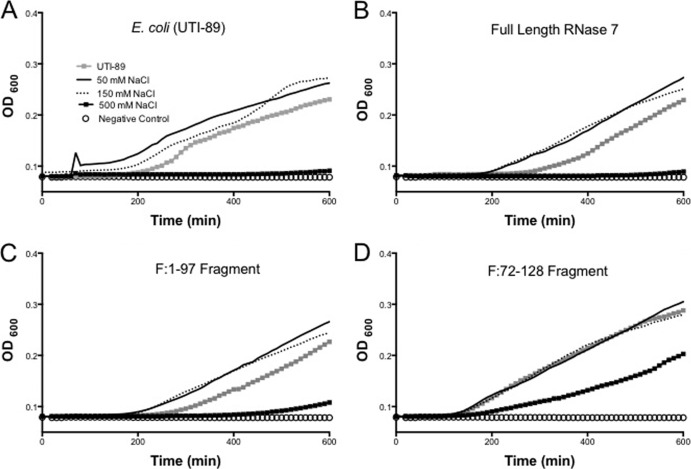
The sodium chloride concentration inhibits activity of RNase 7 against certain uropathogens.
For complete results of the effect of NaCl concentrations, please refer to Fig. 5 and see Fig. S3 and S6 in the supplemental material. The NaCl concentration had little effect on peptides against E. coli (Fig. 5). Full-length RNase 7 and fragment F:1-97 activity against S. saprophyticus was diminished in direct proportion to NaCl concentration (see Fig. S3B and S3C in the supplemental material). Full-length activity against P. mirabilis was diminished significantly with additional of any NaCl titer (see Fig. S6B in the supplemental material). Interestingly, the inhibitory effect of 500 mM NaCl against E. coli was diminished by the addition of F:72-128 (Fig. 5D).
Fig 5.
E. coli growth curves after treatment with different recombinant peptides at various NaCl titers and with no added NaCl (gray squares). (A) E. coli demonstrated improved growth with 50 and 150 mM NaCl. Growth was completely inhibited with NaCl at 500 mM. (B) Full-length RNase 7 activity was not different at 50 and 150 mM NaCl. Bacterial growth was enhanced at low concentrations of NaCl, much like the conditions with no additional peptide (top left panel). (C) F:1-97 has activity pattern comparable to full-length peptide at various NaCl concentrations. (D) F:72-128 has similar activity against E. coli at all NaCl concentrations except 500 mM. Interestingly, growth is enhanced at 500 mM NaCl with F:72-128 compared to all other conditions.
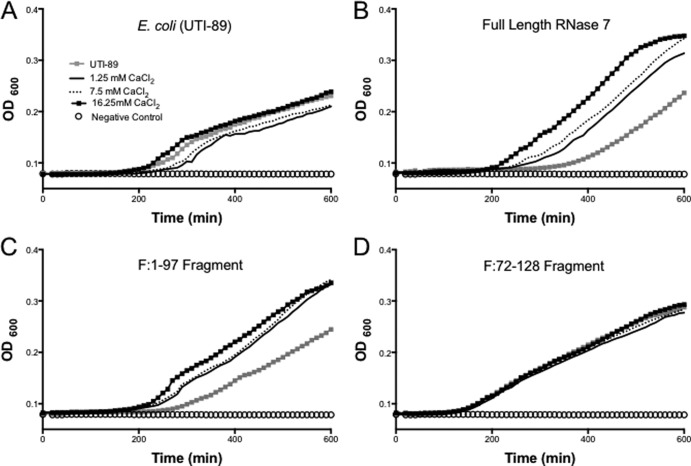
The calcium chloride concentration diminishes the activity of RNase 7 against all uropathogens.
High concentrations of CaCl2 (7.5 and 16.25 mM) significantly reduced full-length RNase 7 activity against all uropathogens (Fig. 6B; see also Fig. S4B and S7B in the supplemental material). The potent fragment F:1-97 was also sensitive to CaCl2 concentrations, with the exception of unchanged activity under all conditions against P. mirabilis (Fig. 6C and see Fig. S4C and S7C in the supplemental material). Interestingly, S. saprophyticus grew significantly better with CaCl2 compared to conditions without CaCl2 (see Fig. S4A in the supplemental material).
Fig 6.
E. coli growth curves after treatment with different recombinant peptides at various CaCl2 titers and with no added CaCl2 (gray squares). (A) E. coli showed similar growth under all conditions. (B) Full-length RNase 7 activity is inversely related to CaCl2 concentration. (C) F:1-97 has activity pattern comparable to full-length peptide at various CaCl2 concentrations. (D) F:72-128 has similar activity against E. coli at all CaCl2.
AMP fragment activity is not entirely dependent on the net charge, hydrophobicity, and hydrophobic moment.
The fragment primary AMP properties are presented in Table 2. The net charge is comparable for the N-terminal and C-terminal fragments despite the lower activity of the C-terminal fragments, and charge is not associated with antimicrobial activity. Within the N-terminal fragments, F:1-107 had a higher net charge than F:1-97 and yet had lower antimicrobial activity. The hydrophobicity was similar between C-terminal and N-terminal fragments despite activity differences. The mean hydrophobic moment was similar between fragments and did not correlate with activity. The maximum hydrophobic moment was higher in the N-terminal fragments than in the C-terminal fragments, which correlates with the overall activity. On the other hand, within the N-terminal fragments the maximum hydrophobic moment was the same within all fragments despite the differential activities. Therefore, this factor may play some role in the overall activity, but it does not completely explain the antimicrobial activity.
Table 2.
Fragment AMP propertiesa
| Peptide or fragment | Net charge | Hydrophobicity | μH |
|
|---|---|---|---|---|
| Maximum | Mean | |||
| RNase 7 | 16 | –0.93 | 0.727 | 0.266 |
| C-terminal fragments | ||||
| F:30-128 | 12 | –0.9 | 0.602 | 0.283 |
| F:46-128 | 9 | –0.88 | 0.468 | 0.229 |
| F:56-128 | 10 | –1.03 | 0.468 | 0.234 |
| F:72-128 | 8 | –0.94 | 0.415 | 0.194 |
| F:84-128 | 7 | –1.23 | 0.415 | 0.23 |
| N-terminal fragments | ||||
| F:1-45 | 7 | –1.16 | 0.727 | 0.271 |
| F:1-71 | 8 | –1.03 | 0.727 | 0.318 |
| F:1-83 | 9 | –0.84 | 0.727 | 0.299 |
| F:1-97 | 13 | –1.02 | 0.727 | 0.28 |
| F:1-107 | 15 | –0.97 | 0.727 | 0.273 |
| Middle fragments | ||||
| F:30-97 | 9 | –1 | 0.602 | 0.275 |
| F:46-97 | 6 | –0.99 | 0.498 | 0.233 |
μH, hydrophobic moment as measure of amphipathicity.
DISCUSSION
Previous studies have demonstrated that RNase 7 is an important AMP in a variety of epithelial tissues, including the urinary tract (9). In the present study, we demonstrate the antimicrobial properties of full-length RNase 7 and recombinant fragments against uropathogenic bacteria. Furthermore, our results support published findings suggesting that the N terminus of RNase 7 was critical to its antimicrobial activity (8). Subsequently, our results expand current knowledge and identify critical regions of the RNase 7 peptide that are crucial to maintaining its antimicrobial properties. In addition, we have identified regions of the RNase 7 molecule that confer differential activity against Gram-positive and Gram-negative uropathogens. Interestingly, the antimicrobial properties do not seem significantly linked to peptide charge, amphipathicity or hydrophobicity. Finally, because the urinary environment is so dynamic, we also evaluated the antimicrobial activity of the RNase 7 peptide under physiologic ranges of pH, osmolarity, and ionic concentration. Our results demonstrate that alterations in these parameters can alter RNase 7's bacteriostatic properties.
To establish the overall structure-function relationship of RNase 7, we have systematically dissected the secondary structures of RNase 7 by creating various recombinant fragments that incorporate various elements of these structures and include either the N terminus or the C terminus. The C terminus of RNase 7 does not appear to contribute significantly to the overall antimicrobial activity. Only the longest C-terminal fragments had activity against S. saprophyticus that were comparable to the full-length peptide. Of note, F:30-128, F:46-128, and F:56-128 displayed preservation of inhibitory activity but diminished killing capability. Furthermore, the fragments that did not include the first 71 amino acids had markedly reduced function, suggesting that this region is extremely critical to overall antimicrobial function. To date, the role of the C-terminal fragment is not completely clear. We speculate that this part of the molecule may be important in peptide stability in eukaryotic cells and possibly even important in trafficking to the cell surface or secretory vesicles during infection. This preservation of inhibitory activity (low MIC) with loss of killing activity (relatively high MBC) against E. coli and S. saprophyticus coupled with the fact that only fragments lacking the C terminus had inhibitory activity against P. mirabilis (MIC = MBC for all other peptides) suggests that the C terminus may be critical for inhibiting microbial growth and division but not critical to actual killing.
Our study documents the importance of the N-terminal portion of RNase 7 in antimicrobial functions. These results confirm other reports that the N-terminal portion of RNase 7 is critical to bacterial killing (8). Interestingly, fragment F:1-71 has markedly reduced activity against E. coli but not against S. saprophyticus and P. mirabilis bacteria compared to the full-length peptide. The addition of 12 amino acids to this fragment (F:1-83) partially restores its antimicrobial activity. These additional 12 amino acids add another secondary motif. The smaller N-terminal fragment (F:1-45) has E. coli activity similar to that of F:1-83. Further studies are necessary to elucidate the mechanisms of fragment F:1-71's decreased antimicrobial activity since this information will offer significant insight into the structure-function relationships of AMPs.
Secondary structure appears to play a critical role in determining the functional properties of RNase 7. Our data suggest that the conservation of β-sheets 1, 3, and 4 is critical to overall antimicrobial activity. Disrupting the peptide domain located at amino acids 71 and 72 leads markedly reduced antimicrobial activity presumably from disrupting the interaction between β-sheets 1 and 3. Fragments F:1-71 and F:72-128 have MICs and MBCs that significantly differ from the fragments that include the amino acids 71 and 72. Furthermore, F:84-128, which does not contain the N-terminal elements or these critical amino acids, has poor antimicrobial function against all uropathogens.
We demonstrate the unique differential activity of recombinant peptides under different conditions against uropathogenic bacteria. By altering the pH and the NaCl and CaCl2 concentrations, we gained additional insight into the specific conditions that are critical to maximizing antimicrobial activity as we potentially consider these peptides as therapeutics. For example, patients with hypercalciuria are at increased risk for UTIs (16). By avoiding excessive sodium excretion in the urine through dietary restriction and avoiding pH extremes in these patients, perhaps the inhibitory effects of calcium on RNase 7's antimicrobial activity can be minimized.
Consequently, hypercalciuric patients with “ideal” urinary conditions may be less susceptible to UTIs by having optimal antimicrobial peptide activity. Further, optimizing these patients' urinary conditions during infections could help with the clearance of uropathogens if infections do occur. Clearly, further investigations are needed to elucidate these relationships and could serve as the foundation for clinical studies in the future.
Another important concept in AMP activity involves peptide properties such as the net charge, isoelectric point, and hydrophobicity. Net charge has been implicated in cationic AMP attraction to the negatively charged bacterial membrane. Classically, AMPs with a higher net positive charge have been associated with increased antimicrobial activity. Our recombinant peptides did not demonstrate any strong correlation between net charge and antimicrobial activity or membrane permeability (Tables 1 and 2). Furthermore, N-terminal fragments and full-length RNase 7 had the highest amphipathicity values, but their values did not differ from one another. These findings suggest that peptide properties may contribute to antimicrobial activity but do not dictate specific activities.
We acknowledge certain limitations of the present study that warrant further investigations in future projects. Our current methodology does not allow us to isolate peptides smaller than 40 amino acids. The studied fragments contained a six-histidine tag (His tag). Although the activity of full-length RNase 7 did not appear to be affected by the His tag, we cannot exclude the possibility that the activity of smaller fragments may have been altered. Areas for future investigations include the creation of His-tag-free fragments, the commercial creation of smaller oligopeptides, the evaluation of the tertiary structure of fragments, and the identification of the precise mechanisms, in addition to membrane disruption, which is responsible for RNase 7 antimicrobial activity.
The oligonucleotide peptides examined here were extracted from cytoplasmic extracts as soluble proteins. The full-length RNase 7 presents potent antimicrobial activity, which is consistent with previously studies by other research teams (6, 8). Without further analysis, we cannot exclude the possibility of portions of unfolded/misfolded oligopeptides existing after our isolation and purification process. However, because they are soluble, we presumed that there are only small portions of unfolded/misfolded oligopeptides. Further, because all experiments were run in triplicate with consistent results, we assume that the same purification methodology would result in the same proportions of folded and unfolded protein in all experiments. Finally, we confirmed that some secondary structure is preserved by performing CD spectroscopy on select recombinant peptides. Future studies can include more sophisticated imaging to ensure that secondary and tertiary structures are maintained. Finally, many of the methodologies presented here offer many insights into the antimicrobial activity of RNase 7. Unfortunately, because of the inherent limitations of each individual assay, we do need to exhibit caution when comparing results between different methodologies or experiments.
Increasing evidence is implicating mechanisms other than microbial membrane disruption in the activity of other AMPs. These mechanisms may include the alteration of cytoplasmic membrane septum formation, cell wall synthesis, nucleic acid synthesis, and protein synthesis, or mechanisms that inhibit enzymatic activity (17). Our findings indicate that multiple mechanisms are potentially responsible for the potent antimicrobial activity of RNase 7. Because Gram-positive and Gram-negative bacteria have very different cell walls, we cannot exclude that the differential activity is secondary to differences in attachment and/or transport through the cell wall into the cytoplasm. Further studies are required to elucidate the specific mechanisms that are outside the scope of the present study. To the best of our knowledge, the finding that different domains within an AMP are responsible for S. saprophyticus, P. mirabilis, and E. coli permeability and antimicrobial activity is novel.
In summary, RNase 7 is an extremely potent antimicrobial peptide that plays a significant role in maintaining the sterility of the kidney and urinary tract. We have previously demonstrated that uropathogenic bacteria are susceptible to RNase 7 at very low concentrations. The susceptibility and broad-spectrum activity described here makes this molecule an interesting candidate to develop as a therapeutic to treat UTIs. The findings presented here will serve as the foundation for ongoing laboratory investigations on the use of recombinant RNase 7 as a future therapeutic against resistant bacterial infections.
Supplementary Material
Footnotes
Published ahead of print 26 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01378-12.
REFERENCES
- 1. Almeida PF, Pokorny A. 2009. Mechanisms of antimicrobial, cytolytic, and cell-penetrating peptides: from kinetics to thermodynamics. Biochemistry 48:8083–8093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rathinakumar R, Walkenhorst WF, Wimley WC. 2009. Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: the importance of interfacial activity. J. Am. Chem. Soc. 131:7609–7617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wimley WC, Hristova K. 2011. Antimicrobial peptides: successes, challenges and unanswered questions. J. Membr. Biol. 239:27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yeaman MR, Yount NY. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27–55 [DOI] [PubMed] [Google Scholar]
- 5. Jiang Z, Vasil AI, Hale JD, Hancock RE, Vasil ML, Hodges RS. 2008. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Biopolymers 90:369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harder J, Schroder JM. 2002. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 277:46779–46784 [DOI] [PubMed] [Google Scholar]
- 7. Torrent M, Badia M, Moussaoui M, Sanchez D, Nogues MV, Boix E. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 277:1713–1725 [DOI] [PubMed] [Google Scholar]
- 8. Huang YC, Lin YM, Chang TW, Wu SJ, Lee YS, Chang MD, Chen C, Wu SH, Liao YD. 2007. The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity. J. Biol. Chem. 282:4626–4633 [DOI] [PubMed] [Google Scholar]
- 9. Spencer JD, Schwaderer AL, Dirosario JD, McHugh KM, McGillivary G, Justice SS, Carpenter AR, Baker PB, Harder J, Hains DS. 2011. Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney Int. 80:174–180 [DOI] [PubMed] [Google Scholar]
- 10. Ahn HS, Cho W, Kang SH, Ko SS, Park MS, Cho H, Lee KH. 2006. Design and synthesis of novel antimicrobial peptides on the basis of alpha helical domain of Tenecin 1, an insect defensin protein, and structure-activity relationship study. Peptides 27:640–648 [DOI] [PubMed] [Google Scholar]
- 11. Soliman W, Wang L, Bhattacharjee S, Kaur K. 2011. Structure-activity relationships of an antimicrobial peptide plantaricins from two-peptide class IIb bacteriocins. J. Med. Chem. 54:2399–2408 [DOI] [PubMed] [Google Scholar]
- 12. Hovelius B, Mardh PA. 1984. Staphylococcus saprophyticus as a common cause of urinary tract infections. Rev. Infect. Dis. 6:328–337 [DOI] [PubMed] [Google Scholar]
- 13. Koten B, Simanski M, Glaser R, Podschun R, Schroder JM, Harder J. 2009. RNase 7 contributes to the cutaneous defense against Enterococcus faecium. PLoS One 4:e6424 doi:10.1371/journal.pone.0006424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas S, Karnik S, Barai RS, Jayaraman VK, Idicula-Thomas S. 2010. CAMP: a useful resource for research on antimicrobial peptides. Nucleic Acids Res. 38:D774–D780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 16. Biyikli NK, Alpay H, Guran T. 2005. Hypercalciuria and recurrent urinary tract infections: incidence and symptoms in children over 5 years of age. Pediatr. Nephrol. 20:1435–1438 [DOI] [PubMed] [Google Scholar]
- 17. Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.