Abstract
Metallo-β-lactamases (MBLs) have rapidly disseminated worldwide among clinically important Gram-negative bacteria and have challenged the therapeutic use of β-lactam antibiotics, particularly carbapenems. The blaGIM-1 gene, encoding one such enzyme, was first discovered in a Pseudomonas aeruginosa isolate from 2002 and has more recently been reported in Enterobacteriaceae. Here, we present crystal structures of GIM-1 in the apo-zinc (metal-free), mono-zinc (where Cys221 was found to be oxidized), and di-zinc forms, providing nine independently refined views of the enzyme. GIM-1 is distinguished from related MBLs in possessing a narrower active-site groove defined by aromatic side chains (Trp228 and Tyr233) at positions normally occupied by hydrophilic residues in other MBLs. Our structures reveal considerable flexibility in two loops (loop 1, residues 60 to 66; loop 2, residues 223 to 242) adjacent to the active site, with open and closed conformations defined by alternative hydrogen-bonding patterns involving Trp228. We suggest that this capacity for rearrangement permits GIM-1 to hydrolyze a wide range of β-lactams in spite of possessing a more constrained active site. Our results highlight the structural diversity within the MBL enzyme family.
INTRODUCTION
β-Lactams have been the most important group of antibiotics used to treat infections (1, 2). However, the dissemination of β-lactamases, hydrolytic enzymes that inactivate β-lactams through cleavage of the amide bond of the β-lactam ring, is threatening their clinical longevity (3–5). Production of β-lactamases has become the most common β-lactam resistance mechanism in bacteria, particularly in opportunistic Gram-negative species. Metallo-β-lactamases (MBLs) hydrolyze all classes of β-lactams except monobactams and are not affected by clinically available β-lactamase inhibitors. Acquired MBL genes are often located on mobile genetic elements (plasmids and transposons) along with other antibiotic resistance genes, potentially resulting in the spread of multidrug resistance. Acquired MBLs reported so far include IMPs, VIMs, NDMs, SPM-1, SIM-1, KHM-1, GIM-1, DIM-1, TMB-1, KHM-1, SMB-1, FIM-1 and AIM-1 (4, 6–9).
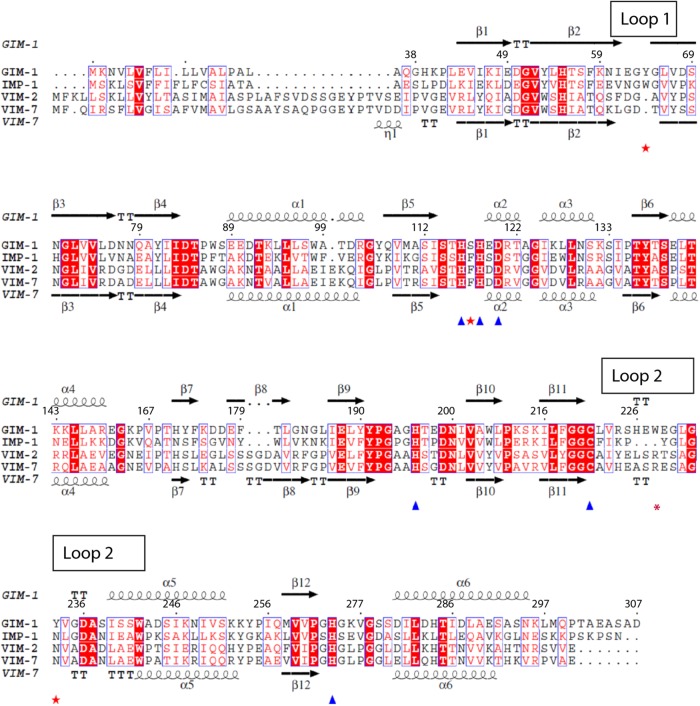
The blaGIM-1 gene was first identified on a 24-kb nontransferable plasmid in 2002 in clinical Pseudomonas aeruginosa isolates from Dusseldorf, Germany (10). blaGIM-1 is embedded in a class 1 integron as a gene cassette and often is accompanied by various aminoglycoside resistance genes. Recent data show that blaGIM-1 has now moved into Enterobacteriaceae (11), and it has also been reported in different clones of multidrug-resistant P. aeruginosa isolates from Germany (12). The reported spread into Serratia marcescens indicates a potential transmission of mobile genetic elements carrying blaGIM-1 between different Gram-negative species. Within the accepted MBL classification scheme, GIM-1 belongs to subclass B1 and most closely resembles the IMP enzymes (39 to 43% sequence identity) while showing significant relatedness to the VIM (28% to 31% identity), NDM (28 to 29% identity), and SPM-1 (28% identity) enzymes. The GIM-1 sequence has some unusual features, within the MBL H116xH118xD120 motif (where x is any residue), containing residues Ser117 and Glu119 that are not frequently found at these positions in other B1 MBLs (10) (Fig. 1).
Fig 1.
Sequence alignment and secondary structure elements of GIM-1 compared to IMP-1, VIM-2, and VIM-7, where the latter also display secondary structure elements (PDB 2Y87). The Zn2+-coordinating residues are marked with triangles and important GIM-1 residue determinants with stars. This figure was generated by EsPript.
Kinetic characterization of GIM-1 revealed a broad activity profile with no clear preference between penicillin, cephalosporin, and carbapenem substrates. Compared to IMP-1, VIM-2, and SPM-1, GIM-1 has reduced affinity (Km) for imipenem, cefoxitin, and cefepime (10). GIM-1 is also unusual among the MBLs in hydrolyzing meropenem more efficiently than imipenem (10).
MATERIALS AND METHODS
Cloning, expression, and purification.
The blaGIM-1 open reading frame (GenBank accession no. Q704V1) beginning at Gln19 (Gln37 according to class B β-lactamase [BBL] numbering [13]), with additional nucleotides encoding an N-terminal hexa-His tag and tobacco etch virus (TEV) cleavage site, was cloned into the expression vector pDEST14 (Gateway Technology, Invitrogen). The following primers (Sigma-Genosys) were used for amplification of the gene: forward GIM-1 Gln (5′-CAT CAC CAT CAC CAT CAC GAA AAC CTG TAT TTC CAG GGA GCA CAG GGT CAT AAA CCG CTA GAA G-3′), reverse GIM-1 attB2 (5′-GGG GAC CAC TTT GTA CAA GAA AGC TGG GTC TTA ATC AGC CGA CGC TTC AGC GG-3′), and forward attB1-His (5′-G GGG ACA AGT TTG TAC AAA AAA GCA GGC TTC GAA GGA GAT AGA ACC ATG CAT CAC CAT CAC CAT CAC-3′). The blaGIM-1 gene was first amplified using forward GIM-1 Gln and reverse GIM-1 attB2 primers, followed by another amplification using forward attB1-His and reverse GIM-1 attB2. The resulting PCR product was used in the BP recombination reaction (to generate an entry clone) with the pDONR221 vector (Invitrogen Life Technologies, Great Britain) followed by transformation into Escherichia coli DH5α (Invitrogen). Sequencing (BigDye Terminator v3.1; Applied Biosystems) was performed to confirm recovery of the correct clone. The entry clone was transferred to the pDEST14 vector by the LR recombination reaction (to generate an expression clone) and transformed into in-house-modified E. coli BL21 Star (DE3) pLysS (Invitrogen) cells that also included the pRARE plasmid (Novagen) for expression and purification.
Expression was induced with 0.5 mM isopropyl-d-thiogalactopyranoside (IPTG) for 20 h at 20°C in Luria broth (LB) medium containing 100 μg/ml of ampicillin and 34 μg/ml of chloramphenicol. The soluble lysate after sonication was purified on a 1-ml His-Trap HP column in buffer A (50 mM HEPES, pH 7.2, and 100 μM ZnCl2), washed with 5% buffer B, and eluted with 5 to 100% buffer B (50 mM HEPES, pH 7.2, 100 μM ZnCl2, and 500 mM imidazole). About 6 mg of in-house-made His-tagged TEV protease was added to GIM-1-containing fractions, and the mixture was dialyzed overnight at 4°C (10-kDa molecular-size-cutoff SnakeSkin [Pierce]) with stirring against 10 mM HEPES, pH 7.2, 100 mM NaCl, 1 mM 2-mercaptoethanol, 1 mM EDTA. A new His-Trap step removed uncleaved protein and the TEV protease. One pure GIM-1 fraction was run on an analytical gel filtration column (Superdex 75 26/600) in buffer C (50 mM HEPES, pH 7.2, 100 μM ZnCl2, and 150 mM NaCl), where it eluted as a single peak corresponding to a molecular mass of approximately 25 kDa.
Crystallization, data collection, and structure determination.
GIM-1 was crystallized by the hanging-drop method, mixing 1 μl protein (10 mg/ml) with 1 μl reservoir solution (0.1 M Tris, pH 7.1, 21% polyethylene glycol monomethyl ether 2000 [PEG MME 2K], 4% glycerol). Crystals grew within 2 days at 22°C. All crystals were cryoprotected with 20 to 25% glycerol, 21% PEG MME 2K, 0.1 M Tris (pH 7.1), and flash frozen in liquid nitrogen. GIM-1-Apo and GIM-1-Ox were obtained after soaking the crystals in the cryoprotectant for about 1.5 h with a pyridine inhibitor. X-ray data collection was performed at the European Synchrotron Radiation Facility (ESRF), France, at beam line ID29. The images were integrated and scaled with XDS (14), and structure factors were obtained using TRUNCATE (15). The phase problem was solved by PHASER (16) using IMP-1 (Protein Data Bank [PDB] entry 1DDK) as a search model (17), and the structures were refined in Phenix (18) and manually built in WinCoot (19).
Protein structure accession numbers.
Coordinates and structure factors of GIM-1, GIM-1-2Mol, GIM-1-Apo, and GIM-1-Ox have been deposited in the Protein Data Bank under accession numbers 2YNT, 2YNW, 2YNU, and 2YNV, respectively.
RESULTS AND DISCUSSION
Here, we report three-dimensional structures of native GIM-1 in two crystal forms (GIM-1 and GIM-1-2Mol), along with the metal-free apo-zinc structure (GIM-1-Apo) and a structure with the zinc ligand Cys221 oxidized in chain A and only one of the two zinc ions present for chains A and B (GIM-1-Ox). The resolutions obtained were 1.60, 1.70, 2.06, and 2.05 Å, respectively (Table 1). All structures were obtained from similar crystallization conditions (Table 1), and no reducing agents were added. The apo- and mono-zinc (oxidized) structures were derived from unsuccessful soaks with candidate inhibitors (pyridines with a carboxyl group[s]). The GIM-1 structure was found to have three molecules in the asymmetric unit, while for the three other structures two molecules were found, resulting in nine independently refined GIM-1 models, all obtained without the use of noncrystallographic symmetry (NCS). The root mean square deviations (RMSD) for CA atoms among the nine models is 0.47 to 0.71 Å, with residues Lys59 to Val67 (in the so-called flapping loop, loop 1 or L3) accounting for the largest main-chain movements (Fig. 2a). The recombinant GIM-1 construct used in this study begins at residue Gln37 according to the BBL numbering scheme (13), which is used throughout this paper, and includes an additional N-terminal serine, Ser36, derived from the TEV protease cleavage site. All GIM-1 models include residues Lys40 and Ser292, with the most complete model containing Ser36-Lys296 (GIM-1 chain A).
Table 1.
X-ray data collection and refinement statistics of GIM-1
| Parameter | Value for: |
|||
|---|---|---|---|---|
| GIM-1 | GIM-1-2Mol | GIM-1-Apo | GIM-1-Ox | |
| PDB entry | 2YNT | 2YNW | 2YNU | 2YNV |
| Data collection statistics | ||||
| Space group | P21212 | P21 | P21 | P21 |
| No. of molecules in asymmetric unit | 3 | 2 | 2 | 2 |
| Unit cell dimensions | ||||
| a, b, c (Å) | 110.92, 140.66, 41.07 | 40.16, 135.53, 38.54 | 38.79, 135.38, 40.51 | 38.65, 131.71, 40.72 |
| γ (°) | 90.83 | 90.23 | 94.78 | |
| Resolution (Å) (highest bin) | 30–1.60 (1.68–1.60) | 45–1.70 (1.79–1.70) | 45–2.06 (2.17–2.06) | 41–2.05 (2.16–2.05) |
| Wavelength (Å) | 1.2400 | 0.97239 | 0.97239 | 0.97239 |
| Completeness (%) | 98.6 (90.7) | 99.7 (99.0) | 97.2 (98.7) | 97.9 (99.1) |
| Avg <I>/<σI> | 16.4 (3.5) | 11.5 (2.7) | 12.1 (2.0) | 10.5 (2.6) |
| Rmerge (%) | 9.4 (46.2) | 4.2 (38.0) | 4.5 (57.4) | 6.0 (42.9) |
| Refinement statistics | ||||
| Resolution range (Å) | 20–1.60 | 20–1.70 | 20–2.06 | 20–2.05 |
| R factor (all reflections) (%) | 16.44 | 16.69 | 18.52 | 19.67 |
| Rfree (%) | 20.51 | 19.86 | 23.47 | 24.72 |
| RMSD | ||||
| Bond length (Å) | 0.006 | 0.013 | 0.018 | 0.015 |
| Bond angle (°) | 1.102 | 1.318 | 1.626 | 1.645 |
| Avg B factor (Å2) | 14.5 | 22.3 | 43.4 | 39.4 |
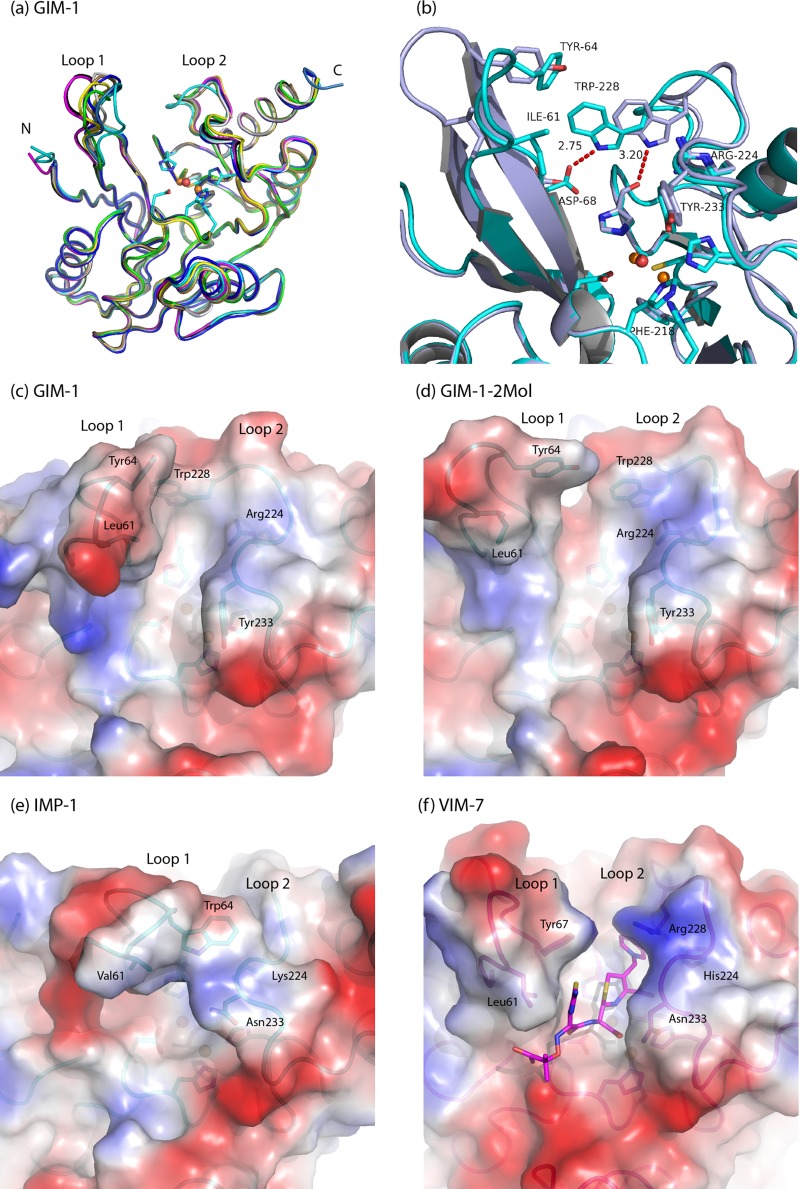
Fig 2.
(a) Superposition of the GIM-1 structures showing the Zn2+ ions and Zn2+ ligands with GIM-1 chains A (cyan), B (dark blue), and C (light blue); GIM-1-2Mol chains A (black) and B (magenta); GIM-1-Apo chains A (yellow) and B (tan); and GIM-1-Ox chains A (gray) and B (green). (b) Interactions involving Trp228 in the closed GIM-1 chain A (cyan) and open GIM-1-2Mol chain A (gray). Calculated electrostatic surfaces of superimposed closed GIM-1 (chain A) (c), open GIM-1-2Mol (chain A) (d), IMP-1 (PDB 1DD6) (e), and VIM-7 (PDB 2Y87) (f), where ceftazidime is docked in VIM-7 (22), are also shown. All figures were made with PyMol (http://www.pymol.org).
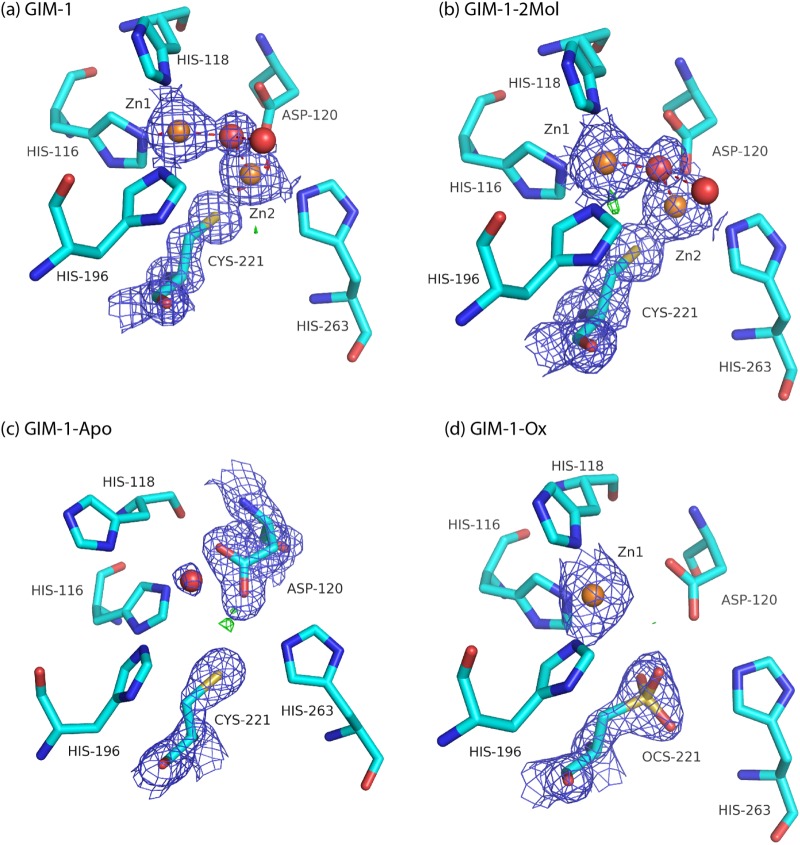
The GIM-1 structure has the typical MBL αβ/βα fold with two Zn2+ ions situated in the groove between the two core β-sheets. The overall GIM-1 structure has an RMSD for CA atoms of 1.01 Å compared to IMP-1 (PDB 1DDK; 44% sequence identity), 1.40 Å compared to VIM-2 (PDB 1KO3; 35% sequence identity), and 1.38 Å compared to VIM-7 (PDB 2Y8B; 36% sequence identity). The native active sites (GIM-1 and GIM-1-2Mol) contain two zinc ions (Zn1, a trihistidine site, and Zn2, a Cys-His-Asp site) bridged by a water molecule and separated by a distance of 3.45 to 3.50 Å, where Zn1 displays a tetrahedral coordination and Zn2 seems to be in a distorted trigonal bipyramidal coordination with one vacant position (Fig. 3a and b) (see Table S1 in the supplemental material). This contrasts with the majority of B1 MBLs where tetrahedral geometry is generally observed for the Zn1 site, while Zn2 often has an additional apical water as a fifth ligand, resulting in a trigonal bipyramidal coordination. In the GIM-1-Apo structure, the electron density indicates an absence of zinc, with the two Zn1 ligands His116 and His118 rotated relative to their positions in the native structures, increasing the space between the three Zn1 His ligands (Fig. 3c). In the oxidized form (GIM-1-Ox) Zn2 is absent, and only Zn1 is present in both chains A and B. However, electron density maps provide strong evidence for oxidation of Cys221 in the A chain only (Osc221) (Fig. 3d). We surmise that the unsuccessful inhibitor soak that preceded collection of the GIM-1-Ox data set resulted in the loss of Zn2, suggesting that the GIM-1 Zn2 site is of lower affinity than the Zn1 site. In other MBLs, the low Zn2 preference has been related to the presence of Cys221, and the Zn2 site is proposed to be essential for substrate binding (30, 31). Oxidation of Cys221 has also been observed in other subclass B1 MBLs (BcII, SPM-1, VIM-7, and VIM-2 [20–23]).
Fig 3.
2Fo-Fc electron density maps of GIM-1 (Mol A at 1.5σ) (a), GIM-1-2Mol (Mol A at 1.5σ) (b), GIM-1-Apo (Mol A at 1.1σ) (c), and GIM-1-Ox (Mol A at 1.5σ) (d). For all panels, Fo-Fc difference maps are displayed at 4.0σ (green) and −4.0σ (red).
In subclass B1 MBLs such as GIM-1, the active site is flanked by two extended loops, loop 1 and loop 2 (Fig. 2a), that connect elements of secondary structure. Of these, the flapping loop (loop 1; residues 60 to 66) is proposed to act as a flexible lid that closes over the active site to promote binding and hydrolysis of substrates. In the various GIM-1 structures this loop adopts a range of conformations, being most open in the native GIM-1-2Mol (chains A and B) structure and most closed in the GIM-1 (chain A) structure, with a maximal difference in the main-chain positions of the two conformations of 7 Å for Glu62 (Fig. 2b). Residues in loop 1 implicated in binding of substrates/ligands to other B1 MBLs include Phe61 and Tyr67 in VIM-2 (20), Val61 and Trp64 in IMP-1 (17), and Leu61 and Tyr67 in VIM-7 (22) (Fig. 2e and f). The presence of isoleucine, tyrosine, and valine at residues 61, 64, and 67, respectively, and the conformational variability of this region in the different structures, are consistent with GIM-1 loop 1 adopting a similar role.
Loop 2 in B1 MBLs (also called L10) comprises residues 223 to 242 and faces loop 1 across the active-site groove. In this region, GIM-1 is distinguished from other B1 MBLs by the presence of arginine at position 224, tryptophan at 228, and tyrosine at 233 (Fig. 1). In eight of the nine GIM-1 models, the Arg224 side chain makes hydrogen bonds, with one or both of the NE and NH1 atoms, to Gly230 O and creates a positively charged R2 binding site (Fig. 2c and d), i.e., the binding site for the C2/C3 substituents of carbapenem or cephalosporin substrates. However, this does not seem to be discriminatory against substrates (cephalosporins) with positively charged C3 substituents, since GIM-1 binds ceftazidime relatively tightly (Km of 31 μM) but cefepime more weakly (Km of 431 μM) (10). GIM-1 is unique among B1 MBLs of known sequence in possessing tryptophan at position 228. Compared to other B1 MBLs of known structure (e.g., VIM-2, VIM-7, IMP-1, and NDM-1), the side chain of GIM-1 Trp228 both limits the size of the active-site groove in the R2 pocket and participates in hydrogen-bonding interactions that vary between the different structures. In the GIM-1-2Mol chain A structure, Trp228 NE1 is hydrogen bonded to the main-chain carbonyl of the Zn2 ligand His263, while in chain A of the GIM structure it instead contacts OD1 of Asp68 at the base of loop 1 (Fig. 2b). The bond of Trp228 NE1 to His263 O is found in five of the nine GIM-1 models. In the GIM-1-Apo structure, Trp228 is partly disordered in chains A and B, so there could be an interplay between Trp228 and the metals. In other B1 MBL structures (e.g., VIM-2, VIM-7, IMP-1, and NDM-1), an interaction from the His263 main-chain oxygen atom with a loop 2 residue is not featured. For other apo MBL structures, e.g., NDM-1 (PDB 3SPU) or L1 (PDB 2FU6), the effects of removing the metal ions are primarily reported in loop 1 and loop 2. However, for GIM-1-Apo, the main effect seems to be for residues 232 to 237 in loop 2, which are disordered (chain B) or have elevated B-factors (chain A). Loop 1 has conformations which are variable, from the most open conformation (GIM-1-2Mol chain A and B) to the most closed conformation (GIM-1 chain A) (Fig. 2c and d) among the GIM-1 structures which both contain two zinc ions, suggesting that the metal does not seem to be determining the loop 1 conformation. Our structures are suggestive of considerable plasticity in this region of the active-site groove, which is consistent with the ability of GIM-1 (like other B1 MBLs) to bind and hydrolyze a wide variety of differently substituted substrates. The presented GIM-1 structures imply that both loop 1 and loop 2 can adapt their conformations during catalysis. Other studies on MBLs have shown a complex interplay between sequence and structure, revealing that loop flexibility, protein stability, and particular mutations are important for biological function and efficiency (24–26). Some of the described effects are also ligand dependent.
Residue 233, normally asparagine in B1 MBLs, has been studied in several such enzymes, but its role(s) in substrate binding and/or as a contributor to a putative oxyanion hole remain unclear and controversial (24–27). In GIM-1, tyrosine is found at position 233 and makes water-mediated hydrogen bonds from the O and N main-chain atoms to His196 and the side-chain hydroxyl to Asp236. We suggest that the Tyr233 side chain provides a hydrophobic surface for interactions with bound substrates through hydrogen bonds, as would be required for a role in an oxyanion hole. Asn233 is conserved in ∼67% (26) of all class B1 MBL sequences, and its alteration, as in a recent study on the IMP-1 enzyme, where 19 mutants of Asn233 were investigated, can significantly affect kinetic parameters (26). However, while expression of the IMP-1 Asn233Tyr mutant reduced the MIC for cefotaxime compared to that for the wild-type enzyme, values for ampicillin, imipenem, or cephalothin were unaffected (26). These data, together with the presence of Tyr233 in B1 MBLs such as BlaB (28) and IMP-14 (GenBank accession no. AY553332), indicate that substitution of tyrosine for asparagine at position 233 is unlikely to have significant consequences for the binding and hydrolysis of substrates by GIM-1. However, our structures suggest that the bulky tyrosine side chain at this position in the GIM-1 structures serves to limit the size of the drug binding pocket and possibly constrain the mobility of loop 2 (Fig. 2c and d).
Superposition of the various GIM-1 structures upon those for other B1 MBLs indicates that compared to, e.g., the IMP or VIM enzymes, the GIM-1 active-site groove is more confined (due to the presence of Ile61 in loop 1 and, in particular, to Trp228 and Tyr233 in loop 2) and is also positively charged (Fig. 2). The area and volume of the antibiotic binding, where ceftazidime is docked into VIM-7 (22), is smaller for GIM-1 (GIM-1 chains A and B, GIM-1-2mol chains A and B, and GIM-1-Ox chain A) than for IMP-1 (PDB 1DD6) and VIM-7 (PDB 2Y87) using the CASTp server (29). The availability of multiple structures from this study also highlights the mobility of both loops 1 and 2, suggesting that, despite the narrower active-site groove of GIM-1, a wide variety of substrates still can be accommodated by reorientation of these regions (that may in particular be associated with alternative hydrogen bonding by the Trp228 side chain). This is consistent with the relatively low Km values obtained for hydrolysis of a broad range of β-lactams by GIM-1. Notably, however, GIM-1 binds relatively weakly (as inferred from elevated Km values) to 7α-methoxy-substituted substrates, such as cefoxitin and moxalactam, that are often reasonably good substrates for related enzymes. This indicates a more limited ability of the restricted GIM-1 active site to accommodate this type of substituent.
In conclusion, our study demonstrates that the GIM-1 MBL contains three features that are typical of B1 MBLs (a binuclear zinc site in an αβ/βα fold, mobile loops 1 and 2 adjacent to the active site, and a lower-affinity Zn2 site) and some unique characteristics, with the presence of Trp228 and Tyr233 creating a narrower and more hydrophobic active-site groove. In particular, the two distinctive hydrogen binding networks involving Trp228 (interacting with either His263 or Asp68) seem to be a novel feature of the GIM-1 enzyme. The availability of nine independently refined models of the GIM-1 MBL provide a more dynamic picture of the active site than is normally provided by crystallographic investigations, highlighting the plasticity of the active-site groove that is a feature of these enzymes. The high-resolution structural information provided here will facilitate the inclusion of GIM-1 in structure-based studies aimed at developing inhibitors that are effective against the range of mobile MBLs and thus aid our efforts to counter the growing threat of MBL-mediated antibiotic resistance.
Supplementary Material
ACKNOWLEDGMENTS
Technical assistance with protein purification by Bjarte Aarmo Lund and Kevin Mathisen and scientific discussions with Ingar Leiros are gratefully acknowledged. Assistance with data collection by Ken A. Johnson is appreciated. Provision of beam time at ID29 at the European Radiation Facility (ESRF), Grenoble, France, is highly valued.
Work on metallo-β-lactamases in the laboratory of J.S. is funded by the United Kingdom Medical Research Council (G1100135) and the U.S. National Institutes of Health (1R01AI100560-01). This work was supported by grants from the Northern Norway Regional Health Authority and Tromsø Research Foundation. The Norwegian Structural Biology Centre (NorStruct) is supported by the National Functional Genomics Program (FUGE) of the Research Council of Norway.
Footnotes
Published ahead of print 3 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02227-12.
REFERENCES
- 1. Essack SY. 2001. The development of β-lactam antibiotics in response to the evolution of β-lactamases. Pharm. Res. 18:1391–1399 [DOI] [PubMed] [Google Scholar]
- 2. Siu LK. 2002. Antibiotics: action and resistance in gram-negative bacteria. J. Microbiol. Immunol. Infect. 35:1–11 [PubMed] [Google Scholar]
- 3. Fisher JF, Meroueh SO, Mobashery S. 2005. Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem. Rev. 105:395–424 [DOI] [PubMed] [Google Scholar]
- 4. Walsh TR. 2010. Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36(Suppl. 3):S8–S14 [DOI] [PubMed] [Google Scholar]
- 5. Walsh TR, Toleman MA, Poirel L, Nordmann P. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect. Dis. 11:381–393 [DOI] [PubMed] [Google Scholar]
- 7. Yong D, Toleman MA, Bell J, Ritchie B, Pratt R, Ryley H, Walsh TR. 2012. Genetic and biochemical characterization of an acquired subgroup B3 metallo-β-lactamase gene, blaAIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. Antimicrob. Agents Chemother. 56:6154–6159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leiros H-KS, Borra PS, Brandsdal BO, Edvardsen KS, Spencer J, Walsh TR, Samuelsen Ø. 2012. Crystal structure of the mobile metallo-β-lactamase AIM-1 from Pseudomonas aeruginosa: insights into antibiotic binding and the role of Gln157. Antimicrob. Agents Chemother. 56:4341–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pollini S, Maradei S, Pecile P, Olivo G, Luzzaro F, Docquier JD, Rossolini GM. 2013. FIM-1, a new acquired metallo-β-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob. Agents Chemother. 57:410–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Castanheira M, Toleman MA, Jones RN, Schmidt FJ, Walsh TR. 2004. Molecular characterization of a β-lactamase gene, blaGIM-1, encoding a new subclass of metallo-β-lactamase. Antimicrob. Agents Chemother. 48:4654–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rieber H, Frontzek A, Pfeifer Y. 2012. Emergence of metallo-β-lactamase GIM-1 in a clinical isolate of Serratia marcescens. Antimicrob. Agents Chemother. 56:4945–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rieber H, Frontzek A, von Baum H, Pfeifer Y. 2012. Emergence of metallo-β-lactamases GIM-1 and VIM in multidrug-resistant Pseudomonas aeruginosa in North Rhine-Westphalia, Germany. J. Antimicrob. Chemother. 67:1043–1045 [DOI] [PubMed] [Google Scholar]
- 13. Garau G, Garcia-Saez I, Bebrone C, Anne C, Mercuri P, Galleni M, Frere JM, Dideberg O. 2004. Update of the standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 48:2347–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 24:795–800 [Google Scholar]
- 15. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Concha NO, Janson CA, Rowling P, Pearson S, Cheever CA, Clarke BP, Lewis C, Galleni M, Frere JM, Payne DJ, Bateson JH, Abdel-Meguid SS. 2000. Crystal structure of the IMP-1 metallo β-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry 39:4288–4298 [DOI] [PubMed] [Google Scholar]
- 18. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 20. Garcia-Saez I, Docquier JD, Rossolini GM, Dideberg O. 2008. The three-dimensional structure of VIM-2, a Zn-β-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form. J. Mol. Biol. 375:604–611 [DOI] [PubMed] [Google Scholar]
- 21. Murphy TA, Catto LE, Halford SE, Hadfield AT, Minor W, Walsh TR, Spencer J. 2006. Crystal structure of Pseudomonas aeruginosa SPM-1 provides insights into variable zinc affinity of metallo-β-lactamases. J. Mol. Biol. 357:890–903 [DOI] [PubMed] [Google Scholar]
- 22. Borra PS, Leiros H-KS, Ahmad R, Spencer J, Leiros I, Walsh TR, Sundsfjord A, Samuelsen Ø. 2011. Structural and computational investigations of VIM-7: insights into the substrate specificity of VIM metallo-β-lactamases. J. Mol. Biol. 411:174–189 [DOI] [PubMed] [Google Scholar]
- 23. Davies AM, Rasia RM, Vila AJ, Sutton BJ, Fabiane SM. 2005. Effect of pH on the active site of an Arg121Cys mutant of the metallo-β-lactamase from Bacillus cereus: implications for the enzyme mechanism. Biochemistry 44:4841–4849 [DOI] [PubMed] [Google Scholar]
- 24. Garau G, Bebrone C, Anne C, Galleni M, Frere JM, Dideberg O. 2005. A metallo-β-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J. Mol. Biol. 345:785–795 [DOI] [PubMed] [Google Scholar]
- 25. Yanchak MP, Taylor RA, Crowder MW. 2000. Mutational analysis of metallo-β-lactamase CcrA from Bacteroides fragilis. Biochemistry 39:11330–11339 [DOI] [PubMed] [Google Scholar]
- 26. Brown NG, Horton LB, Huang W, Vongpunsawad S, Palzkill T. 2011. Analysis of the functional contributions of Asn233 in metallo-β-lactamase IMP-1. Antimicrob. Agents Chemother. 55:5696–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Materon IC, Beharry Z, Huang W, Perez C, Palzkill T. 2004. Analysis of the context dependent sequence requirements of active site residues in the metallo-β-lactamase IMP-1. J. Mol. Biol. 344:653–663 [DOI] [PubMed] [Google Scholar]
- 28. Vessillier S, Docquier JD, Rival S, Frere JM, Galleni M, Amicosante G, Rossolini GM, Franceschini N. 2002. Overproduction and biochemical characterization of the Chryseobacterium meningosepticum BlaB metallo-β-lactamase. Antimicrob. Agents Chemother. 46:1921–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. 2006. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34:W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gonzalez JM, Meini MR, Tomatis PE, Medrano Martin FJ, Cricco JA, Vila AJ. 2012. Metallo-β-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions. Nat. Chem. Biol. 8:698–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jacquin JO, Balbeur D, Damblon C, Marchot P, De Pauw E, Roberts GC, Frere JM, Matagne A. 2009. Positively cooperative binding of zinc ions to Bacillus cereus 569/H/9 β-lactamase II suggests that the binuclear enzyme is the only relevant form for catalysis. J. Mol. Biol. 392:1278–1291 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.