Abstract
Efflux pumps are membrane proteins capable of actively transporting a broad range of substrates from the cytoplasm to the exterior of the cell. Increased efflux activity in response to drug treatment may be the first step in the development of bacterial drug resistance. Previous studies showed that the efflux pump Mmr was significantly overexpressed in strains exposed to isoniazid. In the work to be described, we constructed mutants lacking or overexpressing Mmr in order to clarify the role of this efflux pump in the development of resistance to isoniazid and other drugs in M. tuberculosis. The mmr knockout mutant showed an increased susceptibility to ethidium bromide, tetraphenylphosphonium, and cetyltrimethylammonium bromide (CTAB). Overexpression of mmr caused a decreased susceptibility to ethidium bromide, acriflavine, and safranin O that was obliterated in the presence of the efflux inhibitors verapamil and carbonyl cyanide m-chlorophenylhydrazone. Isoniazid susceptibility was not affected by the absence or overexpression of mmr. The fluorometric method allowed the detection of a decreased efflux of ethidium bromide in the knockout mutant, whereas the overexpressed strain showed increased efflux of this dye. This increased efflux activity was inhibited in the presence of efflux inhibitors. Under our experimental conditions, we have found that efflux pump Mmr is mainly involved in the susceptibility to quaternary compounds such as ethidium bromide and disinfectants such as CTAB. The contribution of this efflux pump to isoniazid resistance in Mycobacterium tuberculosis still needs to be further elucidated.
INTRODUCTION
The World Health Organization (WHO) goal to reduce the global burden of tuberculosis by 2015 (1, 2) faces many challenges, namely, the dissemination of severe cases of drug resistance that reduce the therapeutic efficacy of the available antituberculous drugs. These drug-resistant forms of tuberculosis are known as multidrug and extensively drug-resistant tuberculosis (MDRTB and XDRTB, respectively), with the former defined to be resistance to at least the first-line drugs isoniazid and rifampin and the latter defined to be MDRTB plus resistance to fluoroquinolones and to at least one of the three injectable second-line drugs (kanamycin, amikacin, and capreomycin). Thus, there is a great effort to understand the mechanisms of drug resistance, as well as to develop new drugs and new therapeutic approaches.
Clinically relevant drug resistance in Mycobacterium tuberculosis occurs mainly by the acquisition of spontaneous chromosomal mutations that alter the drug target or the prodrug-activating enzymes, followed by the selection of drug-resistant mutants that may occur in the case of exposure to monotherapy or lower antibiotic doses due to inadequate prescription, poor patient compliance, and patient pharmacokinetic variability (3–5). However, these mutations are not found in many low-level-resistant isolates, suggesting that other mechanisms of resistance may also be involved, such as mechanisms involving the permeability barrier provided by the cell wall and the activity of efflux systems.
Bacterial efflux pumps are membrane proteins that are capable of actively transporting a broad range of substrates, including drugs, from the cytoplasm to the exterior of the cell. They are involved in physiological processes, such as cell wall division, maintenance of the pH homeostasis, and secretion of intracellular metabolites (6–8). Increased expression of efflux pump genes confers a low-level-resistant phenotype, and it has been suggested that under these conditions, bacteria have greater chances of acquiring a chromosomal mutation(s) conferring higher levels of drug resistance (6, 9). A strategy to prevent this chain of events would be the inhibition of efflux pumps, which, in addition, would restore the effectiveness of antimicrobials that are subject to efflux (10–12).
In M. tuberculosis, several efflux pumps have been described, but their contribution to clinical drug resistance remains to be completely clarified (13, 14). One of these pumps is Mmr (Rv3065) of the small multidrug resistance (SMR) family of transporters. Previous studies involving Mmr were performed in the heterologous host Mycobacterium smegmatis and showed that it was involved in the extrusion of several compounds such as tetraphenylphosphonium, ethidium bromide (EtBr), erythromycin, and acriflavine (15, 16). A recent study has shown that Mmr appears to be involved in the efflux of compounds of the pyrrole class in M. tuberculosis (17).
In previous works, we and other authors observed that mmr was one of the efflux pump genes that was significantly overexpressed in a number of M. tuberculosis strains exposed to high levels of isoniazid (9, 18, 19), which suggested that Mmr could be associated with resistance to isoniazid. In the study to be described, we constructed M. tuberculosis mutants lacking or overexpressing Mmr in order to clarify the role of this efflux pump in the development of resistance to isoniazid and other drugs.
MATERIALS AND METHODS
Bacteria and growth conditions.
The strains and plasmids used in this study are listed in Table 1. M. tuberculosis was grown at 37°C in Middlebrook 7H9 broth (Difco, Detroit, MI) supplemented with 10% (vol/vol) Middlebrook albumin-dextrose-catalase (ADC; Difco) and 0.05% (vol/vol) Tween 80 or on Middlebrook 7H10 (Difco) agar plates supplemented with 10% (vol/vol) ADC and 0.05% (vol/vol) Tween 80. Escherichia coli HB101 was grown at 37°C Luria Bertani (LB) broth or on LB agar plates. Plasmids were maintained in E. coli with appropriate antibiotics for selection (50 μg/ml of hygromycin, 20 μg/ml of kanamycin). For the selection of resistance markers in mycobacteria, hygromycin or kanamycin was added to the culture medium at final concentrations of 50 μg/ml and 20 μg/ml, respectively.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| M. tuberculosis | ||
| H37Rv (ATCC 25618) | Wild type | Laboratory collection |
| H37Rv::pLAM12 | Control strain | 20 |
| H37Rv::pJV53 | Recombineering strain | 20 |
| MmrKO | H37Rv knockout for mmr | This study |
| MmrKO::pCRS5 | Strain MmrKO containing pCRS5 | This study |
| H37Rv::pCVZ2 | Strain H37Rv containing pCVZ2 | This study |
| Plasmids | ||
| pLAM12 | Replicative plasmid with a kanamycin resistance gene and an acetamidase expression cassette | 20 |
| pJV53 | Carrying genes for gp60 and gp61 (encoding recombineering enzymes) from mycobacteriophage Che9c cloned into pLAM12, under the control of the acetamidase promoter | 20 |
| pYUB854 | E. coli vector containing the Hygr cassette flanked by multiple-cloning sites and γδ res sites, oriE, and a λcos packaging site | 20, 21 |
| pMmr | pYUB854 containing the DNA regions flanking mmr | This study |
| pCRS5 | Integrative plasmid, mmr cloned into pMV361 (22) | This study |
| pCVZ2 | Replicative plasmid, mmr cloned into pSUM36 (23) | This study |
DNA manipulation.
DNA manipulations were carried out by standard techniques (24). Mycobacterial genomic DNA was isolated as described previously (25). Southern blotting was done with an enhanced chemiluminescence direct nucleic acid labeling and detection system (Amersham Biosciences), according to the manufacturer's instructions. A DNA probe specific for the mmr gene was generated by PCR, based upon an Rv3065 gene sequence from GenBank (accession number NC_000962; region 3430387 to 3430710); primer sequences are available upon request.
E. coli and M. tuberculosis were transformed by electroporation with a Gene Pulser apparatus (Bio-Rad Laboratories Inc., Richmond, CA). Briefly, E. coli competent cells were prepared according to standard protocols (24) and transformed by adding DNA to 40-μl aliquots of cells while incubating on ice. Cells were transferred to chilled 0.2-cm cuvettes (Bio-Rad) and transformed using a Bio-Rad Gene Pulser set at 2.5 kV, 200 Ω, and 25 μF. Cells were recovered in 1 ml LB broth for 1 h at 37°C and plated on selective medium. M. tuberculosis electrocompetent cells were prepared as previously described (21). Briefly, the culture was centrifuged at 2,880 × g for 10 min and the supernatant was discarded. The pellet was resuspended in 1/2 volume of sterile 10% glycerol, and the centrifugation step was repeated. The cells were resuspended in 1/4 volume of sterile 10% glycerol. This process was repeated until in the last step the cells were resuspended in 1/25 volume of sterile 10% glycerol. M. tuberculosis competent cells were transformed by electroporation using a Gene Pulser set at 2.5 kV, 1,000 Ω, and 25 μF. Cells were recovered in 1 ml 7H9 broth for 24 h (for the overexpression and complementation mutants) or 72 h (for the knockout [KO] mutant) at 37°C and plated on selective medium.
Strain construction. (i) Inactivation.
The construction of an M. tuberculosis mmr knockout mutant was performed by allelic exchange, as previously described (20, 21). This method is based on the use of an M. tuberculosis recombineering strain transformed with a plasmid that contains mycobacteriophage Che9c-encoded recombination proteins gp60 and gp61. Expression of these proteins in M. tuberculosis increases the recombination frequency of the bacteria, allowing the recovery of gene replacement mutants following electroporation of linear DNA molecules containing segments homologous to the chromosomal DNA (20, 21).
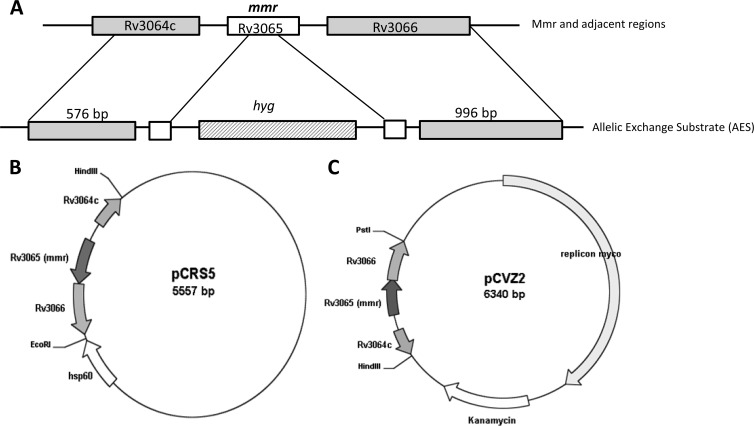
Primers were designed to amplify upstream and downstream DNA regions flanking the mmr gene (576 and 996 bp, respectively). The PCR products were cloned into pYUB854 flanking a hygromycin resistance cassette. The vector obtained, pMmr, was linearized by double digestion with AvrII and HindIII or amplified by PCR in order to obtain the allelic exchange substrate (AES) (Fig. 1A).
Fig 1.
(A) Replacement of the M. tuberculosis mmr gene. An allelic exchange substrate (AES) for replacement of mmr (Rv3065) was generated by cloning 576-bp upstream and 996-bp downstream regions on either side of the hyg resistance cassette of pYUB854. The AES was obtained by PCR or restriction digestion of this segment; (B) integrative plasmid pCRS5 that contains a copy of mmr and that was used to complement the Mmr knockout mutant; (C) replicative plasmid pCVZ2 used to overexpress mmr in H37Rv.
H37Rv::pJV53 (recombineering strain) was grown at 37°C in Middlebrook 7H9 supplemented with 0.05% Tween 80, 20 μg/ml kanamycin, and 0.2% succinate. Once the cells reached an optical density at 600 nm (OD600) of 0.5 to 0.6, acetamide was added to a final concentration of 0.2% and the culture was grown at 37°C overnight. On the following day, electrocompetent cells were prepared and transformed with 100 ng of the AES by electroporation. The transformations were recovered by incubation at 37°C in 7H9 supplemented with ADC and Tween 80 for 72 h. The entire reaction mixture was plated on 7H10 agar plates containing kanamycin, hygromycin, and ADC and incubated at 37°C for 3 to 4 weeks, until colonies (potential mmr knockout mutants) developed. Colonies were inoculated into 7H9 broth containing kanamycin, hygromycin, and ADC and incubated at 37°C until visible growth was obtained (approximately 10 to 20 days). The presence of an inactivated copy of the mmr gene was tested by PCR and Southern blot analysis of each recombinant colony obtained. The mutant strain, namely, M. tuberculosis H37Rv MmrKO, was grown in medium lacking kanamycin until it lost the replicating plasmid pJV53.
As controls, recombineering-deficient strain M. tuberculosis H37Rv::pLAM12 was also transformed with AES and no colony was recovered, and M. tuberculosis H37Rv::pJV53 was used as a negative control for PCR and hybridization analysis.
(ii) Integrative plasmid for complementation.
M. tuberculosis H37Rv MmrKO was complemented by introducing the integrative plasmid pCRS5, a derivative of vector pMV361 (22) carrying a 1,177-bp DNA fragment containing the mmr gene, resulting in M. tuberculosis H37Rv MmrKO::pCRS5 (Fig. 1B).
(iii) Replicative plasmid for overexpression.
An 1,177-bp DNA fragment containing the mmr gene was cloned into the pSUM36 vector (23) using restriction enzymes HindIII and PstI, yielding plasmid pCVZ2. Plasmid pCVZ2 was electroporated into M. tuberculosis H37Rv, resulting in M. tuberculosis H37Rv::pCVZ2 (Fig. 1C).
Neutral red staining.
Neutral red staining was used to ensure that the constructed strains keep free lipids of the cell wall, such as the virulence factor phthiocerol dimycocerosates (PDIMs). Neutral red staining was performed in a test tube, as described previously (26). Briefly, mycobacterial strains were grown at 37°C on Middlebrook 7H10 medium supplemented with 10% ADC and kanamycin (20 μg/ml) or hygromycin B (50 μg/ml) when required. Bacterial cells were placed in 15-ml screw-cap tubes containing 4 ml of methanol-H2O (1:1) and incubated at 37°C for 1 h. After centrifugation at 2,880 × g for 10 min, the methanol was removed and 4 ml of barbital buffer (1% sodium barbital in 5% NaCl, pH 9.8) and 150 μl of neutral red were added. The results were evaluated after 1 h and 24 h of incubation at 37°C.
Drug susceptibility assays. (i) Determination of MICs.
The determination of the MICs for antibiotics, efflux inhibitors, dyes, and biocides against M. tuberculosis H37Rv and Mmr mutant strains was performed by the resazurin microtiter assay, as previously described (27). Ciprofloxacin, ethambutol, erythromycin, kanamycin, isoniazid, rifampin, acriflavine, ethidium bromide, safranin O, tetraphenylphosphonium, cetyltrimethylammonium bromide (CTAB), carbonyl cyanide m-chlorophenylhydrazone (CCCP), chlorpromazine, and verapamil were purchased from Sigma-Aldrich (Madrid, Spain) and used in the MIC determination. Briefly, mycobacterial strains were grown at 37°C in Middlebrook 7H9 broth supplemented with 10% ADC and 0.05% Tween 80 until an OD600 of 0.8. Mycobacterial cultures were diluted in 7H9 supplemented with ADC and 0.5% glycerol in order to obtain a final concentration of 105 CFU/ml. The number of CFU corresponding to aliquots of the inoculum was routinely calculated in order to ensure a constant number of bacterial cells from experiment to experiment. Aliquots of 100 μl were transferred to each well of a 96-well plate that contained 100 μl of each compound at concentrations prepared from 2-fold serial dilutions in 7H9-ADC medium. The inoculated plates were incubated for 6 days at 37°C and for an additional 2 days after the addition of the redox indicator (30 μl of a resazurin solution at 0.1 mg/ml). A change from blue to pink indicates reduction of resazurin and therefore bacterial growth. Thus, the MIC was defined as the lowest concentration of compound that prevented this color change. The MICs of ethidium bromide, tetraphenylphosphonium, acriflavine, safranin O, and CTAB in the presence of the efflux inhibitors chlorpromazine, verapamil, and CCCP were also determined to evaluate the contribution of efflux activity to M. tuberculosis susceptibility to these drugs.
(ii) Determination of isoniazid susceptibility on solid medium.
To determine if there was any difference between the wild type and the knockout mutant when grown in solid medium in the presence of isoniazid, aliquots of 10 μl of 10-fold serial dilutions of 107-CFU/ml exponential-phase cultures were spotted onto 7H10-ADC medium containing concentrations of isoniazid ranging from 0.25 to 0.03 μg/ml. The plates were incubated at 37°C until growth was observed. The assay was performed in duplicate. The results were recorded by the observation of the growth of spots of wild-type and knockout mutant bacterial cells in the isoniazid-containing plates. The highest concentration of isoniazid and the highest dilution that resulted in bacterial growth were recorded for both wild-type and knockout strains and compared. We expected that growth of bacteria at higher isoniazid concentrations than the wild type would correlate with increased ability to tolerate isoniazid.
(iii) Rate of killing.
The rate of killing by isoniazid in M. tuberculosis H37Rv and the MmrKO mutant was determined as previously described (28). Isoniazid was diluted in 10 ml of Middlebrook 7H9 supplemented with 10% ADC to give 2×, 4×, and 8× MIC. A drug-free control was included. Mycobacterial cultures at exponential phase were adjusted to an OD600 of 0.2, and 100 μl was added to each tube containing the isoniazid dilutions. On days 0, 1, 2, 4, 8, and 16, 100-μl aliquots were taken for counting of the numbers of CFU and inoculated in Middlebrook 7H10-ADC medium. The plates were incubated at 37°C until growth was observed. The assay was performed in duplicate.
(iv) Growth competition assay.
The M. tuberculosis wild-type strain H37Rv and the MmrKO strain were used for growth competition testing in the presence of isoniazid. Briefly, three flasks containing 10 ml of Middlebrook 7H9-ADC with isoniazid at 0.25 μg/ml (1/2 MIC) were inoculated independently with H37Rv, MmrKO, and both strains, ensuring that the cell density in each flask was 105 CFU/ml. The three flasks were incubated at 37°C. The three cultures were serially diluted and plated in duplicate on drug-free Middlebrook 7H10-ADC medium on days 0, 2, 7, and 14. In parallel, the culture containing both the wild-type strain H37Rv and the MmrKO strain was serially diluted and plated in duplicate on 7H10-ADC medium with 50 μg/ml of hygromycin. The number of MmrKO cells in the competition assay was determined from the number of colonies grown in hygromycin-containing plates; the number of H37Rv cells in the competition assay was calculated from the number of colonies in the drug-free plates minus the number of MmrKO cells.
Detection of ethidium bromide efflux by fluorometry.
The detection of ethidium bromide efflux on a real-time basis by the M. tuberculosis strains was performed using a fluorometric method previously described (29), but with some adaptations for use in a 96-well plate fluorometer. Briefly, M. tuberculosis strains were grown in 7H9-ADC medium at 37°C until an OD600 of 0.6 to 0.8. Cultures were centrifuged at 2,880 × g for 10 min, the supernatant was discarded, the pellet was washed in phosphate-buffered saline (PBS; pH 7.4), and the OD600 was adjusted to 0.8 with PBS with 0.05% Tween 80. Aliquots of 100 μl of bacterial suspension were transferred into wells of a 96-well plate containing serial dilutions of ethidium bromide at concentrations that ranged from 2 to 0.125 μg/ml. To determine the effect of chlorpromazine, CCCP, and verapamil on the accumulation of ethidium bromide, 10 μl of each compound was added to the corresponding well of the 96-well plate. Each inhibitor was used at 1/2 the MIC in order to not compromise the cellular viability. Relative fluorescence was acquired every 51 s for 60 min at 37°C in a Synergy HT detection microplate reader (Biotek Instruments), using 530/25 nm and 590/20 nm as excitation and detection wavelengths, respectively. For a better comparison of ethidium bromide accumulation experiments, for each assay we determined the relative final fluorescence (RFF) at the last time point (minute 60) of the assay in comparison with reference conditions by using the formula (RFassay − RFref)/RFref, where RFassay is the relative fluorescence at the last time point of the ethidium bromide accumulation assay and RFref is the relative fluorescence at the last time point of the ethidium bromide accumulation assay under the reference conditions (30, 31).
When assaying different strains and mutants, the reference condition was the assay of reference strain H37Rv, whereas when assaying the effect of efflux inhibitors, the reference condition was the assay in the absence of any efflux inhibitor. In both cases, high RFF values indicated that cells accumulated more ethidium bromide under the tested conditions than under the reference conditions and vice versa for negative RFF values. RFF values correlated with the contribution of the Mmr efflux pump to the extrusion of this ethidium bromide or the degree of efflux inhibition. The experiments were repeated three times, and the RFF values presented are the averages of three independent assays.
RESULTS AND DISCUSSION
It is widely accepted that many bacterial pathogens activate efflux pumps in response to drug treatment, providing the bacterial cells with a way of adaptation to a hostile environment. This enables the bacteria to survive under such conditions, facilitating the acquisition of mutations that will confer higher and stable levels of drug resistance (6, 12, 32). Therefore, efflux pumps are becoming attractive for drug discovery programs, with the goal to evaluate if potential new antituberculous compounds may be subject to efflux or to identify efflux inhibitors of currently used antituberculous drugs. This is particularly relevant in a time when MDRTB and XDRTB continue to escalate and fewer alternatives are left to treat the severe forms of drug-resistant tuberculosis.
In this work, we studied the Mmr efflux pump of M. tuberculosis. The basis for this study was provided by previous studies that showed overexpression of mmr in M. tuberculosis strains that had been induced to isoniazid resistance by exposure to this drug for a prolonged period of time (9, 19). Although the mmr gene was significantly overexpressed among other overexpressed efflux pump genes present in the M. tuberculosis genome, a possible association between the Mmr efflux pump and isoniazid resistance was suggested (9, 19). In order to clarify the role of Mmr in the development of isoniazid resistance, as well as in the resistance to other drugs that are substrates of efflux pumps, we constructed an MmrKO mutant in the H37Rv reference strain using a recombineering method (20) and studied the drug susceptibility profile of this mutant (MmrKO), its complemented counterpart (MmrKO::pCRS5), and an H37Rv derivative strain overexpressing the mmr gene from a replicative plasmid (H37Rv::pCVZ2).
We determined the MICs of several compounds against the wild-type strain M. tuberculosis H37Rv and the Mmr mutants (Table 2). For the selected group of antibiotics, only a minor decrease in the MIC of kanamycin was observed in the knockout mutant. The remaining antibiotics presented consistent MICs that were not affected by either the inactivation or overexpression of mmr. In the particular case of isoniazid, no difference in the MICs was observed between the strains (MIC, 0.25 μg/ml for wild type and all Mmr mutants). This was an unexpected result, since in previous studies we found overexpression of Mmr after exposure to isoniazid (9, 18, 19), strongly suggesting that this particular efflux pump could be directly related to the transport of this drug. However, we speculated that the determination of the MIC of isoniazid by 2-fold microdilution assay was possibly not sensitive enough for detecting phenotypic differences between wild type and strains with an mmr gene deleted or strains overexpressing the mmr gene.
Table 2.
MICs of several antimicrobial drugs against M. tuberculosis H37Rv and Mmr mutant strainsd
| Compound | MIC (μg/ml) for M. tuberculosis strain: |
|||
|---|---|---|---|---|
| H37Rv | MmrKOa | MmrKO::pCRS5b | H37Rv::pCVZ2c | |
| Antibiotics | ||||
| Isoniazid | 0.25 | 0.25 | 0.25 | 0.25 |
| Rifampin | 0.06 | 0.06 | 0.06 | 0.06 |
| Ethambutol | 1.25 | 1.25 | 1.25 | 1.25 |
| Kanamycin | 0.15 | 0.07 | >5d | >5d |
| Ciprofloxacin | 0.25 | 0.25 | 0.25 | 0.25 |
| Erythromycin | 25 | 25 | 25 | 25 |
| Efflux inhibitors | ||||
| Chlorpromazine | 5 | 5 | 5 | 5 |
| Verapamil | 150 | 150 | 150 | 150 |
| CCCPe | 1.5 | 1.5 | 1.5 | 1.5 |
| Dyes | ||||
| Ethidium bromide | 2 | 1 | 2 | 4 |
| Tetraphenylphosphonium | 12.5 | 3.1 | 12.5 | 12.5 |
| Acriflavine | 3.1 | 3.1 | 3.1 | 6.2 |
| Safranin O | 1 | 0.5 | 1 | 2 |
| CTABe biocide | 12.5 | 6.2 | 12.5 | 12.5 |
M. tuberculosis H37Rv with mmr gene inactivated.
MmrKO complemented with integrative vector pCRS5.
M. tuberculosis H37Rv containing replicative plasmid pCVZ2.
Increase of the MIC is due to the kanamycin resistance selection marker present in these strains.
CCCP, carbonyl cyanide m-chlorophenylhydrazone; CTAB, cetyltrimethylammonium bromide.
Therefore, other approaches were used in order to clarify if the inactivation of mmr had an effect, even subtle, on the susceptibility to isoniazid. No difference between the wild type and the knockout mutant was observed when grown in solid medium with isoniazid; both strains grew at a dilution of 10−4 until a concentration of 0.06 μg/ml, and no growth was observed at 0.12 and 0.25 μg/ml of isoniazid (data not shown). The rate of killing by inhibitory concentrations of isoniazid was also determined, but no difference between the wild type and knockout strain was observed (data not shown).
Moreover, the growth competition assay showed no difference between the wild-type and mutant strains when simultaneously grown in the presence of isoniazid, with both presenting 106 CFU/ml after 7 days of exposure to isoniazid (data not shown).
Therefore, we can hypothesize that Mmr responds to treatment with isoniazid. Although this efflux pump would not directly transport this drug, it could instead transport any other bacterial metabolite either derived from isoniazid itself or generated in response to isoniazid-induced damage. This indicates that the mmr overexpression observed in the isoniazid-induced strains (9, 19) may be due to a general stress response caused by the prolonged exposure to this drug. In this case, the M. tuberculosis efflux pump systems would present an increased activity in order to extrude any noxious compounds that formed as a direct result of the mechanism of action of isoniazid. This confirms that isoniazid susceptibility was not affected by the inactivation of mmr and that the overexpression results observed in the previous studies may be due to a general response to stress rather than a result of isoniazid being a substrate of Mmr. In fact, a recent study that used a computational approach combined with gene expression data and an interactome network has suggested that the SOS response is upregulated under isoniazid treatment (33). This may affect several cellular processes, namely, the regulation of efflux pumps, and be a trigger for drug resistance. It is also possible that Mmr could be involved in the transport of a cell wall component(s) and that overexpression of this efflux pump would occur to compensate for the cell wall damage caused by isoniazid, which targets InhA, an NADH-dependent enoyl acyl carrier protein reductase involved in the synthesis of mycolic acids (3, 4).
Concerning the other compounds tested, the MmrKO mutant showed an increased susceptibility to ethidium bromide, tetraphenylphosphonium, safranin O, and CTAB. In the particular case of ethidium bromide, the MIC was restored to the wild-type value (2 μg/ml) in the complemented strain MmrKO::pCRS5, whereas the H37Rv strain overexpressing mmr (H37Rv::pCVZ2) presented an increased MIC (4 μg/ml). This strain also showed an increased MIC for safranin O and acriflavine, although there was no change for the latter in the knockout mutant. These data confirm that the mmr gene cloned in either the integrative plasmid pCRS5 or the replicative plasmid pCVZ2 is being expressed and produces a fully functional Mmr protein. To confirm that this phenotype was in fact due to efflux activity and not to any other indirect mechanism, MICs were determined in the presence of efflux inhibitors (Table 3). Most cases of MIC reduction were obtained with CCCP, followed by verapamil. CCCP drastically reduced by more than 30 times the MICs of acriflavine, safranin O, and CTAB for the mmr-overexpressing strain. Since CCCP is a proton gradient uncoupler, these results demonstrate the increased dependency of this efflux pump on the proton motive force, as predicted for efflux pumps of the SMR family of transporters. An MIC reduction was observed for ethidium bromide in all strains in the presence of verapamil, and CCCP caused only a slight reduction (1 dilution) in the strain overexpressing the mmr gene. There was a significant reduction of the MIC of ethidium bromide (8 times) in the presence of verapamil for the knockout mutant, which suggests that other efflux pumps that extrude ethidium bromide may be active in this strain and are affected by verapamil. Chlorpromazine promoted a slight reduction of the MIC of tetraphenylphosphonium and safranin O in the knockout mutant.
Table 3.
Effect of efflux inhibitors on the MICs of potential substrates of Mmr efflux pump
| M. tuberculosis strain | MIC (μg/ml)d |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ethidium bromide |
Tetraphenylphosphonium |
Acriflavine |
Safranin O |
CTAB |
||||||||||||||||
| No inhibitor | CCCP | CPZ | VP | No inhibitor | CCCP | CPZ | VP | No inhibitor | CCCP | CPZ | VP | No inhibitor | CCCP | CPZ | VP | No inhibitor | CCCP | CPZ | VP | |
| H37Rv | 2 | 2 | 2 | 0.5 | 12.5 | 3.1 | 12.5 | 12.5 | 3 | 1.5 | 3 | 3 | 1 | 0.5 | 1 | 0.5 | 12.5 | 3.1 | 12.5 | 12.5 |
| MmrKOa | 1 | 1 | 1 | 0.13 | 3.1 | 3.1 | 1.5 | 1.5 | 3 | 1.5 | 3 | 1.5 | 0.5 | 0.5 | 0.25 | 0.5 | 6.25 | 3.1 | 6.25 | 6.25 |
| MmrKO::pCRS5b | 2 | 2 | 2 | 1 | 12.5 | 3.1 | 12.5 | 12.5 | 3 | 1.5 | 3 | 3 | 1 | 0.5 | 1 | 0.5 | 12.5 | 3.1 | 12.5 | 12.5 |
| H37Rv::pCVZ2c | 4 | 2 | 4 | 1 | 12.5 | 6.2 | 12.5 | 12.5 | 6 | 0.18 | 6 | 6 | 2 | 0.06 | 2 | 2 | 12.5 | 0.4 | 12.5 | 12.5 |
M. tuberculosis H37Rv with mmr gene inactivated.
MmrKO complemented with pCRS5.
M. tuberculosis H37Rv containing pCVZ2.
CCCP, carbonyl cyanide m-chlorophenylhydrazone; CPZ, chlorpromazine; CTAB, cetyltrimethylammonium bromide; VP, verapamil. Data in bold represent at least a 4-fold reduction, considered to denote a significant MIC reduction in the presence of an efflux inhibitor.
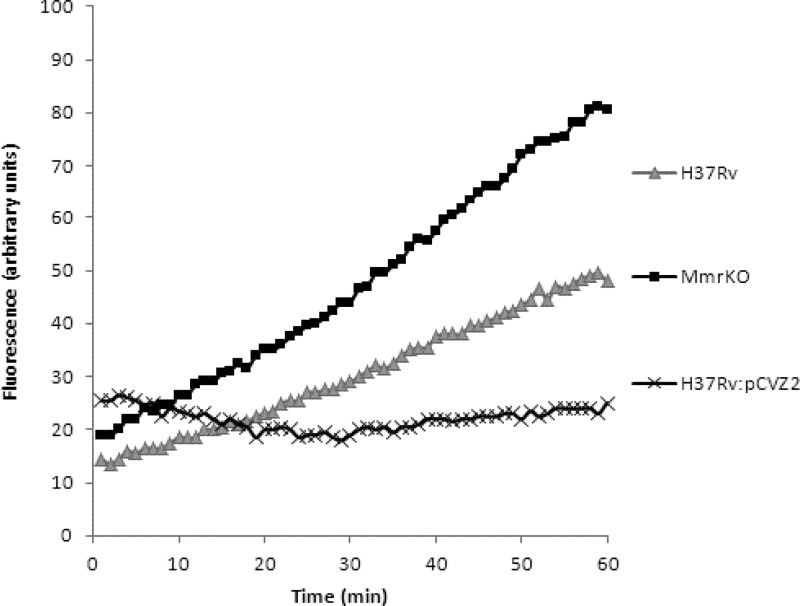
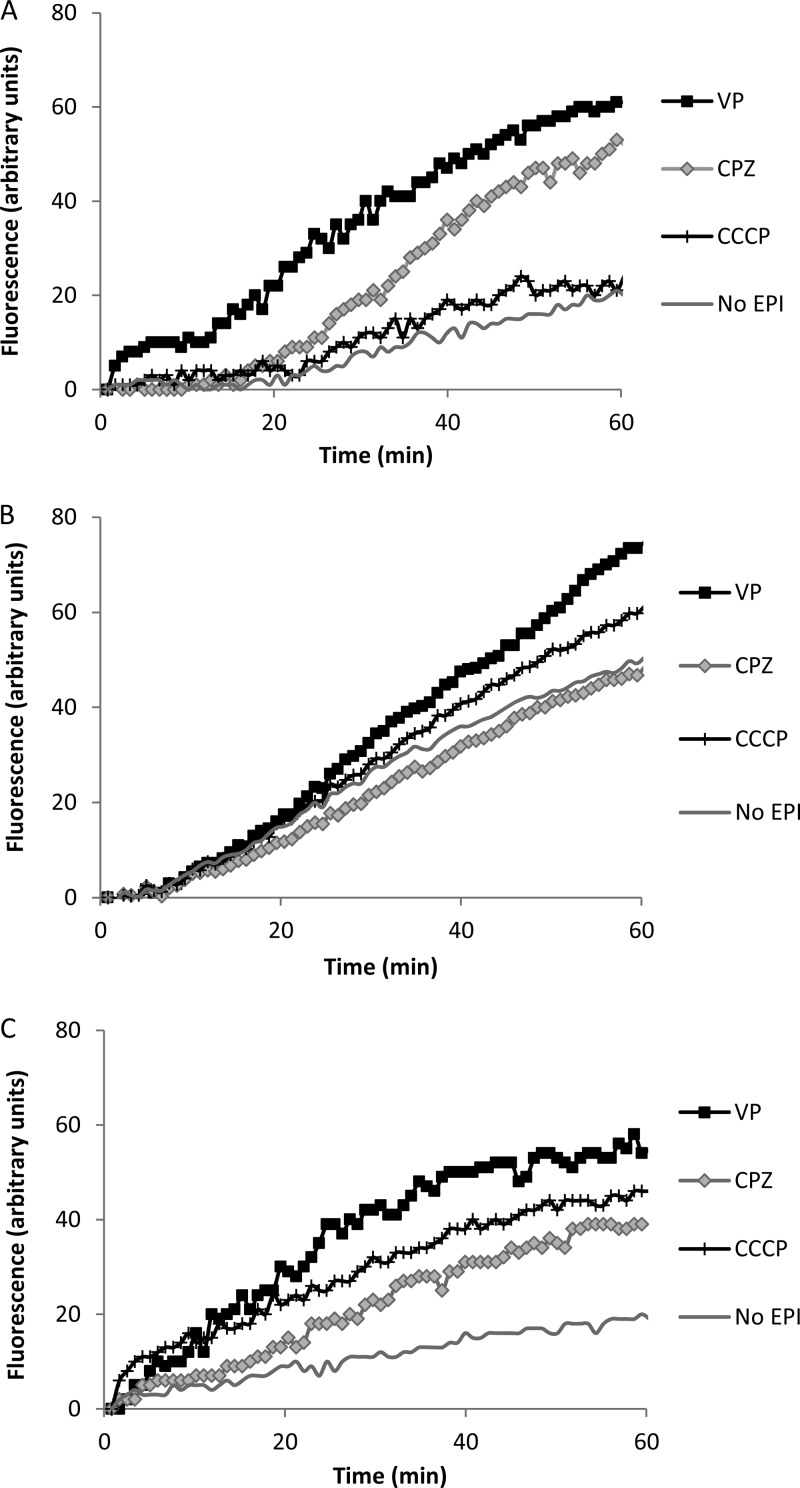
Following the results obtained for the MIC determination, ethidium bromide was selected as a substrate for detecting efflux activity in M. tuberculosis H37Rv and its Mmr mutants. The MmrKO mutant showed a 62% maximum increased accumulation of ethidium bromide at 1 μg/ml relative to wild-type strain H37Rv, whereas overexpressed strain H37Rv::pCVZ2 showed a consistent decreased accumulation of ethidium bromide at all concentrations tested (Table 4; Fig. 2). These results are in agreement with the MIC obtained for ethidium bromide for each strain and further support the possibility that the Mmr efflux activity contributes directly to the susceptibility of M. tuberculosis to this compound and that the absence of Mmr decreases the efflux of quaternary compounds in M. tuberculosis. The effect of efflux inhibitors on the accumulation of ethidium bromide was also tested in all strains using ethidium bromide at 1/2 the MIC (Fig. 3; Table 5). In general, the accumulation of ethidium bromide by wild-type, knockout mutant, and overexpressing strains in the presence of efflux inhibitors (Fig. 3; Table 5) paralleled that of the same strains in the absence of efflux inhibitors (Fig. 2; Table 4). The inhibitor that promoted higher ethidium bromide accumulation for all strains was verapamil, which is in agreement with the MIC results described above (compare the results with the MICs of ethidium bromide in the presence and absence of an inhibitor; Table 3) and with previous studies (19, 34). Surprisingly, despite not having an effect on the MIC of ethidium bromide (Table 3), chlorpromazine caused an increased accumulation of ethidium bromide in the wild-type and overexpressing strains (Fig. 3).
Table 4.
RFF based on the accumulation of ethidium bromide for each mutant strain compared to that for wild-type strain H37Rva
| Strain | RFF at the following ethidium bromide concn (μg/ml): |
||||
|---|---|---|---|---|---|
| 0.125 | 0.25 | 0.5 | 1 | 2 | |
| MmrKO | 0.07 | 0.47 | 0.53 | 0.62 | 0.55 |
| MmrKO::pCRS5 | − 0.21 | −0.16 | −0.20 | −0.25 | −0.11 |
| H37Rv:pCVZ2 | −0.20 | −0.20 | −0.38 | −0.53 | −0.58 |
The values correspond to the last point of measurement at 60 min, when fluorescence had reached a steady state. Data correspond to the averages of three independent assays.
Fig 2.
Accumulation of ethidium bromide at 1 μg/ml by M. tuberculosis H37Rv, H37Rv MmrKO (H37Rv with the mmr gene inactivated), and H37Rv::pCVZ2 (H37Rv containing pCVZ2).
Fig 3.
Effect of efflux inhibitors on the accumulation of ethidium bromide for M. tuberculosis H37Rv (A), H37Rv MmrKO (B), and H37Rv::pCVZ2 (C). Ethidium bromide and efflux inhibitors were used at 1/2 the MIC to not compromise cellular viability. CCCP, carbonyl cyanide m-chlorophenylhydrazone; CPZ, chlorpromazine; EPI, efflux pump inhibitor; VP, verapamil.
Table 5.
RFF based on the accumulation of ethidium bromide for H37Rv and each mutant strain in the presence of efflux inhibitors compared with that for the untreated control (without inhibitor)a
| Strain | RFF for the following efflux inhibitor: |
||
|---|---|---|---|
| CCCP | CPZ | VP | |
| H37Rv | 0.17 | 1.60 | 2.05 |
| MmrKO | 0.21 | −0.05 | 0.46 |
| MmrKO::pCRS5 | 0.08 | 0.53 | 0.61 |
| H37Rv::pCVZ2 | 1.42 | 1.05 | 1.9 |
The values correspond to the last point of measurement at 60 min, when fluorescence had reached a steady state. Data correspond to the averages of three independent assays.
This work was carried out in M. tuberculosis, the natural host of the Mmr efflux pump, and the results largely support those of the previous studies performed in the heterologous host M. smegmatis that reported Mmr to be an efflux pump associated with susceptibility to a variety of compounds, such as ethidium bromide, tetraphenylphosphonium, and safranin O (15, 16). Our findings have been corroborated by the use of ethidium bromide, coupled with the use of real-time monitoring of its efflux by a fluorometric system, which was a good marker for the study of the efflux activity of Mmr and may be used to evaluate the activity of potential efflux inhibitors. In addition, this work has shown that Mmr has an effect on susceptibility to CTAB, a widely used disinfectant, demonstrating the role of efflux systems, in particular, Mmr, in M. tuberculosis tolerance to this class of compounds. In this case, the use of efflux inhibitors would prevent the development of this tolerance. In conclusion, the study of the Mmr efflux pump may be used as proof of concept for the demonstration of efflux activity in M. tuberculosis, and the strategy employed may be used to characterize other efflux pumps of M. tuberculosis and other mycobacteria.
ACKNOWLEDGMENTS
This work was supported by BIO-2009-09405 from the Spanish Ministry of Science and Innovation and FP7-260872 from More Medicines for Tuberculosis. L. Rodrigues was supported by STSM-BM0701-010510-005767 from COST Action BM0701 (Antibiotic Transport and Efflux: New Strategies to Combat Bacterial Resistance [ATENS]) and short-term fellowship ASTF 67/2011 from the European Molecular Biology Organization (EMBO). C. Villellas and R. Bailo were supported by the Spanish Ministry of Education (FPU AP2008-04730) and the Gobierno de Aragón (B047/11), respectively.
We are very thankful to Leonard Amaral and Jean-Marie Pagès for the fruitful discussions and support during Cost Action BM0701 of the European Commission/European Science Foundation.
Footnotes
Published ahead of print 19 November 2012
REFERENCES
- 1. Stop TB Partnership 2010. The global plan to stop TB 2011–2015. World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. World Health Organization 2012. Global tuberculosis report 2012. World Health Organization, Geneva, Switzerland [Google Scholar]
- 3. Almeida Da Silva PE, Palomino JC. 2011. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J. Antimicrob. Chemother. 66:1417–1430 [DOI] [PubMed] [Google Scholar]
- 4. Ramaswamy S, Musser JM. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3–29 [DOI] [PubMed] [Google Scholar]
- 5. Srivastava S, Pasipanodya JG, Meek C, Leff R, Gumbo T. 2011. Multidrug-resistant tuberculosis not due to noncompliance but to between-patient pharmacokinetic variability. J. Infect. Dis. 204:1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Piddock LJ. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19:382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramón-García S, Martín C, Thompson CJ, Aínsa JA. 2009. Role of the Mycobacterium tuberculosis P55 efflux pump in intrinsic drug resistance, oxidative stress responses, and growth. Antimicrob. Agents Chemother. 53:3675–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramón-García S, Mick V, Dainese E, Martín C, Thompson CJ, De Rossi E, Manganelli R, Aínsa JA. 2012. Functional and genetic characterization of the tap efflux pump in Mycobacterium bovis BCG. Antimicrob. Agents Chemother. 56:2074–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Machado D, Couto I, Perdigão J, Rodrigues L, Portugal I, Baptista P, Veigas B, Amaral L, Viveiros M. 2012. Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS One 7:e34538 doi:10.1371/journal.pone.0034538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Amaral L, Viveiros M, Kristiansen JE. 2006. “Non-antibiotics”: alternative therapy for the management of MDRTB and MRSA in economically disadvantaged countries. Curr. Drug Targets 7:887–891 [DOI] [PubMed] [Google Scholar]
- 11. Rodrigues L, Aínsa JA, Amaral L, Viveiros M. 2011. Inhibition of drug efflux in mycobacteria with phenothiazines and other putative efflux inhibitors. Recent Pat. Antiinfect. Drug Discov. 6:118–127 [DOI] [PubMed] [Google Scholar]
- 12. Viveiros M, Martins M, Rodrigues L, Machado D, Couto I, Ainsa J, Amaral L. 2012. Inhibitors of mycobacterial efflux pumps as potential boosters for TB drugs. Expert Rev. Anti Infect. Ther. 10:983–998 [DOI] [PubMed] [Google Scholar]
- 13. De Rossi E, Aínsa JA, Riccardi G. 2006. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol. Rev. 30:36–52 [DOI] [PubMed] [Google Scholar]
- 14. Louw GE, Warren RM, Gey van Pittius NC, McEvoy CR, Van Helden PD, Victor TC. 2009. A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob. Agents Chemother. 53:3181–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Rossi E, Branzoni M, Cantoni R, Milano A, Riccardi G, Ciferri O. 1998. mmr, a Mycobacterium tuberculosis gene conferring resistance to small cationic dyes and inhibitors. J. Bacteriol. 180:6068–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li XZ, Zhang L, Nikaido H. 2004. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48:2415–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Balganesh M, Dinesh N, Sharma S, Kuruppath S, Nair AV, Sharma U. 2012. Efflux pumps of Mycobacterium tuberculosis play a significant role in antituberculosis activity of potential drug candidates. Antimicrob. Agents Chemother. 56:2643–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta AK, Katoch VM, Chauhan DS, Sharma R, Singh M, Venkatesan K, Sharma VD. 2010. Microarray analysis of efflux pump genes in multidrug-resistant Mycobacterium tuberculosis during stress induced by common anti-tuberculous drugs. Microb. Drug Resist. 16:21–28 [DOI] [PubMed] [Google Scholar]
- 19. Rodrigues L, Machado D, Couto I, Amaral L, Viveiros M. 2012. Contribution of efflux activity to isoniazid resistance in the Mycobacterium tuberculosis complex. Infect. Genet. Evol. 12:695–700 [DOI] [PubMed] [Google Scholar]
- 20. van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat. Methods 4:147–152 [DOI] [PubMed] [Google Scholar]
- 21. van Kessel JC, Hatfull GF. 2008. Mycobacterial recombineering. Methods Mol. Biol. 435:203–215 [DOI] [PubMed] [Google Scholar]
- 22. Stover CK, de LA Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Gansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletts RG, Jacobs WR, Jr, Bloom BR. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460 [DOI] [PubMed] [Google Scholar]
- 23. Aínsa JA, Martín C, Cabeza M, De la Cruz F, Mendiola MV. 1996. Construction of a family of Mycobacterium/Escherichia coli shuttle vectors derived from pAL5000 and pACYC184: their use for cloning an antibiotic-resistance gene from Mycobacterium fortuitum. Gene 176:23–26 [DOI] [PubMed] [Google Scholar]
- 24. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25. Parish T, Stoker NG. 1998. Mycobacterial protocols. In Parish T, Stoker NG. (ed), Methods in molecular biology, vol 101 Humana Press, Totowa, NJ [Google Scholar]
- 26. Soto CY, Andreu N, Gibert I, Luquin M. 2002. Simple and rapid differentiation of Mycobacterium tuberculosis H37Ra from M. tuberculosis clinical isolates through two cytochemical tests using neutral red and Nile blue stains. J. Clin. Microbiol. 40:3021–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 46:2720–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reddy VM, Dubuisson T, Einck L, Wallis RS, Jakubiec W, Ladukto L, Campbell S, Nacy CA. 2012. SQ109 and PNU-100480 interact to kill Mycobacterium tuberculosis in vitro. J. Antimicrob. Chemother. 67:1163–1166 [DOI] [PubMed] [Google Scholar]
- 29. Rodrigues L, Wagner D, Viveiros M, Sampaio D, Couto I, Vavra M, Kern WV, Amaral L. 2008. Thioridazine and chlorpromazine inhibition of ethidium bromide efflux in Mycobacterium avium and Mycobacterium smegmatis. J. Antimicrob. Chemother. 6:1076–1082 [DOI] [PubMed] [Google Scholar]
- 30. Machado L, Spengler G, Evaristo M, Handzlik J, Molnár J, Viveiros M, Kiec-Kononowicz K, Amaral L. 2011. Biological activity of twenty-three hydantoin derivatives on intrinsic efflux pump system of Salmonella enterica serovar Enteritidis NCTC 13349. In Vivo 25:769–772 [PubMed] [Google Scholar]
- 31. Paixão L, Rodrigues L, Couto I, Martins M, Fernandes P, de Carvalho CC, Monteiro GA, Sansonetty F, Amaral L, Viveiros M. 2009. Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J. Biol. Eng. 3:18 doi:10.2174/187221209787259910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. da Silva PE, Von Groll A, Martin A, Palomino JC. 2011. Efflux as a mechanism for drug resistance in Mycobacterium tuberculosis. FEMS Immunol. Med. Microbiol. 63:1–9 [DOI] [PubMed] [Google Scholar]
- 33. Chen LC, Yeh HY, Yeh CY, Arias CR, Soo VW. 2012. Identifying co-targets to fight drug resistance based on a random walk model. BMC Syst. Biol. 6:5 doi:10.1186/1752-0509-6-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodrigues L, Ramos J, Couto I, Amaral L, Viveiros M. 2011. Ethidium bromide transport across Mycobacterium smegmatis cell-wall: correlation with antibiotic resistance. BMC Microbiol. 11:35 doi:10.1186/1471-2180-11-35 [DOI] [PMC free article] [PubMed] [Google Scholar]