Abstract
To compare the effects of four antimicrobial peptides (MUC7 12-mer, histatin 12-mer, cathelicidin KR20, and a peptide containing lactoferricin amino acids 1 to 11) on the yeast Saccharomyces cerevisiae, we employed a genomewide fitness screen of combined collections of mutants with homozygous deletions of nonessential genes and heterozygous deletions of essential genes. When an arbitrary fitness score cutoffs of 1 (indicating a fitness defect, or hypersensitivity) and −1 (indicating a fitness gain, or resistance) was used, 425 of the 5,902 mutants tested exhibited altered fitness when treated with at least one peptide. Functional analysis of the 425 strains revealed enrichment among the identified deletions in gene groups associated with the Gene Ontology (GO) terms “ribosomal subunit,” “ribosome biogenesis,” “protein glycosylation,” “vacuolar transport,” “Golgi vesicle transport,” “negative regulation of transcription,” and others. Fitness profiles of all four tested peptides were highly similar, particularly among mutant strains exhibiting the greatest fitness defects. The latter group included deletions in several genes involved in induction of the RIM101 signaling pathway, including several components of the ESCRT sorting machinery. The RIM101 signaling regulates response of yeasts to alkaline and neutral pH and high salts, and our data indicate that this pathway also plays a prominent role in regulating protective measures against all four tested peptides. In summary, the results of the chemical genomic screens of S. cerevisiae mutant collection suggest that the four antimicrobial peptides, despite their differences in structure and physical properties, share many interactions with S. cerevisiae cells and consequently a high degree of similarity between their modes of action.
INTRODUCTION
Cationic antimicrobial peptides (CAMPs) are small positively charged peptides, active against a broad range of microorganisms, including bacteria, fungi, viruses, and parasites (1–3). Because of their broad spectrum of activity, they have been considered promising alternatives to conventional antimicrobial agents. The mechanisms by which CAMPs mediate their effect, however, remain unclear and controversial. Several recent reviews have summarized different models proposed to explain the CAMP mechanism of action (1, 4–7). These models fall into two categories, transmembrane pore formation models (e.g., barrel stave or toroidal) and non-pore-based models (which include the carpet model and the detergent-like model). Both involve peptide-induced membrane permeabilization/disruption of target cells, leading to membrane depolarization, loss of vital ions and other cellular components, and ultimately lysis and cell death. There is, however, a growing acceptance that CAMPs also operate through interactions with intracellular targets or via disruption of key intracellular processes (reviewed in reference 6). In this scenario, the peptides cross the microbial membranes without significant disruption of the membrane. Finally, some peptides have been shown to act at the cell membrane as well as internal sites (8).
In order to provide further insight into the mechanism of action of CAMPs against fungi, we have recently started to use a global approach, namely, a chemical genomic screening (fitness profiling), in yeast. In a previous study (9), we performed this screen with the MUC7 12-mer, the most potent of a series of CAMPs derived from the N-terminal region of the human mucin MUC7. This peptide is active against a broad range of microorganisms, including pathogenic fungi, and has long been studied in our laboratory (10–13). It kills fungal cells by disrupting the plasma membrane, resulting in its depolarization (12) and subsequent release of small molecules such as ATP (our unpublished results), accompanied by rapid accumulation of large quantities of the peptide inside the cell (14). The MUC7 12-mer peptide fitness screen was performed on nearly five thousand homozygous, diploid strains of Saccharomyces cerevisiae containing deletions of nonessential genes (9). The effect of the tested condition on the growth of each strain was measured in parallel via oligonucleotide microarray technology, made possible by built-in, strain-specific DNA tags (15–17). Among the strains exhibiting altered fitness when treated with the MUC7 peptide, those associated with induction of the RIM101 signaling pathway exhibited particularly strong fitness defects. The RIM101 pathway regulates the response to alkaline and neutral pH and other environmental conditions (18–20), and our results suggest that this pathway is also responsive to, and leads to protection from, the type of stress imposed on yeasts by MUC7 peptide (9).
The work presented here extends work already done on the MUC7 screen; in addition to the nonessential-gene-deletion collection, we also screened all essential genes as heterozygous deletions (as a pool of 1,135 heterozygotes). Apart from expanding the coverage of interrogated genes, the inclusion of mutants with heterozygous deletions of essential genes provides a tool to identify potential protein targets, if such targets indeed play roles in the modes of operation of the tested antimicrobial peptides.
Further, the screens were performed with three additional CAMPs that, like the MUC7 12-mer, are derived from proteins found in the human oral cavity (histatin 12-mer, KR20, and lactoferricin amino acids 1 to 11), which have been reported to differ in their modes of action. Histatin 5 and its 12-amino-acid derivative known as P113 (21) are, like the MUC7 peptide, internalized by target yeast cells, leading to concomitant depolarization of plasma membrane and release of small ions, including ATP (12, 22–24). Internalization of histatin is not accompanied by pore formation or by significant destruction of plasma membrane (25, 26). It has been proposed that this peptide acts internally on various targets, including mitochondria (24, 27) and the potassium transporter Trk1p (28). Microbial killing by a human lactoferricin peptide containing amino acids 1 to 11 (hLF1-11) has been reported to involve both intracellular target and pore formation (8). In contrast, KR20, a 20 amino acid peptide derived from the human cathelicidin LL37, is a classical pore forming peptide (29–31). All these peptides are effective growth inhibitors of various bacteria and fungi, including baker's yeast (S. cerevisiae) and the opportunistic yeast pathogen Candida albicans.
The goal of the present work was to determine the degree of similarity between the modes of action of the four tested antimicrobial peptides through the high-throughput fitness screen of the S. cerevisiae mutant collection.
MATERIALS AND METHODS
Strains, reagents, and growth conditions.
Two pools of tagged S. cerevisiae strains, one containing 4,767 mutants with homozygous deletions of nonessential genes and the other consisting of 1,135 mutants with heterozygous deletions of essential mutant genes, were assembled from the S. cerevisiae mutant strain collection of the Donnelly Centre at the University of Toronto. Cells were grown in Sabouraud dextrose broth (SDB) or, when treated with peptides, in 2-fold-diluted SDB (1/2SDB) at 30°C in a rotary shaker. The peptides, described in Table 1, were synthesized by Bio-Synthesis (Lewisville, TX) or by NEO Bioscience (Cambridge, MA). Oligonucleotide primers, described earlier (9), were purchased from BioSynthesis or Eurofins MWG Operon (Huntsville, AL).
Table 1.
Antimicrobial peptides used in this study
Fitness screen.
Frozen stocks of the two S. cerevisiae mutant pools were thawed and grown overnight in SDB. Twenty ml of 1/2SDB was inoculated with overnight culture to an optical density at 600 nm (OD600) of approximately 0.05 and grown for 24 h with or without peptides at the following concentrations (i.e., doses that inhibit wild type growth by 20 to 50%): MUC7, 10 μM; HSN, 20 μM; KR20, 10 μM; hLF, 12 μM. Following two (pool of homozygous nonessential-gene deletion mutants) or four (pool of heterozygous essential-gene deletion mutants) 24-h cycles, cells were collected for DNA isolation.
Tag array preparation, hybridization, and analysis.
The custom tag arrays, in 4×72K (four arrays on one chip, each containing 72,000 features) format, were manufactured by Roche NimbleGen Inc. (Madison, WI). Five replicates of each probe were distributed randomly on the array as described previously (17). DNA isolation, asymmetric PCR, and tag array hybridizations were carried out as described earlier (9), except that the template for PCR consisted of DNA isolated from samples of mutant pools with nonessential and essential-gene deletions mixed at a 2.1:1 ratio. This specific ratio was applied to account for the difference in the number of strains and in the number of tags in each pool. The arrays were scanned using Axon GenePix 4200AL (Molecular Devices, Sunnyvale, CA). For each hybridization, raw intensities of each channel, determined by NimbleScan software (Roche NimbleGen), were quantile normalized (separately for up tags and down tags), and medians of untreated to treated intensity ratios of five replicate features per array were calculated. Fitness scores for each deletion mutant were determined by calculating log2-transformed trimmed means of six values, up tags and down tags for three separate experiments. We selected an arbitrary cutoff, with scores larger than 1 (i.e., a 2-fold change or greater) indicating a fitness defect, or hypersensitivity to the peptides, and scores below −1 indicating a fitness gain, or resistance. From the list of strains meeting the criteria described above, all those whose deleted open reading frames (ORFs) are designated dubious (Saccharomyces Genome Database http[://www.yeastgenome.org/]) were excluded from further analysis.
RESULTS AND DISCUSSION
Fitness profiling and identification of mutants displaying fitness defects.
We performed a parallel fitness screen of 5,902 S. cerevisiae deletion mutants (4,767 with homozygous deletions of nonessential genes and 1,135 with heterozygous deletions of essential genes) grown in the presence of one of the four antimicrobial peptides, MUC7 12-mer, histatin 12-mer, cathelicidin KR20 and lactoferricin amino acids 1 to 11. In this screen, the fitness score of a mutant strain, i.e., the measure of ability of this strain to grow in the presence of a peptide, is expressed as the log2 of the ratio between the number of cells in the untreated control to those exposed to the peptide (see Table S1 in the supplemental material). Thus, positive and negative fitness scores mark hypersensitive and resistant strains, respectively. The abundance of each strain was determined by tag array hybridization.
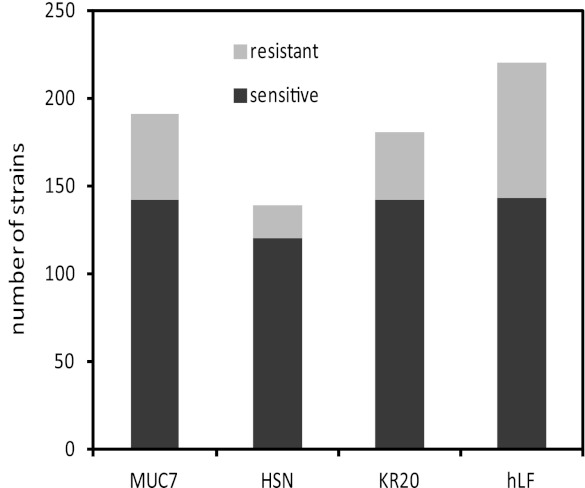
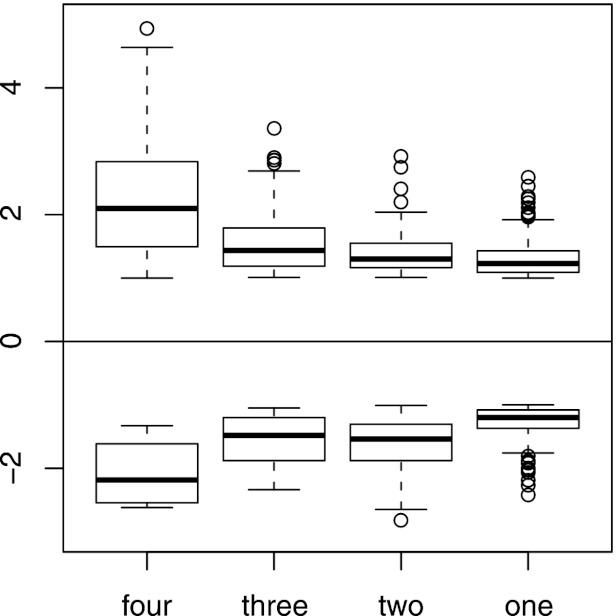
When an arbitrary threshold of fitness scores larger than 1 or smaller than −1 was applied, 425 strains (7.2%) exhibited hypersensitivity or resistance to at least one peptide and were selected for further analysis (see Table S1 in the supplemental material). Among them, members of nonessential-gene deletion and essential-gene deletion pools represented similar fractions (7.1 and 7.8%, respectively). Figure 1 shows number of strains responsive to each peptide treatment within the set threshold. The observed differences between treatments most likely reflect differences in selective pressures applied by the individual peptides. To assess the overall degree of similarity between effects of the four peptides, we divided the selected 425 strains into four categories, those exhibiting altered fitness under treatment with 4, 3, 2, and 1 peptide. The distributions of fitness scores of these groups of mutants are shown in Fig. 2. While strains responsive to only one treatment constitute the largest group (263 of 425, or 62%), strains hypersensitive and resistant to higher number of peptides tend to have significantly higher absolute fitness score values (Fig. 2). Since a high fitness score for a mutant implies importance of the deleted gene for the effectiveness of antimicrobial peptides, this observation indicates that the four tested peptides share crucial aspects of their antifungal modes of operation. On the other hand, the relatively large number of strains with altered fitness with regard to only one treatment may be associated with cellular functions, playing minor but peptide-specific roles in killing of yeasts.
Fig 1.
Strains that were hypersensitive or resistant (having fitness scores larger than 1 or smaller than −1, respectively) to treatment with each peptide tested. The numbers of hypersensitive and resistant strains, respectively, for each peptide are as follows: MUC7, 142 and 49; HSN, 120 and 19; KR20, 142 and 38; hLF, 143 and 77.
Fig 2.
Distributions of fitness scores (ordinate) of strains exhibiting hypersensitivity (top) or resistance (bottom) for treatment with four, three, two, or one peptide. The actual numbers of the strains and data points for each box are as follows: For the hypersensitive strains, 44 and 176 (four peptide treatments), 38 and 114 (three peptide treatments), 46 and 92 (two peptide treatments), and 165 and 165 (one peptide treatment); for the resistant strains, 1 and 4 (four peptide treatments), 7 and 21 (three peptide treatments), 26 and 52 (two peptide treatments) and 106 and 106 (one peptide treatment). Note that some strains may be counted more than once if they are hypersensitive to one peptide and resistant to another peptide.
All 45 strains responsive to all four peptide treatments, and some of those specific to only one peptide treatment are listed in Tables 2 and 3, respectively. Prominent among the 45 strains responsive to all treatments (Table 2) are mutants whose deleted genes are involved in the induction of Rim101p, a transcription factor regulating response to, among others, alkaline and neutral pH (18–20), as well as those encoding several components of the ESCRT sorting machinery (32). These two groups of deletion mutants together constitute 30% of all strains responsive to all four peptides, and both were identified as the most sensitive in our earlier screen of S. cerevisiae nonessential-gene mutants exposed to the MUC7 peptide (9). We argued previously that hypersensitivity of ESCRT deletion mutants was also associated with the inability to induce the RIM101 response, a claim supported by the well-documented fact that, in addition to its major function in sorting proteins for degradation, some elements of ESCRT machinery participate in the process leading to proteolytic activation of the Rim101p (33). The precise role the RIM101 pathway plays in response to CAMPs is unclear, but a stress exerted by peptides on plasma membrane prior to death of the cell may be the trigger activating the signaling and leading to some protection. The notion that cellular membrane is the sensor is supported by the reports demonstrating that the RIM101 pathway is also activated by changes in lipid composition and in physicochemical properties of the plasma membrane (34, 35).
Table 2.
List of deletions of S. cerevisiae genes conferring fitness defect or fitness gain to all four tested peptidesa
| ORF | Gene | Intensity ratiob |
Function(s)c | |||
|---|---|---|---|---|---|---|
| MUC7 | HSN | KR20 | hLF | |||
| YNL183C | NPR1 | 4.03 | 2.64 | 2.50 | 4.94 | Regulation of nitrogen utilization |
| YDR074W | TPS2 | 2.27 | 1.14 | 3.47 | 4.64 | Trehalose biosynthetic process, response to stress |
| YGL045W | RIM8 | 3.14 | 4.20 | 3.60 | 4.15 | RIM101 pathway |
| YPL065W | VPS28 | 3.67 | 3.84 | 3.17 | 4.11 | ESCRT I |
| YCL008C | VPS23 | 2.06 | 3.63 | 3.24 | 3.95 | ESCRT I |
| YJR102C | VPS25 | 2.40 | 3.86 | 2.75 | 3.49 | ESCRT II |
| YOR275C | RIM20 | 2.90 | 3.29 | 2.09 | 3.43 | RIM101 pathway |
| YLR025W | SNF7 | 3.24 | 2.88 | 2.77 | 3.43 | ESCRT III |
| YCR052W | RSC6 | 3.35 | 3.18 | 2.31 | 2.21 | Chromatin remodeling |
| YPL002C | VPS22 | 1.83 | 3.27 | 3.07 | 3.34 | ESCRT II |
| YLR417W | VPS36 | 1.98 | 2.66 | 3.30 | 2.33 | ESCRT II |
| YFL008W | SMC1 | 2.21 | 3.26 | 1.07 | 1.12 | Double-strand break repair, mitotic sister chromatid segregation |
| YNL294C | RIM21 | 2.60 | 2.84 | 2.47 | 3.19 | RIM101 pathway |
| YOR043W | WHI2 | 3.19 | 2.41 | 3.15 | 2.71 | Response to stress |
| YER157W | COG3 | 3.03 | 3.10 | 1.39 | 2.21 | ER to Golgi vesicle-mediated transport |
| YKL180W | RPL17A | 3.10 | 2.89 | 1.32 | 1.00 | 60S ribosomal subunit component |
| YMR077C | VPS20 | 2.09 | 2.79 | 2.80 | 3.08 | ESCRT III |
| YJL159W | HSP150 | 1.53 | 1.65 | 1.91 | 3.05 | Cell wall organization |
| YIL065C | FIS1 | 2.33 | 2.35 | 2.97 | 2.30 | Mitochondrial fission |
| YOR030W | DFG16 | 1.72 | 2.94 | 1.80 | 1.95 | RIM101 pathway |
| YMR154C | RIM13 | 2.23 | 2.93 | 2.22 | 2.61 | RIM101 pathway |
| YJL186W | MNN5 | 1.32 | 2.24 | 2.86 | 1.82 | Protein glycosylation |
| YIL077C | YIL077C | 2.65 | 2.57 | 2.20 | 2.83 | Putative protein of unknown function |
| YNR052C | POP2 | 1.44 | 1.91 | 1.38 | 2.80 | Regulation of transcription from RNA polymerase II promoter |
| YOR179C | SYC1 | 2.77 | 1.19 | 1.30 | 1.16 | Termination of RNA polymerase II transcription |
| YDL047W | SIT4 | 2.76 | 1.88 | 1.89 | 2.76 | Actin cytoskeleton organization, response to oxidative stress |
| YJR043C | POL32 | 2.30 | 1.50 | 1.42 | 2.76 | DNA replication, mismatch repair |
| YCR087C-A | LUG1 | 2.19 | 1.35 | 1.51 | 2.63 | Putative protein of unknown function |
| YBR006W | UGA2 | 1.68 | 1.02 | 1.59 | 2.56 | Cellular response to oxidative stress |
| YHL027W | RIM101 | 1.72 | 2.45 | 2.39 | 1.93 | RIM101 pathway |
| YNL166C | BNI5 | 1.66 | 1.59 | 2.40 | 1.58 | Septin ring assembly |
| YGR122W | YGR122W | 1.04 | 1.45 | 2.32 | 1.20 | RIM101 pathway |
| YIL098C | FMC1 | 1.05 | 2.10 | 2.18 | 1.96 | Mitochondrial proton-transporting ATP synthase complex assembly |
| YIL053W | RHR2 | 1.63 | 1.43 | 2.10 | 1.26 | Glycerol biosynthetic process, response to osmotic stress |
| YEL003W | GIM4 | 1.04 | 1.72 | 2.02 | 1.98 | Tubulin complex assembly |
| YIL041W | GVP36 | 1.05 | 1.84 | 1.95 | 1.40 | Endocytosis, polarization of actin cytoskeleton, vacuole organization |
| YIL110W | HPM1 | 1.54 | 1.95 | 1.51 | 1.57 | Peptidylhistidine methylation, to form telemethylhistidine |
| YHR021C | RPS27B | 1.41 | 1.89 | 1.53 | 1.91 | 40S ribosomal subunit component |
| YBR283C | SSH1 | 1.03 | 1.27 | 1.39 | 1.88 | SRP-dependent cotranslational protein targeting to membrane |
| YCL001W-A | YCL001W-A | 1.46 | 1.26 | 1.85 | 1.16 | Putative protein of unknown function |
| YBR286W | APE3 | 1.30 | 1.80 | 1.65 | 1.33 | Vacuolar protein catabolic process |
| YCR028C | FEN2 | 1.59 | 1.49 | 1.35 | 1.05 | Endocytosis, pantothenate transmembrane transport |
| YCR034W | FEN1 | 1.29 | 1.47 | 1.28 | 1.30 | Fatty acid elongation, vesicle-mediated transport |
| YNR044W | AGA1 | 1.21 | 1.07 | 1.46 | 1.03 | Agglutination involved in conjugation with cellular fusion |
| YER174C | GRX4 | −2.62 | −1.33 | −1.90 | −2.47 | Response to oxidative stress, actin cytoskeleton organization |
Essential genes are in bold.
Log2 hybridization intensity ratio between untreated and treated samples.
Biological process or component of a complex.
Table 3.
List of deletions of S. cerevisiae genes conferring altered fitness scores to only one of the four tested peptidesa
| ORF | Gene | Intensity ratiob |
Biological process | |||
|---|---|---|---|---|---|---|
| MUC7 | HSN | KR20 | hLF | |||
| YLR360W | VPS38 | 1.96 | −0.18 | 0.30 | 0.45 | Late endosome to vacuole transport |
| YOR310C | NOP58 | 1.34 | −0.66 | −0.33 | 0.05 | Pre-rRNA processing |
| YIL153W | RRD1 | −1.69 | −0.05 | −0.15 | 0.91 | Response to stress |
| YIR012W | SQT1 | 0.29 | 1.38 | −0.33 | 0.31 | Ribosomal assembly |
| YMR231W | PEP5 | 0.29 | 1.35 | 0.26 | 0.27 | Late endosome to vacuole transport |
| YLR376C | PSY3 | 0.14 | −1.89 | −0.33 | −0.13 | Recombinatorial repair |
| YDR495C | VPS3 | −0.30 | −1.92 | −0.46 | 0.45 | Protein targeting to vacuole |
| YOR089C | VPS21 | −0.14 | −2.07 | 0.22 | 0.58 | Protein targeting to vacuole |
| YFR009W | GCN20 | 0.10 | −0.10 | 1.48 | 0.36 | Regulation of translational elongation |
| YIL064W | SEE1 | 0.49 | 0.39 | 1.34 | 0.32 | Vesicle-mediated transport |
| YOR005C | DNL4 | −0.14 | −0.24 | 1.31 | −1.07 | Double-strand break repair |
| YOR270C | VPH1 | −0.01 | −0.42 | −1.35 | 0.61 | Vacuolar acidification |
| YPR187W | RPO26 | 0.00 | 0.34 | −1.35 | −0.16 | Transcription |
| YJR033C | RAV1 | 0.82 | 0.11 | −1.37 | 0.04 | Vacuolar acidification |
| YNL248C | RPA49 | −0.17 | −0.32 | −1.43 | 0.27 | RNA polymerase I subunit A49 |
| YNL262W | POL2 | 0.27 | −0.21 | −1.88 | 0.53 | DNA synthesis involved in DNA repair |
| YKL143W | LTV1 | 0.14 | −0.28 | −0.92 | 1.66 | Response to oxidative and osmotic stress |
| YOR233W | KIN4 | 0.36 | 0.00 | 0.07 | 1.61 | Mitotic spindle orientation checkpoint |
| YPR133W-A | TOM5 | 0.32 | −0.24 | 0.14 | 1.38 | Protein targeting to mitochondrion |
| YJL130C | URA2 | 0.33 | −0.81 | 0.14 | 1.31 | Pyrimidine base biosynthetic process |
| YBL097W | BRN1 | 0.93 | 0.37 | 0.09 | −1.35 | Mitotic chromosome condensation |
| YGR185C | TYS1 | 2.11 | 0.15 | 0.96 | −1.37 | Tyrosyl-tRNA aminoacylation |
| YDR310C | SUM1 | −0.19 | 0.20 | 0.53 | −1.41 | Negative regulation of transcription, |
| YPR094W | RDS3 | 0.03 | −0.03 | −0.16 | −1.44 | spliceosome assembly |
| YCR036W | RBK1 | 0.46 | 0.40 | 0.58 | −1.76 | D-ribose metabolic process |
| YDL235C | YPD1 | 0.68 | −0.15 | −0.11 | −2.00 | osmosensory signaling pathway |
| YER125W | RPS5 | 0.11 | 0.23 | 0.18 | −2.27 | E3 ubiquitin ligase |
| YIL051C | MMF1 | 0.24 | 0.63 | 0.70 | −2.42 | mitochondrial translation |
The list is limited to deletions exhibiting the following fitness scores: for sensitive strains, at least 1.3 for one peptide and less than 0.5 for all other three peptides; for resistant strains, at least −1.3 for one peptide and more than −0.5 for all other three peptides. Fitness scores specific to one peptide treatment are in bold.
Log2 hybridization intensity ratio between untreated and treated samples.
Several other deletion strains with high fitness scores and which were responsive to all treatments are presented in Table 2. Some of these deletions belong to the functional groups found to be enriched by the DAVID analysis (RPS27B, POP2, and COG3; see below). Others are associated with response to stress, among other functions (WHI2, SIT4, RHR2, and GRX4). Among the most sensitive strains in the present screen is a deletion mutant in NPR1. Npr1p is a protein kinase that indirectly regulates plasma membrane localization of amino acid permease Gap1p (36) but also affects other cellular functions, including RAS/cAMP pathway (37).
To obtain better insights into cellular processes affecting action of antimicrobial peptides, and to further evaluate the extent of similarities and differences between treatments, we employed two global analytical approaches, DAVID and GSEA.
DAVID analysis.
Identification of functional groups of genes by DAVID (the Database for Annotation, Visualization and Integrated Discovery) (38) analysis aims at identification of functional groups of genes overrepresented among those selected in genomics experiments. The results of such analysis probing all 425 identified mutants are shown in Table 4. One of the functional gene groups identified by DAVID, described by the Gene Ontology (GO) term “vacuolar transport,” includes all of the ESCRT genes described above and listed in the Table 2. When this group of genes, combined with those associated with the RIM101 pathway activation, was subjected to cluster analysis (39), all ESCRT and RIM101 genes clustered together (Fig. 3), supporting the notion that deletions in these genes confer sensitivity to antimicrobial peptides via the same mechanism.
Table 4.
Functional groups of genes identified by DAVID
| Functional group | Fold enrichment | P value | Genesa |
|---|---|---|---|
| Ribosomal subunit | 21 | 2.5E-32 | MRPL4, MRPS5, RSM7, IMG1, RPL15B, RPL9A, RPS27B, RPS21B, RPS20, RPL34B, RPS13, RPS29A, RPS25A, YGR054W, RPS24A, RPS8A, MRPL8, NIP7, RPL17A, RPL2B, RPS14A, RPL40A, RPL34A, MRP1 |
| Ribosome biogenesis | 13 | 5.8E-25 | UTP6, RRP8, LCB1, IMP4, UAF30, PWP1, NOP12, RSA1, NOP14, NOP8, RPP1, LSM7, LSM3, UTP9, NOP58, NOP15, LSM4, POP6, NIP7, RPS14A, LSM8, FAF1 |
| Vacuolar transport | 37 | 7.2E-22 | VPS23, VPS38, VPS30, VPS24, VPS60, SNF7, VPS25, MVB12, VPS22, VPS20, VTA1, DID4, VPS36, VPS28 |
| Negative regulation of transcription | 26 | 8.9E-17 | MKS1, YGR122W, SPT6, SSN2, DOT6, SSN8, OPI1, CCL1, MOT3, SRB8, SUM1, DLS1 |
| RNA splicing | 30 | 1.6E-11 | LSM8, LSM3, SLU7, LSM4, RDS3, BUD31, MUD2, SMD2 |
| Protein amino acid glycosylation | 50 | 6.4E-11 | ALG7, GDA1, ALG6, ALG3, ALG5, ERD1, OST1 |
| Golgi vehicle transport | 23 | 9.8E-11 | SYS1, PEP12, SEC22, COG3, VPS52, YKT6, GOS1, COG1 |
| Transcription from RNA polymerase I promoter | 105 | 6.7E-9 | RPA49, RPB8, RPO26, UAF30, RPA135 |
| Mitochondrial transport | 57 | 3.5E-8 | TIM18, TIM12, AFG3, TOM22, TOM5 |
| Mitochondrion inner membrane | 29 | 5.8E-7 | MRS3, MDM31, QRI5, GGC1, CTP1 |
| Transcription regulation | 11 | 2.9E-5 | NOT3, POP2, MED4, SSN2, SRB4 |
Essential genes are in bold.
Fig 3.

Clustering of all deletions in genes belonging to functional group represented by GO term “vacuolar transport” and those associated with RIM101 pathway (red). Names of genes encoding components of ESCRT known to be required for proteolytic processing of Rim101p are indicated in blue. In the heatmap, red and green correspond to fitness defect and gain, respectively.
Three functional groups of mutations (Table 4), together encompassing 48 genes, are represented by the GO terms “ribosomal subunit,” “ribosomal biogenesis,” and “transcription from RNA polymerase I promoter.” A recently reported meta-analysis of a large number of transcriptome and fitness profiling studies of S. cerevisiae has shown that deletions of ribosomal genes generally confer resistance to many stress conditions (40). This trend is consistent with noted downregulation of ribosomal genes under similar stress conditions (40–42). It was proposed that decreased ribosomal function led to protection from many stresses by lowering growth rates (40) or by enabling redirection of cellular resources to other defense mechanisms (41). In contrast, in our screen, the majority of deletions in genes encoding cytoplasmic ribosomal proteins led to fitness defects (sensitivity) under the treatment with the four peptides (Fig. 4). A possible explanation for this difference is that defensive measures protecting yeasts from antimicrobial peptides may require increase in the levels of de novo protein synthesis. This notion is consistent with very strong effect of mutations disrupting signaling via the RIM101 pathway also observed in this fitness screen. In addition, the observed effect of deletions of ribosomal genes may be exacerbated by the fact that in the present screen, at each cycle, cells were exposed to the peptides at the stationary phase when content of ribosomal proteins is significantly reduced (43). Interestingly, closer examination of the data presented by Zakrzewska et al. (40) reveals that deletion of ribosomal genes leads to diminished fitness under conditions known to induce RIM101 pathway, i.e., alkaline pH and high concentrations of NaCl. In contrast to cytoplasmic ribosomes, deletions of genes encoding mitochondrial ribosomal proteins tend to confer some measure of resistance to the peptides in our screen (Fig. 4). Such deletions may affect the mitochondrial function, leading to lowered energy production, a condition known to protect against many antimicrobial peptides (14, 27, 44, 45).
Fig 4.
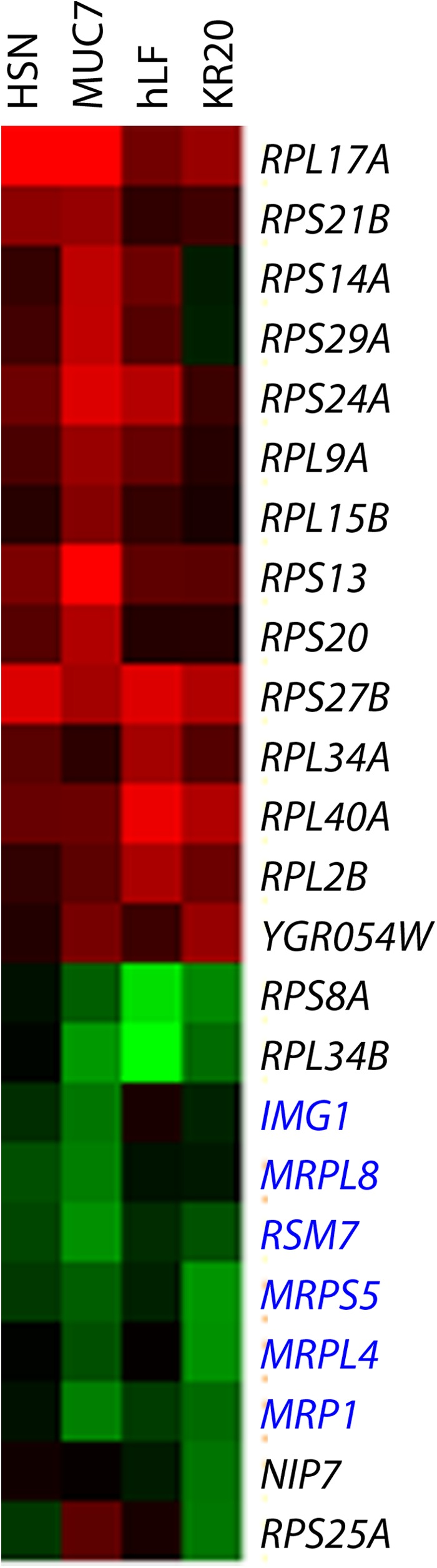
Clustering of all deletions in genes belonging to functional group represented by GO term “ribosomal subunit.” Names of genes encoding mitochondrial proteins are in blue. In the heatmap, red and green correspond to fitness defect and gain, respectively.
Another functional group of deletions identified in the screen consists of genes classified by GO terms “Golgi vesicle transport” and “protein amino acid glycosylation” (Table 4). The majority of the former encode subunits of tethering complex conserved oligomeric Golgi (COG) and elements of SNARE interacting with this complex. The COG is involved in intra-Golgi trafficking and is required for proper functioning of the process of protein glycosylation (46, 47). Those in the latter group are associated with earlier stages of oligosaccharide chain assembly which take place in endoplasmic reticulum (48). Deletions of genes belonging to these groups may, like deletions of ribosomal genes, disrupt the supply of new proteins needed for the required protective response. In addition, defective protein glycosylation may also lead to changes in the cell wall structure and, in consequence, affect initial interaction between the peptides and yeast cell surface.
“Negative regulation of transcription,” “transcription regulation,” and “RNA splicing,” are GO terms describing 25 genes whose deletions also affect S. cerevisiae growth in the presence of the tested antimicrobial peptides. These gene sets, together with ribosomal functions noted above, suggest the importance of changes in gene expression to enable defense mechanisms against antimicrobial peptides.
GSEA analysis.
While the number of genes above the fitness score threshold for individual peptides was too small to obtain meaningful results by the DAVID analysis, another method, GSEA (Gene Set Enrichment Analysis) (49), could be employed to examine individual peptide treatments. This technique relies on ranking of the magnitudes of phenotypic responses of all genes rather than on a list of genes exhibiting fitness scores above a fixed threshold. It estimates to what degree genes belonging to a given functional group are concentrated near the upper or lower end of the ranked data set (49). Gene sets exhibiting highest enrichment in GSEA are shown in Table 5. The results point to some differences between treatments, with a number of gene sets, e.g., “putative OCA complex” and “death,” as well as those associated with ribosomal functions, limited to only some peptides. In most cases, differences between the peptide treatments are quantitative and may indicate differences in relative importance of various biological processes to the effectiveness of individual peptides, rather than fundamental differences in their modes of action.
Table 5.
GSEA analysisa
| Gene set | Sizeb | MUC7 |
HSN |
KR20 |
Hlf |
||||
|---|---|---|---|---|---|---|---|---|---|
| NES | FDR | NES | FDR | NES | FDR | NES | FDR | ||
| Cytosolic small ribosomal subunit | 61 | 2.335 | 0.000 | 1.767 | 0.147 | — | — | 1.752 | 0.137 |
| rRNA metabolic process | 245 | 2.025 | 0.003 | 1.529 | 0.491 | — | — | 1.280 | 0.721 |
| Endosome transport | 73 | 1.918 | 0.021 | 1.348 | 0.748 | 1.405 | 0.669 | 1.460 | 0.504 |
| tRNA wobble base modification | 25 | 1.754 | 0.120 | 1.407 | 0.644 | 1.923 | 0.022 | 1.564 | 0.362 |
| Actin cytoskeleton organization | 85 | 1.872 | 0.033 | 1.278 | 0.777 | 1.050 | 0.881 | 1.602 | 0.292 |
| Putative OCA complex | 6 | 1.573 | 0.330 | — | — | 1.603 | 0.355 | 1.889 | 0.053 |
| Oligosaccharide metabolic process | 36 | 1.510 | 0.425 | 1.463 | 0.568 | 1.618 | 0.338 | 1.896 | 0.054 |
| mRNA cleavage factor complex | 20 | 1.379 | 0.537 | 1.877 | 0.073 | 1.176 | 0.888 | 0.717 | 0.985 |
| Small nuclear ribonucleoprotein complex | 57 | 1.393 | 0.520 | 1.843 | 0.077 | 1.182 | 0.885 | 0.951 | 0.945 |
| Death | 27 | — | — | 1.821 | 0.098 | 1.261 | 0.828 | — | — |
NES, normalized enrichment score; FDR, false discovery rate q value (49). Gene set listed in the order of the lowest to the highest FDR (the lowest FDR for each gene set shown in bold); —, gene set not detected by GSEA.
Number of genes in each gene set.
Comparison of the results of the current and previous MUC7 screens.
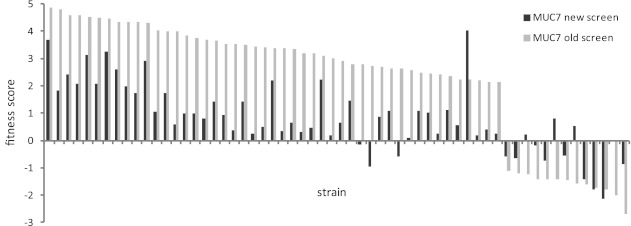
We were also interested in how the present fitness screen of the nonessential set of mutants treated with MUC7 peptide compares to the analogous screen we performed earlier (9). To account for the level of noise typical in such high-throughput experiments, we adjusted the threshold fitness scores to 0.5 and −0.5 in the present data set. With these relaxed criteria, of the 60 strains identified in the old screen, two-thirds, or 40 strains, showed hypersensitivity or resistance in the new. Four strains exhibited scores with opposite signs, and 16 had no altered fitness. A graph showing fitness scores of all 60 strains in both experiments is presented in Fig. 5.
Fig 5.
Comparison of the fitness scores of the 60 strains identified in the old screen of the S. cerevisiae strains with deletions of homozygous nonessential genes treated with MUC7 peptide (9) with the fitness scores of the same strains obtained in the present screen. Strains are sorted according to the fitness score values of the old screen.
Conclusion.
The comparative fitness screen presented in this report reveals several cellular functions important for survival and growth of S. cerevisiae cells in the presence of the four antimicrobial peptides tested. It confirms that the previously identified RIM101 signaling pathway plays a prominent role in regulating protective measures against these CAMPs. It adds other functions, including protein synthesis and glycosylation, as well as regulation of transcription, that are consistent with the presence of active response to the peptides requiring changes in the pattern of gene expression. Importantly, the most relevant elements of this response are shared by all four peptides tested in this screen, despite differences in their structures and physicochemical properties.
In addition to the homozygous nonessential mutant collection, in the present study we also screened a set of essential heterozygous mutants. Such mutants are commonly employed in haploinsufficiency profiling, a powerful tool for identification of protein targets of drugs (50). The presented results do not indicate the existence in S. cerevisiae of a protein acting as a specific target of any of the four tested antimicrobial peptides. Deletion mutants in essential genes with significant fitness scores appear to be distributed among most of the gene categories identified by the DAVID analysis (Table 4). Therefore, their effects on fitness seem to be associated with the mutant phenotypes and not with physical interactions between peptides and protein products of the deleted genes.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by NIH/NIDCR grants RO1DE009820 (L.A.B.) and R03DE019880 (M.L.), AADR Student Research Fellowship (N.E.S.), and the UB Dental Medicine Centennial Fund (S.B. and N.E.S.) and from the Canadian Cancer Society and the NHGRI (C.N. and A.Y.L.).
Footnotes
Published ahead of print 3 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01439-12.
REFERENCES
- 1. Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
- 2. Reddy KV, Yedery RD, Aranha C. 2004. Antimicrobial peptides: premises and promises. Int. J. Antimicrob. Agents 24:536–547 [DOI] [PubMed] [Google Scholar]
- 3. Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395 [DOI] [PubMed] [Google Scholar]
- 4. Bechinger B. 2011. Insights into the mechanisms of action of host defence peptides from biophysical and structural investigations. J. Pept. Sci. 17:306–314 [DOI] [PubMed] [Google Scholar]
- 5. Huang HW. 2006. Molecular mechanism of antimicrobial peptides: the origin of cooperativity. Biochim. Biophys. Acta 1758:1292–1302 [DOI] [PubMed] [Google Scholar]
- 6. Nicolas P. 2009. Multifunctional host defense peptides: intracellular-targeting antimicrobial peptides. FEBS J. 276:6483–6496 [DOI] [PubMed] [Google Scholar]
- 7. Wimley WC, Hristova K. 2011. Antimicrobial peptides: successes, challenges and unanswered questions. J. Membr. Biol. 239:27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lupetti A, Paulusma-Annema A, Welling MM, Senesi S, van Dissel JT, Nibbering PH. 2000. Candidacidal activities of human lactoferrin peptides derived from the N terminus. Antimicrob. Agents Chemother. 44:3257–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lis M, Fuss JR, Bobek LA. 2009. Exploring the mode of action of antimicrobial peptide MUC7 12-mer by fitness profiling of Saccharomyces cerevisiae genomewide mutant collection. Antimicrob. Agents Chemother. 53:3762–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bobek LA, Situ H. 2003. MUC7 20-Mer: investigation of antimicrobial activity, secondary structure, and possible mechanism of antifungal action. Antimicrob. Agents Chemother. 47:643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Situ H, Bobek LA. 2000. In vitro assessment of antifungal therapeutic potential of salivary histatin-5, two variants of histatin-5, and salivary mucin (MUC7) domain 1. Antimicrob. Agents Chemother. 44:1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Situ H, Wei G, Smith CJ, Mashhoon S, Bobek LA. 2003. Human salivary MUC7 mucin peptides: effect of size, charge and cysteine residues on antifungal activity. Biochem. J. 375:175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wei GX, Bobek LA. 2005. Human salivary mucin MUC7 12-mer-L and 12-mer-D peptides: antifungal activity in saliva, enhancement of activity with protease inhibitor cocktail or EDTA, and cytotoxicity to human cells. Antimicrob. Agents Chemother. 49:2336–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lis M, Bobek LA. 2008. Proteomic and metabolic characterization of a Candida albicans mutant resistant to the antimicrobial peptide MUC7 12-mer. FEMS Immunol. Med. Microbiol. 54:80–91 [DOI] [PubMed] [Google Scholar]
- 15. Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391 [DOI] [PubMed] [Google Scholar]
- 16. Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A, Garrett-Engele P, Rush CM, Bard M, Schimmack G, Phillips JW, Roberts CJ, Shoemaker DD. 2004. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116:121–137 [DOI] [PubMed] [Google Scholar]
- 17. Pierce SE, Davis RW, Nislow C, Giaever G. 2007. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat. Protoc. 2:2958–2974 [DOI] [PubMed] [Google Scholar]
- 18. Lamb TM, Mitchell AP. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lamb TM, Xu W, Diamond A, Mitchell AP. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276:1850–1856 [DOI] [PubMed] [Google Scholar]
- 20. Penalva MA, Tilburn J, Bignell E, Arst HN., Jr 2008. Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 16:291–300 [DOI] [PubMed] [Google Scholar]
- 21. Rothstein DM, Spacciapoli P, Tran LT, Xu T, Roberts FD, Dalla Serra M, Buxton DK, Oppenheim FG, Friden P. 2001. Anticandida activity is retained in P-113, a 12-amino-acid fragment of histatin 5. Antimicrob. Agents Chemother. 45:1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koshlukova SE, Lloyd TL, Araujo MW, Edgerton M. 1999. Salivary histatin 5 induces non-lytic release of ATP from Candida albicans leading to cell death. J. Biol. Chem. 274:18872–18879 [DOI] [PubMed] [Google Scholar]
- 23. Mochon AB, Liu H. 2008. The antimicrobial peptide histatin-5 causes a spatially restricted disruption on the Candida albicans surface, allowing rapid entry of the peptide into the cytoplasm. PLoS Pathog. 4:e1000190 doi:10.1371/journal.ppat.1000190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruissen AL, Groenink J, Helmerhorst EJ, Walgreen-Weterings E, Van't Hof W, Veerman EC, Nieuw Amerongen AV. 2001. Effects of histatin 5 and derived peptides on Candida albicans. Biochem. J. 356:361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. den Hertog AL, van Marle J, van Veen HA, Van't Hof W, Bolscher JG, Veerman EC, Nieuw Amerongen AV. 2005. Candidacidal effects of two antimicrobial peptides: histatin 5 causes small membrane defects, but LL-37 causes massive disruption of the cell membrane. Biochem. J. 388:689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Helmerhorst EJ, Wvan'T Hof Breeuwer P, Veerman EC, Abee T, Troxler RF, Amerongen AV, Oppenheim FG. 2001. Characterization of histatin 5 with respect to amphipathicity, hydrophobicity, and effects on cell and mitochondrial membrane integrity excludes a candidacidal mechanism of pore formation. J. Biol. Chem. 276:5643–5649 [DOI] [PubMed] [Google Scholar]
- 27. Helmerhorst EJ, Breeuwer P, Wvan'T Hof Walgreen-Weterings E, Oomen LC, Veerman EC, Amerongen AV, Abee T. 1999. The cellular target of histatin 5 on Candida albicans is the energized mitochondrion. J. Biol. Chem. 274:7286–7291 [DOI] [PubMed] [Google Scholar]
- 28. Vylkova S, Li XS, Berner JC, Edgerton M. 2006. Distinct antifungal mechanisms: beta-defensins require Candida albicans Ssa1 protein, while Trk1p mediates activity of cysteine-free cationic peptides. Antimicrob. Agents Chemother. 50:324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Braff MH, Hawkins MA, Di Nardo A, Lopez-Garcia B, Howell MD, Wong C, Lin K, Streib JE, Dorschner R, Leung DY, Gallo RL. 2005. Structure-function relationships among human cathelicidin peptides: dissociation of antimicrobial properties from host immunostimulatory activities. J. Immunol. 174:4271–4278 [DOI] [PubMed] [Google Scholar]
- 30. Henzler Wildman KA, Lee DK, Ramamoorthy A. 2003. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry 42:6545–6558 [DOI] [PubMed] [Google Scholar]
- 31. Mendez-Samperio P. 2010. The human cathelicidin hCAP18/LL-37: a multifunctional peptide involved in mycobacterial infections. Peptides 31:1791–1798 [DOI] [PubMed] [Google Scholar]
- 32. Williams RL, Urbe S. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8:355–368 [DOI] [PubMed] [Google Scholar]
- 33. Xu W, Smith FJ, Jr, Subaran R, Mitchell AP. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 15:5528–5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ikeda M, Kihara A, Denpoh A, Igarashi Y. 2008. The rim101 pathway is involved in rsb1 expression induced by altered lipid asymmetry. Mol. Biol. Cell 19:1922–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mattiazzi M, Jambhekar A, Kaferle P, Derisi JL, Krizaj I, Petrovic U. 2010. Genetic interactions between a phospholipase A2 and the Rim101 pathway components in S. cerevisiae reveal a role for this pathway in response to changes in membrane composition and shape. Mol. Genet. Genomics 283:519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Donnell AF, Apffel A, Gardner RG, Cyert MS. 2010. Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell 21:3552–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnston SD, Enomoto S, Schneper L, McClellan MC, Twu F, Montgomery ND, Haney SA, Broach JR, Berman J. 2001. CAC3(MSI1) suppression of RAS2(G19V) is independent of chromatin assembly factor I and mediated by NPR1. Mol. Cell. Biol. 21:1784–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zakrzewska A, Boorsma A, Beek AT, Hageman JA, Westerhuis JA, Hellingwerf KJ, Brul S, Klis FM, Smits GJ. 2010. Comparative analysis of transcriptome and fitness profiles reveals general and condition-specific cellular functions involved in adaptation to environmental change in Saccharomyces cerevisiae. Omics 14:603–614 [DOI] [PubMed] [Google Scholar]
- 41. Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437–440 [DOI] [PubMed] [Google Scholar]
- 44. Gyurko C, Lendenmann U, Helmerhorst EJ, Troxler RF, Oppenheim FG. 2001. Killing of Candida albicans by histatin 5: cellular uptake and energy requirement. Antonie Van Leeuwenhoek 79:297–309 [DOI] [PubMed] [Google Scholar]
- 45. Lehrer RI, Ganz T, Szklarek D, Selsted ME. 1988. Modulation of the in vitro candidacidal activity of human neutrophil defensins by target cell metabolism and divalent cations. J. Clin. Invest. 81:1829–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brocker C, Engelbrecht-Vandre S, Ungermann C. 2010. Multisubunit tethering complexes and their role in membrane fusion. Curr. Biol. 20:R943–R952 [DOI] [PubMed] [Google Scholar]
- 47. Smith RD, Lupashin VV. 2008. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr. Res. 343:2024–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Helenius A, Aebi M. 2001. Intracellular functions of N-linked glycans. Science 291:2364–2369 [DOI] [PubMed] [Google Scholar]
- 49. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith AM, Ammar R, Nislow C, Giaever G. 2010. A survey of yeast genomic assays for drug and target discovery. Pharmacol. Ther. 127:156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.