Abstract
Vibrio fischeri proliferates in a sessile, stable community known as a biofilm, which is one alternative survival strategy of its life cycle. Although this survival strategy provides adequate protection from abiotic factors, marine biofilms are still susceptible to grazing by bacteria-consuming protozoa. Subsequently, grazing pressure can be controlled by certain defense mechanisms that confer higher biofilm antipredator fitness. In the present work, we hypothesized that V. fischeri exhibits an antipredator fitness behavior while forming biofilms. Different predators representing commonly found species in aquatic populations were examined, including the flagellates Rhynchomonas nasuta and Neobodo designis (early biofilm feeders) and the ciliate Tetrahymena pyriformis (late biofilm grazer). V. fischeri biofilms included isolates from both seawater and squid hosts (Euprymna and Sepiola species). Our results demonstrate inhibition of predation by biofilms, specifically, isolates from seawater. Additionally, antiprotozoan behavior was observed to be higher in late biofilms, particularly toward the ciliate T. pyriformis; however, inhibitory effects were found to be widespread among all isolates tested. These results provide an alternative explanation for the adaptive advantage and persistence of V. fischeri biofilms and provide an important contribution to the understanding of defensive mechanisms that exist in the out-of-host environment.
INTRODUCTION
Biofilms form at almost every surface that is in contact with water, and they are a natural part of aquatic ecosystems (1, 2). Moreover, biofilm communities offer a refuge toward diverse stresses, such as antibiotics (3–5), dehydration and osmotic stress (5, 6), UV light exposure (6, 7), and starvation (3). The pronounced stress resistance of biofilms has been observed to be prevalent in marine communities, particularly ones comprised of environmental Vibrio biofilms (2, 8, 9).
Biofilms formed by Vibrio species are ubiquitous in aquatic ecosystems, although no study has specified the prevalence of Vibrio fischeri biofilms. V. fischeri (and other marine bacteria) in its planktonic state are found over broad geographical ranges, and their biofilms are subjected to multiple physiological stresses that lead to alterations in bacterial physiology (promoting bacterial fitness and bacterial speciation) (10). Thus, the survival of planktonic cells and biofilms in the environment is not only defined by the capacity to overcome abiotic pressures but also by the ability to serve as a protective niche against natural protozoan consumers (11, 12). Thus, an alternative function for the prevalence and relative fitness of V. fischeri biofilms is that they serve as refuges to combat a range of predators, including protozoan grazers. Grazing is one of the most common mortality factors of bacterial populations (13–15) and causes rapid changes in the morphology and species composition of microbial communities (13, 16–18). Interactions between bacteria and protozoa within biofilm communities remain largely unexplored; however, recent studies have revealed the impact of grazing on the dynamics of natural biofilm communities. Quorum sensing is an important factor for antipredatory activity in many bacterial species (19–22), and observations have suggested that bacterial genetic diversity enhances grazing resistance (23). Grazing resistance was observed in biofilms of Pseudomonas aeruginosa when early and late biofilm communities exhibited antipredatory behavior against two flagellates (Bodo saltans and Rhynchomonas nasuta) and the ciliate Tetrahymena sp. (18). Similar results were also observed when communities of Vibrio cholerae prevented predation through an antiprotozoal factor regulated by the response regulator HapR (14, 20). Moreover, V. cholerae biofilms exhibit widespread grazing resistance among toxigenic and nontoxigenic isolates, which has an impact on strain distribution and cholera epidemics (20). Previous studies also reported that some bacterial communities synthesize chemical compounds (e.g., violacein) that inhibit protozoan feeding by inducing cell lysis. These specific chemical defenses are prevalent in the tropical aquatic and soil bacterium Chromobacterium violaceum and other marine bacteria, such as Janthinobacterium lividum and Pseudoalteromonas luteoviolacea (24).
The protozoan community in marine ecosystems is cosmopolitan and follows a succession pattern depending on the nature of the grazer. For example, early biofilm colonizers (or generalists, including flagellates and ciliates) are highly motile, allowing fast surface feeding, while intermediate late colonizers (some ciliates and amoebas) are classified as specialists and are abundant in mature biofilms (25, 26). Therefore, the aim of our study was to investigate whether V. fischeri biofilms are resistant to protozoan grazing, and if any differences exist in predator avoidance between various free-living and symbiotic strains. We tested symbiotic strains isolated from the light organ of two different squid genera, Euprymna (Indo-West Pacific) and Sepiola (Mediterranean), along with free-living strains isolated directly from seawater. Three different protozoan predators among the 20 most commonly reported species of predators were chosen for these studies (20, 25) and included two early-feeding flagellates, Neobodo designis and Rhynchomonas nasuta, and the late ciliate colonizer Tetrahymena pyriformis. This experimental setting allowed us to identify biofilm survival depending on (i) strain type, (ii) age of biofilm, (iii) protozoan colonizer, and (iv) protozoan feeding type, and these factors have implications for understanding how V. fischeri survives, persists, and diversifies in the presence of grazers.
MATERIALS AND METHODS
Bacterial and protozoan strains used in this study.
Bacterial strains used in this study are listed in Table 1. V. fischeri strains were grown on Luria-Bertani high-salt agar (LBS; with a per liter composition of 10 g tryptone, 5 g yeast extract, 20 g sodium chloride, 50 ml 1 M Tris [pH 7.5], 3.75 ml 80% glycerol, 15 g agar, and 950 ml distilled water) and incubated for 24 h at 28°C. V. fischeri strains were isolated from different squid light organs captured live (Euprymna or Sepiola) or directly from seawater (27). All strains were subsequently subcultured in LBS liquid medium (no agar) and incubated with moderate shaking (200 rpm) for 18 h. The benthic flagellate grazers R. nasuta and N. designis were isolated from Chowder Bay at the Sydney Institute for Marine Science (SIMS), New South Wales, Australia, treated with an antibiotic cocktail (kanamycin, gentamicin, streptomycin, ampicillin, and trobamycin at 150 μg/ml each), and diluted through many generations (∼15) to remove the natural bacterial community (14). R. nasuta and N. designis were maintained axenically in 0.5× nine salts solution medium (NSS; with a per liter composition of 8.8 g NaCl, 0.735 g Na2SO4, 0.04 g NaHCO3, 0.125 g KCl, 0.02 g KBr, 0.935 g MgCl2 · 6H2O, 0.205 g CaCl2 · 2H2O, 0.004 g SrCl2 · 6H2O, 0.004 g H3BO3). Additional cultures were supplemented with heat-killed P. aeruginosa, which served as prey. The ciliate T. pyriformis (CCAP 1630/1W; Culture Collection of Algae and Protozoa, Windmere, United Kingdom) was maintained in PPY medium (with a per liter composition of 20 g proteose peptone and 2.5 g yeast extract). Cultures were incubated at room temperature (20 to 23°C) for 2 weeks.
Table 1.
Vibrio fischeri strains used in this study
| Strain | Host | Location |
|---|---|---|
| WH1 | Free living | USA (Woods Hole, MA) |
| MDR7 | Free living | USA (Marina del Rey, CA) |
| CB37 | Free living | Australia (Coogee Bay, Sydney, NSW) |
| CB21 | Free living | Australia (Coogee Bay, Sydney, NSW) |
| CHB8 | Free living | Australia (Chowder Bay, Sydney, NSW) |
| CHB12 | Free living | Australia (Chowder Bay, Sydney, NSW) |
| CHB30 | Free living | Australia (Chowder Bay, Sydney, NSW) |
| BSM40 | Free living | France (Banyuls sur mer) |
| BSM46 | Free living | France (Banyuls sur mer) |
| BSM50 | Free living | France (Banyuls sur mer) |
| PP3 | Free living | USA (Kaneohe Bay, O'ahu, HI) |
| PP42 | Free living | USA (Kaneohe Bay, O'ahu, HI) |
| VLS2 | Free living | USA (Kaneohe Bay, O'ahu, HI) |
| SR5 | Sepiola robusta | France (Banyuls sur mer) |
| SL518 | Sepiola lingulata | France (Banyuls sur mer) |
| SA1G | Sepiola affinis | France (Banyuls sur mer) |
| SA18 | Sepiola affinis | France (Banyuls sur mer) |
| SA25 | Sepiola affinis | France (Banyuls sur mer) |
| SA70 | Sepiola affinis | France (Banyuls sur mer) |
| SI66 | Sepiola intermedia | Italy (Bari) |
| SI1D | Sepiola intermedia | France (Banyuls sur mer) |
| EM17 | Euprymna morsei | Japan (Tokyo Bay) |
| ET101 | Euprymna tasmanica | Australia (Townsville, QLD) |
| ETWW | Euprymna tasmanica | Australia (Woy Woy, NSW) |
| ETBB20 | Euprymna tasmanica | Australia (Botany Bay, Sydney, NSW) |
| ETBB45 | Euprymna tasmanica | Australia (Botany Bay, Sydney, NSW) |
| ETBB67 | Euprymna tasmanica | Australia (Botany Bay, Sydney, NSW) |
| ETSB1 | Euprymna tasmanica | Australia (Shark Bay, WA) |
| EB12 | Euprymna berryi | Japan (Tosa Bay) |
| ES114 | Euprymna scolopes | USA (Kaneohe Bay, O'ahu, HI) |
| ESP915 | Euprymna scolopes | USA (Paiko, O'ahu, HI) |
| ESL5 | Euprymna scolopes | USA (Kaneohe Bay, O'ahu, HI) |
| ESC9 | Euprymna scolopes | USA (Kaneohe Bay, O'ahu, HI) |
Grazing assays.
Grazing experiments were completed in 24-well microtiter plates as previously described (20) for both early and late biofilms. For the early setup, overnight cultures of all bacterial strains were incubated at a dilution of 106 cells per ml in a total of 1 ml of fresh Väätänen-NSS medium (VNSS; with a composition of 1 g peptone, 0.5 g yeast extract, 0.5 g dextrose, 0.01 g FeSO4 · 7H2O, 0.01 g Na2HPO4, mixed with 1 liter of NSS medium) (14), which allowed growth of both bacteria and protozoa. As a negative control, sterile uninoculated VNSS medium was used. Strains were grown for 6 h (to let the early biofilm to become established) at room temperature (28°C) in the 24-well microtiter plates, and subsequently the planktonic population was removed and replaced with 1 ml of fresh VNSS medium that contained overnight cultures of either R. nasuta or N. designis (early grazers) at an abundance of 102 cells per ml. In the late biofilm setup, overnight cultures of all bacterial strains were inoculated as described for the early biofilm setup, but with the bacterial biofilms forming for 24 h prior to addition of the late biofilm grazer T. pyriformis. After formation of mature (late) biofilms, the planktonic suspension was removed and Tetrahymena was added at a concentration of 102 cells per ml. Plates were incubated for 24 h at 20°C with shaking at 50 rpm. Additionally, as a positive control, bacterial cultures were inoculated in parallel (106 cells per ml in 1 ml of VNSS medium) and incubated without protozoan grazers for 6 h (early biofilms) or 24 h (late biofilms) with shaking (50 rpm) at 20°C. For each experiment, all samples were inoculated in triplicate (3 wells), and assays were repeated three times (3 independent studies), for a total of 9 wells per bacterial strain.
Enumeration of protozoans and quantification of biofilm formation.
Numbers of grazers and grazer growth rates were calculated from direct cell counts using light microscopy. The number of grazers was calculated when 5 μl of supernatant was fixed with acid Lugol's solution (5%, final concentration) and enumerated using direct microscopy. Bacterial biofilms were measured using a colorimetric assay, where the supernatant from each plate was removed and wells were washed three times with 1 ml of sterile VNSS medium. One milliliter of 0.2% crystal violet solution was added to each well, and the mixture was incubated for 30 min at room temperature. After incubation, the crystal violet solution was removed and plates were washed five times with distilled water and dried. One milliliter of ethanol (95%) was added, and plates were incubated for 30 min to allow the dye to solubilize. The contents of each well (including negative controls) were transferred to a new plate, and the optical density was measured at 562 nm using a plate reader (Bio-Tek FLX 800; MTX Lab systems Inc., VA). The optical density observed is directly proportional to the amount of biofilm formed, and values were corrected by blank readings (uninoculated VNSS wells). For biofilm biomass quantifications, pairwise comparisons were performed using two-factor analysis of variance and Tukey's post hoc comparisons in order to test for significant differences between treatments (grazing versus nongrazing). Three plates were used for evaluation of statistical significance (3 independent studies).
Scanning electron microscopy.
To observe toxic effects or morphological changes in the protozoan cultures, light microscopy and scanning electron microscopy (SEM) were performed. Overnight cultures of all strains were reinoculated in triplicate in glass test tubes containing 5 ml of VNSS with an immersed sterile coverslip. Early (2 sets of tubes) or late (1 set of tubes) biofilms were allowed to form. After 6 or 24 h of incubation, grazers were added (either Rhynchomonas or Neobodo for the early biofilms and Tetrahymena for the late biofilms) and incubated at room temperature for 24 h. After incubation, coverslips were washed with sterile VNSS, fixed with a 0.5% solution of glutaraldehyde, and gold coated for SEM with a Hitachi S34000 SEM apparatus (Hitachi, Schaumburg, IL) as previously described (19).
RESULTS
Resistance to grazing of early and late biofilms.
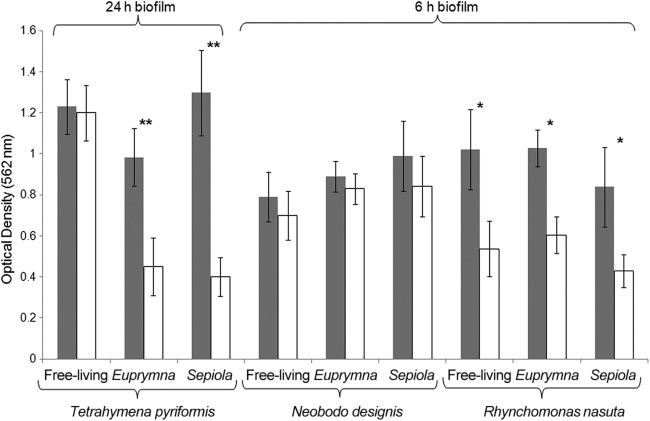
To determine potential inhibitory effects of biofilms (early and late) against three different protozoans with contrasting feeding modes, we compared the quantity of bacterial biofilm that was formed with and without exposure to each predator (Fig. 1). For early symbiotic strains, a significant reduction (P < 0.001) in biofilm biomass was observed after addition of the ciliated predator T. pyriformis, whereas biofilms formed by free-living strains (Table 1; Fig. 1) did not show any significant differences from nongrazed biofilms. In late biofilms, the raptorial feeder R. nasuta had no effect on the biomass of any of the strains examined; however, the flagellate N. designis was able to reduce the biofilm biomass of all strains by more than 30% (P < 0.05).
Fig 1.
Quantification of biofilm biomass of free-living and symbiotic (Euprymna and Sepiola) V. fischeri strains before grazing (gray bars) and after grazing (white bars) on early (R. nauta and N. designis) and late (T. pyriformis) biofilms. The optical densities were measured after solubilization of crystal violet, with the abundance of biofilm considered directly proportional to the optical density reading. Error bars represent the standard deviations. *, P < 0.05 for the difference between grazed and nongrazed bacteria; **, P < 0.01. The composite data are from 13 free-living strains, 8 Sepiola strains, and 12 Euprymna strains (see Table 1).
Biofilms cause a decrease in predator number.
In this study, we hypothesized that a reduction in the number of grazers would occur if the biofilm biomass did not decrease compared to cells that were consumed. Our experiments demonstrated that antiprotozoal effects occurred in both early and late biofilms and that variations in toxicity exist between the two early biofilm flagellates, consistent with other protists that have similar feeding modes (28, 29).
Protist abundance decreased in every case that the biofilms persisted (Fig. 2). Growth rates of T. pyriformis (ciliate, late grazer) were significantly reduced when exposed to biofilms formed by free-living strains (isolated from France, Australia, and the United States) (Table 1; Fig. 1), whereas high growth rates were observed when symbiotic biofilms were grazed. The same pattern was observed with N. designis (flagellate, early grazer), where numbers increased after grazing of early biofilms formed by both symbiotic and free-living strains. Interestingly, R. nasuta (flagellate, early grazer) numbers decreased when grazing on both symbiotic and free-living early biofilms.
Fig 2.
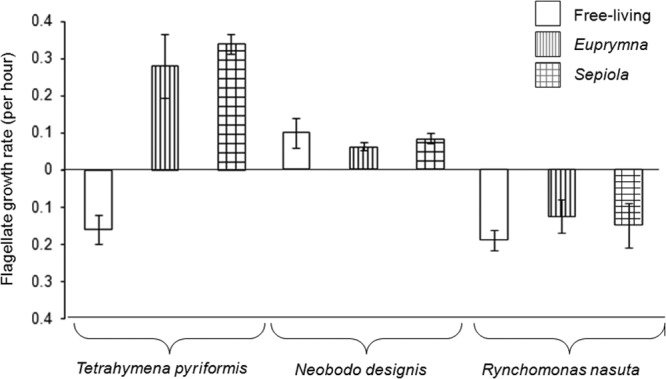
Protozoan growth rates after grazing on early and late biofilms. Negative numbers represent mortality, and positive numbers represent growth. Different bar patterns represent different groups of V. fischeri isolates (free-living or symbiotic with Euprymna or Sepiola). Errors bars are the standard deviations for each treatment. Each protozoan group was significantly different from the others (P < 0.05), and within the Tetrahymena group there was a significant difference in growth rate when free-living strains were grazed (P < 0.05).
Microscopy studies.
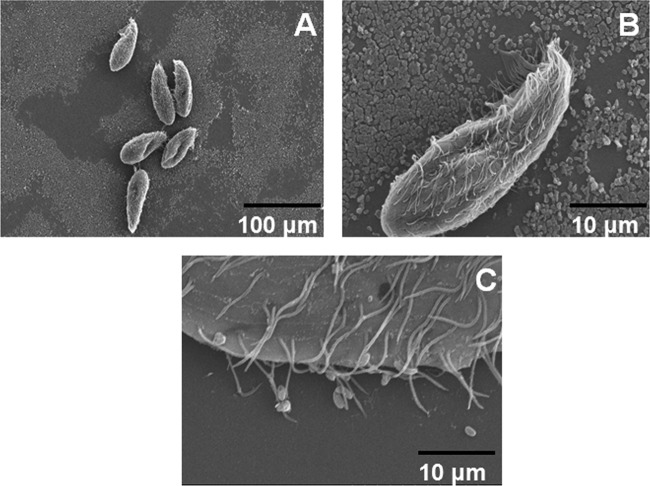
Biofilms that were preformed on a glass coverslip were exposed to all grazers for 24 h and then examined via SEM in order to visualize possible toxic effects after grazing. Vibrio strains that were successfully grazed by the free-swimming filter feeder T. pyriformis (including most of the symbiotic strains) were found on glass coverslips with T. pyriformis (Fig. 3). The small amount of glass-associated biofilm was distinguishable from the larger ciliated T. pyriformis (late grazer). Additionally, evidence of grazing activity was present and included signs of active feeding by a large number of ciliated protozoa (Fig. 3A and B). Discernible changes in grazer morphology were observed in T. pyriformis after grazing on free-living strains (Fig. 4). Two different phenotypes in protozoan structure were observed, including grazing morphology (Fig. 4A and B) and cell lysis (Fig. 4C and D). Late biofilms (24 h) were formed at the air-liquid interface of coverslips. Immediately after biofilm formation, Tetrahymena was added to the cultures, and after 24 h coverslips were analyzed using SEM. We observed two different coverslips per strain (for the protozoa Tetrahymena), and 60% of them exhibited a phenotype change for Tetrahymena, with protozoan lysis being the most common phenotype (after grazing biofilms formed by free-living strains).
Fig 3.
SEM images of T. pyriformis grazing dynamics. (A) T. pyriformis cells on a mature V. fischeri biofilm. (B) Magnification of T. pyriformis and the biofilm. (C) T. pyriformis grazing through a filter feeding strategy.
Fig 4.
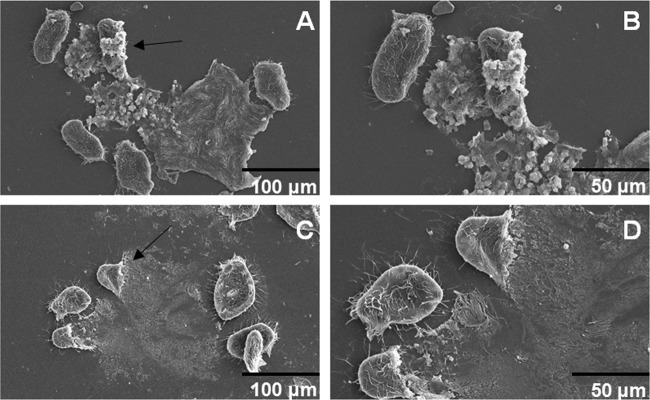
SEM images of the antiprotozoan effects on T. pyriformis. (A and B) Morphological changes in the grazer and the bacterial growth around the predator. (C and D) Cell lysis of the grazer. Arrows in panels A and C point to the corresponding grazers that are shown in panels B and D.
DISCUSSION
Vibrios are a ubiquitous diverse group of heterotrophic bacteria that are found in oceans, estuaries, and marine sediments worldwide. The diversity and dynamics of cooccurring populations has frequently been linked to environmental changes, including fluctuations in temperature, salinity, ocean hydrodynamics, and nutrient composition (10, 30). Therefore, environmental fluctuations lead to considerable bacterial microdiversity and evolution, including evolution of a wide variety of pathogenic and facultative mutualistic strains. The latter include the bioluminescent bacterium V. fischeri, which exists in a free-living stage (seawater) or as a mutualist of sepiolid squids and monocentrid fishes (31–33).
Understanding the influences of abiotic factors on V. fischeri biofilm populations has become a central theme of our research (10, 27); however, success in the environment is also dictated by the ability of the biofilm to tolerate natural protozoan consumers. Recent studies have emphasized the effects of protozoan grazers (17, 23, 34, 35), particularly in Vibrio communities, such as V. cholerae (12, 14), but not in communities of other species, including mutualists, such as V. fischeri and V. logei. In this study, we investigated the effects of common bacteriovores T. pyriformis (ciliate, late grazer), N. designis (flagellate, early grazer), and R. nasuta (flagellate, early grazer) on 33 V. fischeri isolates from different origins, including seawater and two squid genera: Euprymna, found in Indo-West Pacific waters, and Sepiola, found in the Mediterranean Sea. Symbiotic strains were chosen based on their similar growth and infection capabilities (all symbiotic strains could infect juvenile sepiolid squids) despite being from different host squids and geographical locations, whereas free-living strains were chosen based on their inability to infect juvenile squids yet their being found ubiquitously in the ocean.
Our results indicated that biofilms may protect V. fischeri from predation. For example, when the generalist ciliate T. pyriformis (late grazer) was added to late biofilms, a selective resistance to grazing was exhibited by free-living strains only and not symbiotic strains. Earlier work showed that free-living strains of V. fischeri are able to grow under a wider variety of conditions than their symbiotic relatives (10). This demonstrated that free-living strains have better adaptive mechanisms, which may be the result of genetic changes responsible for shifts in bacterial phenotypes (formation of biofilms) (2, 3).
It has been reported that biofilms of different species of environmental isolates secrete antiprotozoan factors, such as violacein (24) and others of an unknown nature (15, 20). These components may be present in V. fischeri biofilms, but they have not been identified thus far. T. pyriformis is a ciliated protozoa that grazes mature biofilms, similar to the ones found in the environment. Conversely, biofilm-like structures formed in the squid's light organ are expelled daily due to the diurnal cycle of the squid. This cycle allows exponential growth of V. fischeri followed by an expulsion of 99% of the symbionts into seawater every day at dawn (33, 36, 37). Thus, diel expulsion of a large number (1014) of bacteria possibly disrupts the preformed biofilm and therefore does not select for production of antiprotozoan compounds. Moreover, these strains may be exposed less frequently to predation and therefore are less capable of responding to predators that are only found in the aquatic environment, as in the case of the protozoan grazers used in this study.
Early biofilms were exposed to the flagellates N. designis and R. nasuta, which have different feeding modes and are considerably smaller in size than T. pyriformis (200 times bigger than V. fischeri). Our results revealed that early biofilms exposed to N. designis were consumed, resulting in a pronounced increase in flagellate numbers, which was opposite to observations for the flagellate R. nasuta. These contrasting results indicate that flagellate size is not necessarily correlated to grazing dynamics. For example, N. designis is a direct interception feeder, creating currents that carry suspended or loosely attached bacteria toward the mouth (28). R. nasuta is a raptorial feeder that grasps its prey with a proboscis-like structure (26). Another major difference between these two flagellates is that R. nasuta has a lower grazing (ingestion) success in sparsely populated biofilms due to its slower moving speed, and it is more successful in waters with higher prey densities (38). These different feeding modes may be crucial to the grazing success regarding V. fischeri biofilms. Symbiotic V. fischeri strains also exhibit resistance to R. nasuta predation (Fig. 1), indicating that some defensive mechanism still exists within biofilms formed by mutualistic strains. These defenses may be coopted to avoid the squid host innate immune response, which is mostly comprised of hemocytes and macrophages (39, 40). Future studies will focus on whether receptor-mediated phagocytosis in protozoa affects immune response evasion through biofilm formation.
In this study, protozoan abundance was reduced after grazing, and antiprotozoan effects on T. pyriformis (ciliate, late grazer) were observed (Fig. 2). Morphological changes of T. pyriformis occurred with bacteria possibly colonizing the protozoan predator, resulting in subsequent lysis of T. pyriformis cells. These two effects may be the result of inactivation of the predator or synthesis of lytic compounds that are released before or after ingestion of V. fischeri cells.
Quorum sensing controls biofilm formation (41) and has been described as one possible mechanism for antiprotozoal activity (20). In the case of V. fischeri, quorum sensing is under the control of the transcriptional regulator LuxR-LuxI (8). In addition to genes involved in light production, the LuxR regulon activates another number of genes involved in synthesis of efflux proteins, transporters, permeases, and proteases (8). The role of these quorum-sensing proteins in antipredator activity is unknown; however, they may contribute to the competitive fitness of biofilms under grazing pressure. Future studies will help determine the genetic factors responsible for protozoan death and may provide important clues of how survival of Vibrio biofilms is linked to quorum-sensing mechanisms.
The nature of the protozoan predator and its feeding characteristics may influence the impact of grazing on V. fischeri biofilms. Late/early and generalist/specialist protists have distinct grazing preferences, which are related to the type of V. fischeri strain examined (free living or symbiotic). Here we showed that V. fischeri biofilms have the ability to quickly adapt to grazing pressure, possibly by releasing antigrazing compounds and products that negatively influence protist survival. The observation that free-living strains are considerably more resistant to grazing pressure than symbiotic strains suggests that host selection may compromise the fitness of V. fischeri strains that are more amenable to a stable (and predatorless) environment than the external environment. Future research will address various adaptive mechanisms of bacteria-protist interactions that share fundamental processes with host immune responses.
ACKNOWLEDGMENTS
We thank P. Cooke for helping with the SEM images and two anonymous reviewers who provided helpful suggestions, ideas, and future studies.
This project was partially supported by The Centre for Marine Bio-Innovation and the Australian Research Council, grant DPDP1096481, and NSF IOS-0744498, NIH NIAID 1SC1AI081659-01, and NIH ARRA-3SC1AI081659-02S1 to M.K.N. A.C.-D. was supported by RISE (NIH NIGMS R25GM061222) and NASA (NMSGC).
We have no conflicts of interests to declare.
Footnotes
Published ahead of print 9 November 2012
REFERENCES
- 1. Watnick P, Kotler R. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yildiz FH, Visick KL. 2008. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chavez-Dozal A, Nishiguchi MK. 2011. Variation in biofilm formation among symbiotic and free-living strains of Vibrio fischeri. J. Basic Microbiol. 51:452S–458S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 5. Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 6. Espeland EM, Wetzel RG. 2001. Complexation, stabilization, and UV photolysis of extracellular and surface-bound glucosidase and alkaline phosphatase: implications for biofilms microbiota. Microb. Ecol. 42:572–585 [DOI] [PubMed] [Google Scholar]
- 7. Dalesman S, Karnik V, Lukowiak K. 2011. Sensory meadiation of memory blocking stressors in the pond snail Lymnaea stagnalis. J. Exp. Biol. 214:2528–2533 [DOI] [PubMed] [Google Scholar]
- 8. Antunes LCM, Schaefer AL, Ferreira RBR, Qin N, Stevens AM, Ruby EG, Greenberg EP. 2007. Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J. Bacteriol. 189:8387–8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ariyakumar DS, Nishiguchi MK. 2009. Characterization of two host specific genes, mannose sensitive hemagglutinin (mshA) and uridyl phosphate dehydrogenase (UDPDH) that are involved in the Vibrio fischeri-Euprymna tasmanica mutualism. FEMS Microbiol. Lett. 299:65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soto W, Gutierrez J, Remmega MD, Nishiguchi MK. 2009. Salinity and temperature effects on physiological responses of Vibrio fischeri from diverse ecological niches. Microb. Ecol. 57:140–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kadouri D, O'Toole GA. 2005. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol. 71:4044–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matz C, Kjelleberg S. 2005. Off the hook: how bacteria survive protozoan grazing. Trends Microbiol. 13:302–307 [DOI] [PubMed] [Google Scholar]
- 13. Adiba S, Nizak C, Van Baalen M, Denamur E, Depaulis F. 2010. From grazing resistance to pathogenesis: the coincidental evolution of virulence factors. PLoS One 5:e11882 doi:10.1371/journal.pone.0011882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Erken M, Weitere Kjelleberg M, McDougald SD. 2011. In situ grazing resistance of Vibrio cholerae in the marine environment. FEMS Microbiol. Ecol. 76:504–512 [DOI] [PubMed] [Google Scholar]
- 15. Matz C, Jurgens K. 2001. Effects of hydrophobic and electrostatic cell surface properties of bacteria on feeding of heterotrophic nanoflagellates. Appl. Environ. Microbiol. 67:814–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hazlett BA. 2003. Predator recognition and learned irrelevance in the crayfish Orconectes virilis. Ethology 109:765–780 [Google Scholar]
- 17. Miki T, Jacquet S. 2008. Complex interactions in the microbial world: underexplored key links between viruses, bacteria and protozoan grazers in aquatic environments. Aquat. Microb. Ecol. 51:195–208 [Google Scholar]
- 18. Weitere M, Bergfeld T, Rice SA, Matz C, Kjelleberg S. 2005. Grazing resistance of Pseudomonas aeruginosa biofilms depends on type of protective mechanism, developmental stage and protozoan feeding mode. Environ. Microbiol. 7:1593–1601 [DOI] [PubMed] [Google Scholar]
- 19. Greiner LL, Edwards JL, Shao J, Rabinak C, Entz D, Apicella MA. 2005. Biofilm formation by Neisseria gonorrhoeae. Infect. Immun. 73:1964–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S. 2005. Biofilm formation and phenotype variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 102:16819–16824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Queck SY, Weitere M, Moreno AM, Rice SA, Kjelleberg S. 2006. The role of quorum sensing mediated developmental traits in the resistance of Serratia marcescens biofilms against protozoan grazing. Environ. Microbiol. 8:1017–1025 [DOI] [PubMed] [Google Scholar]
- 22. Turovskiy Y, Rosenberg L, Chikindas M. 2007. Autoinducer-2-mediated quorum sensing is not involved in Listeria monocytogenes adaptive responses to the food preservatives lactic acid and nisin. J. Food Saf. 27:386–399 [Google Scholar]
- 23. Koh KS, Matz C, Tan CH, Le HL, Rice SA, Marshall DJ, Steinberg PD, Kjelleberg S. 2012. Minimal increase in genetic diversity enhances predation resistance. Mol. Ecol. 21:1741–1749 [DOI] [PubMed] [Google Scholar]
- 24. Matz C, Webb JS, Schupp PJ, Phang SY, Penesyan A, Egan S, Steinberg P, Kjelleberg S. 2008. Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS One 3:e2744 doi:10.1371/journal.pone.0002744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arndt H, Schmidt-Denter K, Auer B, Weitere M. 2003. Protozoans and biofilms, p 173–189 In Krumbein WE, Paterson DM, Zavarzin GA. (ed), Fossil and recent biofilms. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 26. Bohme A, Risse-Buhl U, Kusel K. 2009. Protists with different feeding modes change biofilm morphology. FEMS Microbiol. Ecol. 69:158–169 [DOI] [PubMed] [Google Scholar]
- 27. Zamborsky DJ, Nishiguchi MK. 2011. Phylogeographical patterns among sympatric populations of sepiloid squids and their Vibrio symbionts in the Mediterranean Sea. Appl. Environ. Microbiol. 77:642–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Artolozaga I, Ayo B, Latatu A, Azua I, Unanue M, Irriberri J. 2000. Spatial distribution of protists in the presence of macroaggregates in a marine system. FEMS Microbiol. Ecol. 33:191–196 [DOI] [PubMed] [Google Scholar]
- 29. Boenigk J, Arndt H. 2002. Bacterivory by heterotrophic flagellates: community structure and feeding strategies. Antonie Van Leeuwenhoek 81:465–480 [DOI] [PubMed] [Google Scholar]
- 30. Thompson JR, Randa MA, Marcelino LA, Tomita-Mitchell A, Lim E, Polz MF. 2004. Diversity and dynamics of a north atlantic coastal Vibrio community. Appl. Environ. Microbiol. 70:4103–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nishiguchi MK, Lopez JE, Boletzky SV. 2004. Enlightenment of old ideas from new investigations: the evolution of bacteriogenic light organs in squids. Evol. Dev. 6:41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nyholm SV, Nishiguchi MK. 2008. The evolutionary ecology of a sepiolid squid-Vibrio association: from cell to environment. Vie Milieu Paris 58:175–184 [PMC free article] [PubMed] [Google Scholar]
- 33. Ruby EG, McFall-Ngai MJ. 1999. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends Microbiol. 7:414–420 [DOI] [PubMed] [Google Scholar]
- 34. Jurgens K, Matz C. 2002. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuweenhoek 81:413–434 [DOI] [PubMed] [Google Scholar]
- 35. Kicklighter CE, Hay ME. 2006. Integrating prey defensive traits: contrasts of marine worm from temperate and tropical habitats. Ecol. Monogr. 76:195–215 [Google Scholar]
- 36. Ruby EG. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50:591–624 [DOI] [PubMed] [Google Scholar]
- 37. Ruby EG, Lee K. 1998. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl. Environ. Microbiol. 64:805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Erken M, Farrenschon N, Speckmann S, Arndt H, Weitere M. 2012. Quantification of individual flagellate-bacteria interactions within semi-natural biofilms. Protist 163:632–642 [DOI] [PubMed] [Google Scholar]
- 39. Castillo MG, Goodson MS, McFall-Ngai M. 2009. Identification and molecular characterization of a complement C3 molecule in lophotrochozoan, the Hawaiian bobtail squid Euprymna scolopes. Dev. Comp. Immunol. 33:69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McFall-Ngai MJ, Nyholm S, Castillo M. 2010. The role of the immune system in the initiation and persistence of the Euprymna scolopes-Vibrio fischeri symbiosis. Semin. Immunol. 22:48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101–104 [DOI] [PubMed] [Google Scholar]