Abstract
Positively charged gold nanoparticles (0.8-nm core diameter) reduced yeast survival, but not growth, at a concentration of 10 to 100 μg/ml. Among 17 resistant deletion mutants isolated in a genome-wide screen, highly significant enrichment was observed for respiration-deficient mutants lacking genes encoding proteins associated with the mitochondrion.
TEXT
The increasing use of nanomaterials in industrial processes and commercial products has generated a need for systematic assessment of potential biological and environmental risks (1, 2). This task is complicated by the sheer number and variety of nanomaterials and by the multitude of assays available to assess deleterious effects. Gold nanomaterials have received significant attention because of their unique physical and chemical properties that make them well suited for both basic biological research and biomedical applications (3, 4). A number of studies have evaluated the toxicity of a variety of gold nanoparticles (reviewed in references 5 and 6). Although the multiplicity of both gold nanoparticle type and toxicity assay complicate direct comparisons, a number of reports indicate that the type of particle tested in the present study (∼1 nm, positively charged) can elicit toxicity (7–10). While the yeast Saccharomyces cerevisiae has been a prominent and highly informative biological model (11), its use for evaluating the effects of nanomaterials appears to be limited based on few published reports (12–15). Here, we asked whether use of the yeast model could be informative with respect to determining the toxicity of the same functionalized gold nanoparticle previously found to cause significant mortality in embryonic zebrafish (8).
Functionalized gold nanoparticles.
Synthesis and characterization of the gold nanoparticles (AuNPs) have been described (16, 17). The particles (Au11[ligand]10) comprised a 0.8-nm 11-atom gold core, functionalized with either (i) positively charged N,N,N trimethylammoniumethanethiol (TMAT) as the iodide salt, (ii) negatively charged 2-mercaptoethanesulfonate (MES) as the sodium salt, or (iii) neutral 2-[2-(2-mercaptoethoxy)ethoxy]ethanol (MEEE) (Fig. 1A). Throughout this report, these functionalized gold nanoparticles are referred to as 0.8-nm AuTMAT, 0.8-nm AuMES, and 0.8-nm AuMEEE, respectively, to distinguish them from other gold nanoparticles described. Analogs of the TMAT functional group (Fig. 1B), tetramethylammonium iodide, tetramethylammonium chloride, and choline chloride, were purchased from Sigma-Aldrich.
Fig 1.
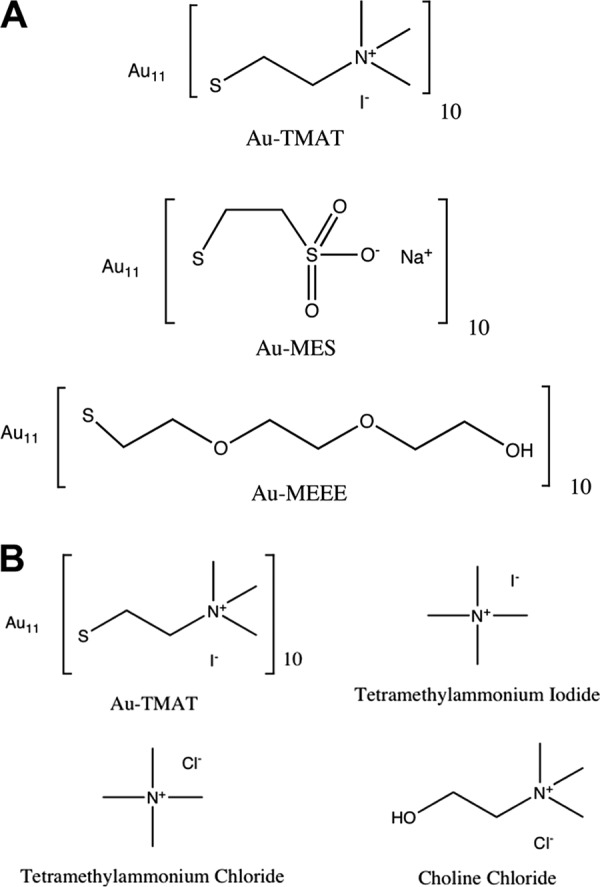
Structures of the 0.8-nm AuNPs and analogs. (A) Functional groups used to derivatize the nanogold particles. Positively charged trimethylammoniomethanethiol (TMAT), negatively charged 2-mercaptoethanesulfonate (MES), and neutral 2-[2-(2-mercaptoethoxy)ethoxy]ethanol (MEEE). (B) Structures of TMAT functional analogs. The molecular weights of AuTMAT, tetramethylammonium chloride, tetramethylammonium iodide, and choline chloride are 4,625.67, 109.6, 201.05, and 139.63 Da, respectively.
Yeast, media, toxicity assays.
Saccharomyces cerevisiae BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) was used to assess the effects of the functionalized 0.8-nm AuNPs on yeast survival and growth. Strain KK86 is a rho0 derivative of BY4742 lacking mitochondrial DNA (18). A nonessential yeast gene deletion library (19) constructed in the BY4742 genetic background (YSC1054; Open Biosystems, Inc.) was screened for resistance to these gold nanoparticles. S. cerevisiae was grown in yeast nitrogen base without amino acids (Difco) containing 2% glucose and supplemented with 20 μg/ml histidine, 30 μg/ml each of leucine and lysine, and 10 μg/ml of uracil (YNB and supplements) to satisfy auxotrophic requirements or in 1% yeast extract, 2% peptone, and 2% glucose (YEPD).
Fluorescence microscopy.
Yeast strains were grown and prepared for fluorescence microscopy essentially as described previously (20). Briefly, cells were grown statically for 24 h at 30°C in 200 μl of YNB in a 96-well plate, after which they were pelleted by centrifugation and fixed by resuspension in 70% ethanol at room temperature for 30 min. The fixed cells were washed once with sterile distilled water and resuspended in 20 μl of sterile distilled water. Five microliters of cell suspension were mixed with 5 μl of mounting medium containing DAPI (4′,6-diamidino-2-phenylindole; Molecular Probes, Invitrogen; catalog no. P36935). The mixture (2.5 μl) was spotted onto a slide, covered with a coverslip, and allowed to dry in the dark 1 to 2 h before sealing with transparent nail polish. Slides were held in the dark at room temperature for up to 3 days before being visualized using a Zeiss Axiovert S100 microscope equipped with a 100× objective, 2× zoom lens, and a Photometrics CoolSNAP HQ charge-coupled-device (CCD) camera controlled by MetaMorph 6.3 imaging software. For visualizing DAPI, excitation and emission wavelengths were 350 and 460 nm, respectively. Autofluorescence was used to visualize cell shape; excitation and emission wavelengths were 480 and 535 nm, respectively.
Yeast growth inhibition was assessed as a reduction in cell yield (A600) in treated versus control cells. Cells were grown overnight in YNB and supplements, washed twice in sterile distilled water, and then diluted 1,000-fold in duplicate 250-μl aliquots of YNB and supplements (control) or YNB, supplements, and AuNPs. Cells were incubated for 48 h at 30°C and 200 rpm in 1.5-ml screw-cap polypropylene tubes in triplicate, after which A600 values were measured.
To assess survival, cultures grown overnight in either YNB and supplements or YEPD were washed twice with sterile distilled water and then incubated in 100-μl aliquots of sterile distilled water at 105 to 107 cells/ml in 500-μl screw-cap polypropylene tubes containing each of the functionalized 0.8-nm AuNPs. At least three replicates were performed per strain at each AuNP dose. After 24 h at 30°C and 200 rpm, cells were plated on YEPD agar in duplicate, and colonies were counted after 48 h at 30°C to determine survival relative to control cells incubated in parallel under identical conditions in sterile distilled water lacking AuNPs. The number of cells killed in the water-only control was subtracted from the number killed in the parallel AuNP exposures. No differences in survival were observed between BY4742 that had been grown overnight in either YNB and supplements or YEPD.
Screening of yeast deletion library for mutants with enhanced survival in the presence of AuNPs.
The nonessential yeast gene deletion library was screened for mutants that exhibited better survival than the parent strain, BY4742, after 24 h of incubation in sterile distilled water containing AuNPs. The library was screened in pools consisting of about 100 mutants each, suspended in sterile distilled water. To minimize the possibility of underrepresentation of slower-growing mutants, all deletion mutants were initially grown individually in 96-well plates in YEPD for 24 h at 30°C, only after which cells were pooled (1 to 2 plates per pool), washed twice in sterile distilled water, and resuspended in sterile distilled water at about 3 × 107 cells/ml. Pools of cells were then incubated in 100-μl aliquots of sterile distilled water at about 105 cells/ml in 500-μl screw-cap polypropylene tubes (1 tube per pool) containing up to 20 ppm AuNPs. After 24 h at 30°C and 200 rpm, cells were plated on YEPD agar which was incubated 48 h at 30°C. All colonies of survivors were restreaked on YEPD and were retested individually for survival after 24 h of incubation in sterile distilled water in the presence of AuNPs. The survival of mutants that retested positive relative to BY4742 (P ≤ 0.05, two-sided Student's t test) was then evaluated at multiple concentrations of the AuNPs. The AuNP exposure protocol used to screen the library was developed based on preliminary experiments to determine conditions that the parental strain, BY4742, was unable to survive.
Mutant identification.
Deletion mutants that exhibited enhanced survival were identified by sequencing mutant-specific oligonucleotide tag sequences within a PCR product generated using primers complementary to sequences shared by all mutants. Colony PCR (21) was performed using the polymerase pfx (Invitrogen) according to the manufacturer's instructions. PCR products were purified (Qiaquick spin columns; Qiagen) by following the PCR cleanup protocol and sequenced at the Oregon State University Center for Genome Research and Biocomputing.
Assessment of toxicity.
Possible deleterious interactions between the 0.8-nm AuNPs and yeast were assessed initially as the ability to inhibit growth, measured as cell yield (A600) after incubation in YNB and supplements for 48 h. No reduction in yield was observed between BY4742 grown in the absence (control) and presence of the 0.8-nm AuTMAT, AuMES, or AuMEEE at concentrations as high as 100 μg/ml (data not shown). While we are not aware of published data on the response of yeast to these 0.8-nm AuNPs, a recent study reported that the same 0.8-nm AuTMAT NP and a 1.5-nm AuTMAT NP (a larger particle with the same surface coating), at concentrations ranging from 80 ppb to 250 ppm, induced significantly greater lethality in embryonic zebrafish than either the negatively charged AuMES or neutral AuMEEE NPs of the same sizes (8). A related but somewhat larger cationic AuNP (1.4-nm diameter) used to monitor endocytosis in log-phase S. cerevisiae spheroplasts at 5 to 10 μM was not reported to cause growth inhibition (22–24). This particle is quite different than the particles used in the present study. It is coated with phosphine ligands easily displaced in biological medium in the presence of thiols and has only 6 positive charges in the ligand shell compared to at least 30 positive charges in the 1.5-nm AuTMAT particle. Because the incubation period was not longer than 90 min prior to fixation of cells for microscopic analysis, the possibility of inhibition cannot be ruled out. Nonetheless, the process of endocytosis during the incubation was not found to be abnormal. The highest 0.8-nm AuTMAT concentration tested in the present study with no apparent deleterious effect on yeast growth was 21.6 μM (100 μg/ml).
We next tested whether the functionalized 0.8-nm AuNPs could reduce survival of nongrowing stationary-phase cells incubated in water over 24 h. While a reduction in survival was not observed for the cells incubated with the 0.8-nm AuMES or AuMEEE nanoparticles, reduced survival of BY4742 was observed at 10 ppb of positively charged 0.8-nm AuTMAT, the lowest concentration tested (Fig. 2). Within the range of 10 ppb and 100 ppm, a linear relationship was observed between the log of the AuTMAT dose and the log of the number of BY4742 cells killed. That is, a fixed number of cells were killed at a given AuTMAT concentration regardless of the number of cells treated, consistent with a requirement for direct and irreversible interaction between cells and some minimum number of AuTMAT particles. In order to distinguish between toxicity of the 0.8-nm AuTMAT nanoparticles and the positively charged quaternary ammonium functional group contained in the TMAT group alone, cells were exposed independently to the TMAT analogs tetramethylammonium iodide, tetramethylammonium chloride, and choline chloride (Fig. 1B) at functional group concentrations ranging from 500 to 900 μM or 2 to 4 times greater on a molar basis than the concentration of TMAT groups (216 μM) at the highest 0.8-nm AuTMAT dose tested, 100 μg/ml. No reduction in survival of BY4742 exposed to these TMAT analogs was observed relative to untreated control cells incubated in parallel (data not shown).
Fig 2.
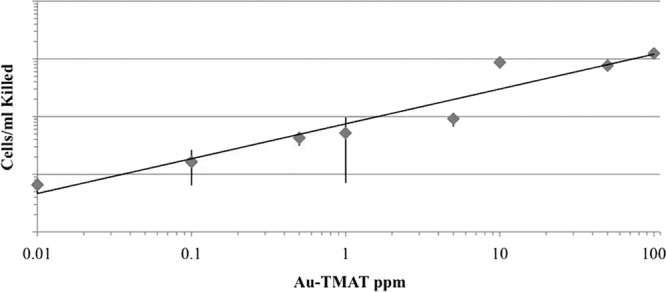
Stationary-phase BY4742 cells killed as a function of AuTMAT concentration. Cells were grown in either YEPD or YNB, washed, and suspended in sterile distilled water alone (control) or sterile distilled water supplemented with AuTMAT at 200 rpm and 30°C. Survival was assessed by plating onto duplicate YEPD plates after a 24-h incubation. The number of cells killed in the water-only control were subtracted from the number of cells killed in the AuTMAT exposures. Data are means of four experiments with three to 16 replicates performed at each concentration. Error bars are standard deviations.
Screen of yeast deletion library for resistant mutants.
In order to determine which genes might predispose yeast to 0.8-nm AuTMAT-induced damage during stationary phase, mutants better able to survive the exposure were sought by screening a library of nonessential yeast deletion mutants as described above, “Screening of yeast deletion library for mutants with enhanced survival in the presence of AuNPs.” Initially, 230 putative resistant mutants were isolated following exposure of pools of nongrowing deletion mutants to a concentration of 0.8-nm AuTMAT that killed an equal number of cells of the parent strain, BY4742. As described above, the screen for resistant mutants involved exposing about 105 CFU/ml cells to a concentration of AuTMAT that killed about 105 CFU/ml of the parent strain, BY4742. Upon retesting, 32 mutants were found to be reproducibly resistant and were identified by sequence analysis of mutant-specific oligonucleotide tags. Among the 32 mutants, 17 unique gene deletions were identified from among the total 4,750 mutants screened, indicating that multiple clones of the same mutants had been isolated (Table 1). As indicated in Table 1, 12 mutants were isolated once, two were isolated twice, one was isolated four times, and two were isolated six times.
Table 1.
List of genesa whose loss confers resistance to 0.8-nm AuTMAT
| Locus | Gene name (no. of times mutant was isolated) | Function (Saccharomyces Genome Database 25 December 2011) | RDb |
|---|---|---|---|
| YBR268w | MRPL37 (2) | Mitochondrial ribosomal protein of the large subunit | Yes |
| YCR046c | IMG1 (1) | Mitochondrial ribosomal protein of the large subunit; required for respiration and maintenance of mitochondrial genome | Yes |
| YER114c | BOI2 (1) | Protein implicated in polar growth, functionally redundant with Boi1p; interacts with bud-emergence protein Bem1p; contains an SH3 (src homology 3) domain and a PH (pleckstrin homology) domain | Yes |
| YGL166w | CUP2 (1) | Cu-binding transcription factor; activates transcription of CUP1-1 and CUP1-2 metallothionein genes in response to elevated Cu concentrations | No |
| YGR150c | CCM1 (2) | Mitochondrial 15s rRNA-binding protein; required for intron removal of COB and COX1 pre-mRNAs; has pentatricopeptide repeat (PPR) motifs; mutant has defective plasma membrane electron transport | Yes |
| YGR207c | CIR1 (4) | Mitochondrial protein that interacts with frataxin (Yfh1p); putative ortholog of mammalian electron transfer flavoprotein complex subunit ETF-beta; may play role in oxidative stress response | No |
| YHR006w | STP2 (6) | Transcription factor; activated by proteolytic processing in response to signals from the SPS sensor system for external amino acids; activates transcription of amino acid permease genes | Yes |
| YJL129c | TRK1 (1) | Component of Trk1p-Trk2p K transport system; 180-kDa high-affinity K transporter; phosphorylated in vivo and interacts physically with phosphatase Ppz1p, suggesting Trk1p activity is regulated by phosphorylation | No |
| YLR382c | NAM2 (6) | Mitochondrial leucyl-tRNA synthetase; also has direct role in splicing of several mitochondrial group I introns; indirectly required for mitochondrial genome maintenance | Yes |
| YLR439w | MRPL4 (1) | Mitochondrial ribosomal protein of the large subunit; homolog of prokaryotic L29 ribosomal protein; located at the ribosomal tunnel exit | Yes |
| YMR072w | ABF2 (1) | Mitochondrial DNA-binding protein involved in mitochondrial DNA replication and recombination; member of HMG1 DNA-binding protein family; activity may be regulated by protein kinase A phosphorylation | Yes |
| YMR155w | (1) | Unknown | No |
| YMR173w | DDR48 (1) | DNA damage-responsive protein; expression increases in response to heat-shock stress or treatments that produce DNA lesions; contains multiple repeats of amino acid sequence NNNDSYGS | No |
| YMR192w | GYL1 (1) | Putative GTPase-activating protein (GAP) with role in exocytosis; stimulates Gyp5p GAP activity on Ypt1p; colocalizes with Gyp5p at sites of polarized growth; interacts with Gyp5p, Rvs161p, and Rvs167p | No |
| YMR223w | UBP8 (1) | Ubiquitin-specific protease; member of SAGA (Spt-Ada-Gcn5-acetyltransferase) acetylation complex; required for SAGA-mediated deubiquitination of histone H2 | No |
| YPL183w-A | RTC6 (1) | Protein involved in translation; mutants have defects in biogenesis of nuclear ribosomes; sequence similar to prokaryotic ribosomal protein L36; may be a mitochondrial ribosomal protein encoded in the nucleus | Yes |
| YPR100w | MRPL51 (1) | Mitochondrial ribosomal protein of the large subunit | Yes |
Eight of the 17 gene deletions were confirmed independently to be responsible for resistance. Five deletion mutants were crossed to deletion strains of the opposite mating type to generate homozygous diploids that exhibited the same phenotype (GYL1Δ, ABF2Δ, STP2Δ, NAM2Δ, BOI1Δ mutants). Two were transformed with wild-type alleles of the deleted genes, which restored sensitivity (DDR48, CIR1). One deletion mutant obtained in a different strain background was also found to be resistant (TRK1Δ mutants).
A gene ontology (GO) analysis was undertaken to correlate loss of the genes that resulted in increased cell survival with specific components, processes, and functions (Table 2). Highly significant enrichment was observed for genes whose products localize to the mitochondrion and to the mitochondrial large ribosomal subunit in particular. The process of “mitochondrial organization” and the function “structural constituent of ribosome” were also enriched significantly. Of the 17 genes, loss of 10 has been reported to result in respiration deficiency, equivalent to a frequency of ∼60%, compared to ∼7% for the original library (319 among a total of 4,750 deletion mutants) (4). This represents an approximate 9-fold enrichment (P < 10−4, chi-square test). Among the 17 AuNP-resistant mutants reported here, five (the mrpl37Δ, ccm1Δ, nam2Δ, img1Δ, rtc6Δ mutants) were found to be sensitive to hydrogen peroxide (27) and seven (the abf2Δ, img1Δ, mrpl4Δ, mrpl51Δ, boi2Δ, stp2Δ, trk1Δ mutants) were found sensitive to either or both hydrogen peroxide and menadione (28) in previous genome-wide screens. Nine of these mutants are missing genes encoding products associated with the mitochondrion, the mrpl37Δ, ccm1Δ, nam2Δ, img1Δ, rtc6Δ, abf2Δ, img1Δ, mrpl4Δ, and mrpl51Δ mutants. While one study reported that a 1.4-nm gold nanoparticle coated with a negatively charged triphenylphosphine ligand generated significant oxidative stress and mitochondrial damage in HeLa cells (9), loss of functional mitochondria in yeast has not always been reported to increase oxidative stress (7, 29).
Table 2.
GO analyses comparing the 17 genes deleted in the AuTMAT-resistant mutants with the collection of 4,750 genes represented in the deletion library from which the 17 were derived
| Term | Frequency (%) among 17 AuTMAT-resistant mutants | Library frequency (%) (among 4,750 genes) | Corrected P valuea | Genes annotated to term |
|---|---|---|---|---|
| GO component terms | ||||
| Mitochondrial matrix | 8 genes (47.1) | 174 genes (3.7) | 1.92 × 10−6 | YBR268w, YCR046c, YGR207c, YLR382c, YLR439w, YMR072w, YPL183w-A, YPR100w |
| Mitochondrial large ribosomal subunit | 5 genes (29.4) | 41 genes (0.9) | 8.12 × 10−6 | YBR268w, YCR046c, YLR439w, YPL183w-A, YPR100w |
| Organelle lumen | 9 genes, 52.9) | 563 genes (11.9) | 0.00166 | YBR268w, YCR046c, YGR207c, YLR382c, YLR439w, YMR072w, YMR223w, YPL183w-A, YPR100w |
| GO process term | ||||
| Mitochondrial organization | 8 genes (47.1) | 238 genes (5.0) | 3.83 × 10−5 | YBR268w, YCR046c, YGR150c, YLR382c, YLR439w, YMR072w, YPL183w-A, YPR100w |
| GO function term | ||||
| Structural molecule activity | 6 genes (35.3) | 254 genes (5.3) | 0.00248 | YBR268w, YCR046c, YLR439w, YMR223w, YPL183w-A YPR100w |
| Structural constituent of ribosome | 5 genes (29.4) | 177 genes (3.7) | 0.00437 | YBR268w, YCR046c, YLR439w, YPL183w-A, YPR100w |
The corrected P values indicate the significance of the enrichment of genes with the associated terms. Analysis was performed 7 August 2011 (http://go.princeton.edu/cgi-bin/GOTermFinder/GOTermFinder).
As noted above, yeast cells were observed to take up a positively charged triphenylphosphine-stabilized 1.4-nm AuNP via endocytosis (23). Uptake was blocked in an end3 endocytosis mutant, while AuNPs accumulated in the early endosome of a sec18 secretory mutant. No apparent signs of toxicity were noted over the short 15- to 90-min exposure of spheroplasts to 5 μM of these 1.4-nm gold nanoparticles. Although potential toxicity was not a specific focus of subsequent studies of endocytosis using the same positively charged, triphenylphosphine-stabilized AuNPs and similar conditions, no reduction in cell viability was noted (22, 24). These results are consistent with our finding that yeast cell growth, assessed as cell yield, was unaffected by exposing intact cells to as much as 22 μM of the 0.8-nm AuTMAT nanoparticles, corresponding to the 100-μg/ml dose. The observation that a trk1Δ mutant was resistant suggests an alternative route for AuTMAT uptake. TRK1 encodes a potassium channel, and in Candida albicans, this same channel was found to mediate the toxicity of the cationic protein, histatin 5, whose molecular weight, 3,036 Da, is close to that of the 0.8-nm AuTMAT, 4,626 Da (30). The authors proposed a model whereby binding of histatin 5 by Tkr1 distorted channel shape, allowing leakage of ATP and potassium. We speculate that “jamming” of the S. cerevisiae Trk1 channel by the 0.8-nm AuTMAT might lead to similar leakage of essential cytoplasmic constituents. On the other hand, it is possible that AuTMAT could reduce cell survival in the absence of uptake through interaction with negatively charged phosphomannans in the cell wall or phospholipids in the cell membrane.
Five of the 17 resistant mutants (the ccmΔ, nam2Δ, mrpl4Δ, rtc6Δ, and mrpl51Δ mutants) were reported elsewhere to excrete significant amounts of glutathione after 48 h of growth in a YNB-based medium in an independent screen of the corresponding homozygous diploids (31). Whether these mutants accumulated or excreted excess glutathione during the 0.8-nm AuTMAT exposure performed in water at 30°C over 24 h in the present study is unknown. If so, thiol exchange with glutathione may have resulted in detoxification of the original AuTMAT NP.
Dose-response analysis of resistant mutants.
Once the 17 deletion mutants were identified, dose-response analysis was undertaken on clones retrieved from the original library rather than the clones that had been exposed to the 0.8-nm AuTMAT to minimize the possibility that second-site mutations may have been selected during exposure. Table 3 lists the mutants in decreasing order of resistance to 100 ppm of 0.8-nm AuTMAT. The most resistant mutant, the mrpl51Δ mutant, exhibited about 6-fold better survival than the parent strain, while the least resistant mutant, the YMR155wΔ mutant, exhibited only 16% better survival. At the 10-ppm 0.8-nm AuTMAT dose, four mutants exhibited the same (the boi2Δ, img1Δ, and rtc6Δ mutants) or worse (the abf2Δ mutant) survival than the parent. The other 13 mutants had better survival than the parent at both the 10- and 100-ppm 0.8-nm AuTMAT doses. One explanation for the observed enrichment in mutants impaired in mitochondrial protein synthesis is that such activity is critical for maintenance of the mitochondrial genome (32) and that the subsequent likely loss of the mitochondrial genome in these mutants accounts for their resistance. To test this possibility, we assayed survival of KK86, a BY4742 derivative lacking mitochondrial DNA (18) following treatment with AuTMAT. At the 10-ppm dose, the survival of KK86 was found to be the same as that of the parent strain BY4742, while at the 100-ppm AuTMAT dose, survival of BY4742 was 1.8 times better (P = 0.032). This indicates that the rho0 strain is actually more sensitive than the wild-type strain. We conclude that loss of the mitochondrial genome alone cannot explain the AuTMAT resistance. All mutants were also subjected to fluorescence microscopy using DAPI to determine the presence of mitochondrial DNA with BY4742 and KK86 serving as positive and negative controls, respectively. Among the 12 to 27 cells scored per strain, 25 to 100% contained mitochondrial DNA based on observation of a single spot of prominent fluorescence, with weaker spots of fluorescence throughout a cell (20) (see Table S1 and Fig. S1 in the supplemental material).
Table 3.
Resistant mutant cells killed as a function of the 0.8-nm AuTMAT dosea
| Deletion mutant | No. of mutant cells killed as % of BY4742 cells killed at: |
|
|---|---|---|
| 10 ppm AuTMAT | 100 ppm AuTMAT | |
| MRPL51Δ | 20.0 | 15.5 |
| CUP2Δ | 45.3 | 25.1 |
| TRK1Δ | 24.3 | 25.5 |
| UBP8Δ | 62.5 | 30.7 |
| ABF2Δ | >100 | 30.7 |
| DDR48Δ | 48.2 | 37.3 |
| STP2Δ | 37.5 | 38.9 |
| GYL1Δ | 69.7 | 39.5 |
| CCM1Δ | 47.2 | 42.5 |
| MRPL4Δ | 66.4 | 43.1 |
| CIR1Δ | 52.4 | 51.2 |
| MRPL37Δ | 66.5 | 52.3 |
| BOI2Δ | 90.3 | 52.6 |
| IMG1Δ | 93.5 | 54.6 |
| RTC6Δ | 86.9 | 65.3 |
| NAM2Δ | 68.8 | 72.7 |
| YMR155wΔ | 50.7 | 86.0 |
Data are mutant cells killed expressed as a percentage of BY4742 cells killed at the same AuTMAT dose. Approximately 1 × 107 to 2 × 107 cells/ml were exposed to AuTMAT at the indicated doses for 24 h in 100-μl aliquots. At the 10- and 100-ppm doses, 7.20 × 106 and 1.50 × 107 BY4742 cells/ml were killed, respectively. Data are means of three replicates for mutants and four to seven replicates for BY4742. Values in bold indicate that significantly fewer mutant than BY4742 cells were killed (P < 0.05, Student's two-sided t test).
Was the yeast model informative with respect to assessing the potential toxicity of the 0.8-nm AuNPs tested? The observation that growing cells were insensitive to the damage observed in higher eukaryotes at similar concentrations suggests major differences in uptake or response. On the other hand, the observed susceptibility of nongrowing stationary-phase cells to 0.8-nm AuTMAT-mediated killing indicates similarities. Through the ability to readily link phenotype with genotype by mutational analysis, it was possible to determine that functional mitochondria appear to predispose yeast to damage. The finding that about 1/3 of the resistant mutants had previously been found to excrete significant amounts of glutathione suggests the possibility of 0.8-nm AuTMAT detoxification by thiol exchange.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alex Hadduck for technical assistance, Brian K. Kennedy for providing strain KK86, Qilin Li and Lisa Truong for helpful discussions, and anonymous reviewers for helpful criticisms.
This study was funded by U.S. Environmental Protection Agency (EPA) STAR program grant R833325 to A.T.B. The Oregon State University Environmental Health Sciences Center (grant number P30 ES000210; NIEHS, NIH) provided the library of yeast deletion mutants. The Safer Nanomaterials and Nanomanufacturing Initiative provided the AuNPs through a grant from the Air Force Research Laboratory under agreement number FA8650-05-1-5041. Additional funding was provided by the W. M. Keck Foundation.
Footnotes
Published ahead of print 9 November 2012
Supplemental material for this article may be found at 10.1128/AEM.01737-12.
REFERENCES
- 1. Nel A, Xia T, Madler L, Li N. 2006. Toxic potential of materials at the nanolevel. Science 311:622–627 [DOI] [PubMed] [Google Scholar]
- 2. Shaw SY, Westley EC, Pittet MJ, Subramanian A, Schreiber SL, Weissleder R. 2008. Pertubational profiling of nanomaterial biologic activity. Proc. Natl. Acad. Sci. U. S. A. 105:7387–7392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lévy R, Shaheen U, Cesbron Y, Sée V. 2010. Gold nanoparticles delivery in mammalian live cells: a critical review. Nano Rev. 1:4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma X, Wu Y, Shubin J, Tian Y, Zhang X, Zhao Y, Yu L, Liang X-J. 2011. Gold nanoparticles induce autophagosome accumulation through size-dependent nano uptake and lysosome impairment. ACS Nano 5:8629–8639 [DOI] [PubMed] [Google Scholar]
- 5. Alkilany AM, Murphy CJ. 2010. Toxicity and cellular uptake of gold nanoparticles: what we have learned so far? J. Nanopart. Res. 12:2313–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murphy CJ, Gole AM, Stone JW, Sisco PN, Alkilany AM, Goldsmith EC, Baxter SC. 2008. Gold nanoparticles in biology: beyond toxicity to cellular imaging. Acc. Chem. Res. 41:1721–1730 [DOI] [PubMed] [Google Scholar]
- 7. Evans MV, Turton HE, Grant CM, Dawes IW. 1998. Toxicity of linoleic acid hydroperoxide to Saccharomyces cerevisiae: involvement of a respiration-related process for maximal sensitivity and adaptive response. J. Bacteriol. 180:483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harper S, Usenko C, Hutchison JE, Maddux BLS, Tanguay RL. 2008. In vivo biodistribution and toxicity depends on nanomaterial composition, size, surface functionalisation and route of exposure. J. Exp. Nanosci. 3:195–206 [Google Scholar]
- 9. Pan Y, Leifert A, Ruau D, Neuss S, Bornemann J, Schmid G, Brandau W, Simon U, Jahnen-Dechent W. 2009. Gold nanoparticles of diameter 1.4 nm trigger necrosis by oxidative stress and mitochondrial damage. Small 5:2067–2076 [DOI] [PubMed] [Google Scholar]
- 10. Truong L, Tilton SC, Zaikova T, Richman E, Waters KM, Hutchison JE, Tanguay RL. 20 January 2012. Surface functionalities of gold nanoparticles impact embryonic gene expression responses.. Nanotoxicology. [Epub ahead of print.] doi:10.3109/17435390.2011.648225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Botstein D, Fink GR. 2011. Yeast: an experimental organism for 21st century biology. Genetics 189:695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiron JP, Lamandé J, Moussa F, Trivin F, Céolin R. 2000. Effect of “micronized” C60 fullerene on the microbial growth in vitro. Ann. Pharm. Fr. 58:170–175 [PubMed] [Google Scholar]
- 13. Hadduck AN, Hindagolla V, Contreras A, Li Q, Bakalinsky AT. 2010. Does aqueous fullerene inhibit growth of Saccharomyces cerevisiae or Escherichia coli? Appl. Environ. Microbiol. 76:8239–8242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kasemets K, Ivask A, Dubourguier Kahru H-CA. 2009. Toxicity of nanoparticles of ZnO, CuO and TiO2 to yeast Saccharomyces cerevisiae. Toxicol. In Vitro 23:1116–1122 [DOI] [PubMed] [Google Scholar]
- 15. Maheshwari V, Fomenko DE, Singh G, Saraf RF. 2010. Ion mediated monolayer deposition of gold nanoparticles on microorganisms: discrimination by age. Langmuir 26:371–377 [DOI] [PubMed] [Google Scholar]
- 16. Harper SL, Carriere JL, Miller JM, Hutchison JE, Maddux BLS, Tanguay RL. 2011. Systematic evaluation of nanomaterial toxicity: utility of standardized materials and rapid assays. ACS Nano 5:4688–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Woehrle GH, Hutchison JE. 2005. Thiol-functionalized undecagold clusters by ligand exchange: synthesis, mechanism, and properties. Inorg. Chem. 44:6149–6158 [DOI] [PubMed] [Google Scholar]
- 18. Kaeberlein M, Hu D, Kerr EO, Tsuchiya M, Westman EA, Dang N, Fields S, Kennedy BK. 2005. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 1:e69 doi:10.1371/journal.pgen.0010069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Véronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906 [DOI] [PubMed] [Google Scholar]
- 20. Williamson DH, Fennell DJ. 1975. The use of fluorescent DNA-binding agent for detecting and separating yeast mitochondrial DNA, p 335–351 In Prescott DM. (ed), Methods in cell biology, vol XII. Yeast cells. Academic Press, New York, NY: [DOI] [PubMed] [Google Scholar]
- 21. Burke D, Dawson D, Stearns T. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22. Griffith J, Reggiori F. 2009. Ultrastructural analysis or nanogold-labeled endocytic compartments of yeast Saccharomyces cerevisiae using a cryosectioning procedure. J. Histochem. Cytochem. 57:801–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prescianotto-Baschong C, Riezman H. 1998. Morphology of the yeast endocytic pathway. Mol. Biol. Cell 9:173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prescianotto-Baschong C, Riezman H. 2002. Ordering of compartments in the yeast endocytic pathway. Traffic 3:37–49 [DOI] [PubMed] [Google Scholar]
- 25. Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, Neupert W, Westermann B. 2002. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Merz S, Westermann B. 2009. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 10:R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. 2004. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. U. S. A. 101:6564–6569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tucker CL, Fields S. 2004. Quantitative genome-wide analysis of yeast deletion strain sensitivities to oxidative and chemical stress. Comp. Funct. Genomics 5:216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grant CM, MacIver FH, Dawes IW. 1997. Mitochondrial function is required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. FEBS Lett. 410:219–222 [DOI] [PubMed] [Google Scholar]
- 30. Baev D, Rivetta A, Vylkova S, Sun JN, Zeng Slayman G-FCL, Edgerton M. 2004. The TRK1 potassium transporter is the critical effector for killing of Candida albicans by the cationic protein, histatin 5. J. Biol. Chem. 279:55060–55072 [DOI] [PubMed] [Google Scholar]
- 31. Perrone GG, Grant CM, Dawes IW. 2005. Genetic and environmental factors influencing glutathione homoeostasis in Saccharomyces cerevisiae. Mol. Biol. Cell 16:218–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Myers AM, Pape LK, Tzagoloff A. 1985. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 4:2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


