Abstract
The gastrointestinal microbiota affects the metabolism of the mammalian host and has consequences for health. However, the complexity of gut microbial communities and host metabolic pathways make functional connections difficult to unravel, especially in terms of causation. In this study, we have characterized the fecal microbiota of hamsters whose cholesterol metabolism was extensively modulated by the dietary addition of plant sterol esters (PSE). PSE intake induced dramatic shifts in the fecal microbiota, reducing several bacterial taxa within the families Coriobacteriaceae and Erysipelotrichaceae. The abundance of these taxa displayed remarkably high correlations with host cholesterol metabolites. Most importantly, the associations between several bacterial taxa with fecal and biliary cholesterol excretion showed an almost perfect fit to a sigmoidal nonlinear model of bacterial inhibition, suggesting that host cholesterol excretion can shape microbiota structure through the antibacterial action of cholesterol. In vitro experiments suggested a modest antibacterial effect of cholesterol, and especially of cholesteryl-linoleate, but not plant sterols when included in model bile micelles. The findings obtained in this study are relevant to our understanding of gut microbiota-host lipid metabolism interactions, as they provide the first evidence for a role of cholesterol excreted with the bile as a relevant host factor that modulates the gut microbiota. The findings further suggest that the connections between Coriobacteriaceae and Erysipelotrichaceae and host lipid metabolism, which have been observed in several studies, could be caused by a metabolic phenotype of the host (cholesterol excretion) affecting the gut microbiota.
INTRODUCTION
The mammalian gastrointestinal tract is colonized by trillions of microorganisms (the gut microbiota), a large fraction of which are bacteria. This microbial community has an extensive impact on host metabolism with important implications for health (1–3). The contribution of the gut microbiota to energy harvest from the diet and to fat storage constitutes a key beneficial trait that underlies host-microbiota symbiosis in mammals (4). However, this contribution has likely become detrimental to modern humans living in societies with excess food resources, as it increases susceptibility to metabolic disorders, such as obesity, type 2 diabetes, and coronary heart disease. Accordingly, the gut microbiota is increasingly being accepted as an important factor that contributes to pathological conditions associated with obesity (5), and in humans, metabolic pathologies often are associated with alterations in the gut microbiota (which is referred to as dysbiosis) (6–9). Unfortunately, there is still little consensus on the bacterial groups that are linked to obesity-related diseases and metabolic phenotypes (3). In addition, although comparisons between germ-free and conventional mice and rats have clearly established a role of the microbiota in modulating host lipid metabolism (2, 10–12), it remains unclear whether dysbioses contribute to metabolic pathologies. However, such basic information is essential for our understanding of the diet-microbiota-host metabolism interplay, especially for the development of dietary strategies to prevent metabolic disorders through a modulation of the gut microbiome (3, 13, 14).
Novel molecular technologies based on massive parallel sequencing have enabled the identification of associations between host lipid metabolism and gut microbial community structure in both humans and animals. Two bacterial families, the Erysipelotrichaceae and Coriobacteriaceae, have been repeatedly linked to the host lipid metabolism and associated with the dyslipidemic phenotypes. Spencer and coworkers (8) showed that levels of Erysipelotrichaceae were positively associated with changes in liver fat in humans, and higher proportions of this bacterial group have been also identified in morbidly obese individuals (9). Erysipelotrichaceae have been also linked to lipidemic imbalances in mice and in a hamster model of hypercholesterolemia (15, 16). For Coriobacteriaceae, strong positive links have been determined with plasma non-high-density lipoprotein (non-HDL) in hamsters (15). Moreover, Claus and colleagues (2) showed an association between Coriobacteriaceae, in particular the genus Eggerthella, with host metabolism and especially hepatic triglyceride levels in mice. The recurrent identification of associations between Coriobacteriaceae and Erysipelotrichaceae, and specific taxa within these families, with host lipid and cholesterol phenotypes in different host species (humans, mice, and hamsters) suggests a genuine link between these bacterial groups and the host lipid metabolism (2, 14, 15).
The important similarities between hamsters and humans in terms of lipid profiles, enzymatic pathways in lipoprotein and bile metabolism, and susceptibility to diet-induced atherosclerosis pose advantages in using these animals to investigate functional interactions between cholesterol metabolism and the gut microbiota (17). In a previous study, we used the hamster model of hypercholesterolemia and investigated the interplay between grain sorghum lipid extract in the diet, gut microbial ecology, and cholesterol metabolism. This study showed that specific bacterial groups in the fecal samples were tightly linked to diet-induced improvements in host cholesterol metabolism (15). In particular, Coriobacteriaceae and unclassified members of the Erysipelotrichaceae were negatively correlated with non-HDL cholesterol and cholesterol absorption, while bifidobacteria showed positive correlations with HDL cholesterol. Some of these correlations were highly significant, but the directionality of these interactions was not established. Unfortunately, hamsters cannot be reared germ free (18), which precludes the study of causation between specific bacterial taxa and host cholesterol metabolism employing gnotobiotic approaches. However, it is possible to specifically modulate the hamster's cholesterol metabolism and study the effects on the gut microbiota. For example, plant sterols and their esters offer an opportunity to modulate cholesterol metabolism in hamsters (19). These compounds reduce cholesterol absorption in the intestine by a displacement of cholesterol by the plant sterol in intestinal micelles, by cocrystallization between plant sterols and cholesterol leading to the formation of insoluble crystals, and by impeding cholesterol hydrolysis by lipases and cholesterol esterases (20–26). The chemical processes by which plant sterols exert their actions have been extensively studied in vitro and do not require the participation of intestinal bacteria.
In this study, we have characterized the fecal microbiota of hamsters whose cholesterol metabolism was extensively modulated by dietary addition of plant sterol esters (19). In these hamsters, plant sterol ester (PSE) intake reduced cholesterol absorption and increased cholesterol excretion and, consequently, decreased plasma non-HDL cholesterol and liver esterified cholesterol levels. Pyrosequencing of 16S rRNA tags revealed that PSE also induced dramatic shifts in the fecal microbiota with remarkably high correlations with host cholesterol metabolites. Most importantly, the associations between several bacterial taxa with fecal and biliary cholesterol excretion showed an excellent fit to a nonlinear sigmoidal inhibitory model used to describe dose-response relationships between bacteria and inhibitory compounds (27, 28), suggesting that host cholesterol excretion can shape microbial community structure through the antimicrobial action of cholesterol excreted in the gut.
MATERIALS AND METHODS
Animal experiments and diets.
The fecal samples analyzed were obtained in a previous study that determined the effect of dietary PSE on hamsters' lipid metabolism (19). The handling of animals, feed composition, plant sterol composition of the diets, sample collection, and metabolic analysis were described in that report. Briefly, Bio-F1B male Syrian hamsters (Bio Breeders, Watertown, MA) were individually caged and randomly assigned to four dietary treatments throughout a 4-week period, of which three were included in the present study: a modified AIN-93 M diet containing no PSE (C) and diets containing 5% (wt/wt) plant sterols esterified with fatty acids from beef tallow (BT) or stearic acid (SA). The final energy distribution of each diet was 36% fat, 35% carbohydrate, and 29% protein. The animals were housed in a facility with controlled atmosphere (25°C) under 12-h light/dark cycles and had access to food and water ad libitum. The animals were euthanized by CO2 asphyxiation after 4 weeks of dietary intervention. Blood was collected by cardiac puncture, and plasma was obtained by centrifugation (2,000 × g for 30 min at 4°C). Total and HDL cholesterol were enzymatically quantified in the plasma samples. Livers were excised and immediately frozen in liquid nitrogen. Total cholesterol, triglycerides, free cholesterol, esterified cholesterol, and phospholipids were measured in the livers. Cholesterol absorption was quantified in fecal samples collected at week 3 with radiolabeled sterols as previously described (29). The complete fecal output was collected during week 4 and stored frozen (−80°C). Fecal concentration of neutral sterols, bile acids, cholesterol, dihydrocholesterol, coprostan-3-one, and coprostan-3-ol was determined as previously described (19).
Characterization of the fecal microbiota.
The gut microbial composition was determined in fecal samples of hamsters fed a control diet (C) (n = 7) or plant sterols esterified with SA (n = 9) and BT (n = 6). DNA was extracted using a standard method that combined enzymatic and mechanical cell lysis with phenol-chloroform extractions (15). The fecal microbial community was characterized by massive parallel sequencing of the V3 region of the 16S rRNA. PCRs were performed with the forward primer (A-338F) 5′-gcctccctcgcgccatcagACTCCTACGGGAGGCAGCAG-3′ and the reverse primer (B-518R) 5′-gccttgccagcccgctcagNNNNNNNNATTACCGCGGCTGCTGG-3′ (with the A and B adaptors indicated in lowercase and an 8-nucleotide barcode shown as Ns), and products were sequenced using the Roche Genome Sequencer GS-FLX (454 Life Sciences) as described previously (15). The sequence data set is available upon request.
The sequences obtained were subjected to quality control using the QIIME pipeline (30). Sequences <150 or >350 bp in length were removed, as were sequences containing one or more ambiguous nucleotides or mismatches to the primer or barcode, an average quality score below 25, and homopolymer runs longer than 6 bp. Chimera removal was performed using the BLAST Fragments algorithm in QIIME. An average of 1,700 sequences per sample was obtained after quality control. Taxonomical characterization of the sequences was done with the Classifier tool from the Ribosomal Database Project (RDP) (31), which classified the sequences from the phylum to the genus level. Additionally, operational taxonomic units (OTU) were determined using a 97% sequence similarity cutoff to characterize the microbiota at a lower taxonomic level roughly equivalent to bacterial species. OTUs were generated by aligning the quality-controlled sequences with the Infernal Alignment algorithm of RDP, followed by clustering with the Complete Linkage Clustering tool of RDP. The exact abundance of OTUs determined to be significantly affected by PSE consumption or associated with host physiological parameters was determined using BLASTn as described previously (32). Briefly, 5 representative sequences of the selected OTUs were taxonomically assigned and aligned by ClustalW within their respective phyla. Phylogenetic trees were constructed for each phylum with the neighbor-joining algorithm, and distance matrices were generated (MEGA 4.0) (33). Sequences that clustered in the tree with >97% similarity were combined into a single OTU, and consensus sequences were generated within these clusters. The consensus sequences were used to assign all sequences in the entire sequence set to respective OTUs by aligning them in BioEdit (34) with the BLASTn algorithm (>97% similarity and at least 95% overlap) against a local database composed of all of the quality-controlled sequences. UniFrac analysis was performed using the QIIME pipeline to investigate the beta diversity of microbial communities (30).
In vitro inhibition bacterial assays.
Representative bacterial strains originating from the mammalian gastrointestinal tract were selected to test for antibacterial activity of cholesterol, cholesteryl-linoleate, and the plant sterols β-sitosterol and stigmastanol when incorporated into micelles containing bile salts and lecithin. The bacteria and growth media used were Bifidobacterium longum subsp. infantis ATCC 15697T (in MRS medium supplemented with 0.5 mg/liter l-cysteine), Lactobacillus reuteri Lpuph-1 (in MRS medium), Eggerthella lenta ATCC 25559T (in MRS medium), Slackia heliotrinireducens ATCC 29202T (in MRS medium), Collinsella intestinalis ATCC 13228T (in peptone yeast glucose [PYG] medium), two human isolates of Collinsella aerofaciens, KD-D8-5 and IM-D3-18 (in PYG medium), and Clostridium histolyticum ATCC 19401T (in PYG medium). All media were prereduced for 24 h, and bacterial inocula for inhibition tests were prepared as follows under anaerobic conditions at 37°C (Bactron IV anaerobic chamber; Shel Laboratory). Bacterial cultures started from one single colony were grown for 24 h, transferred to fresh media (1% inoculum), and then grown for another 16 h.
Micelles containing cholesterol (>98% purity; Sigma), cholesteryl-linoleate (>99% purity; Sigma), or plant sterols (stigmastanol and β-sitosterol, synthesized as described in reference 21) were prepared as described by Brown et al. (21), with minor modifications. Briefly, 48 μl of a solution containing either cholesterol, cholesteryl-linoleate, stigmastanol, or β-sitosterol (161 mg/ml in chloroform) was mixed with 41 μl of a lecithin solution (221.3 mg/ml in chloroform), and chloroform was removed under a stream of nitrogen. The resulting mixture of lecithin and cholesterol/sterols was dissolved in a solution containing 5.4 mg/ml of the bile salt sodium taurocholate (>97% purity; Sigma) in distilled water. The solutions were sonicated for 3 to 6 min with 30% amplitude using a Branson 450 Sonifier (Danbury, CT) and afterwards filter sterilized with 0.45-μm pore filters (Fisherbrand; Fisher Scientific). To test for the antibacterial capacity of cholesterol, cholesteryl-linoleate, or the plant sterols, 5 μl of a 1:10 dilution of the bacterial inocula was transferred into a solution made of 500 μl of prereduced micelle suspensions and 500 μl of the appropriate prereduced double-concentrated (2×) media. Given the antibacterial activity of bile acids, micelle suspensions containing bile acids but no sterols were inoculated and used as controls. Cultures were incubated anaerobically at 37°C for 12 h (mid-log phase), and spectrophotometric optical density (OD) quantifications were performed at 600 nm (BioMate3; Thermo Scientific). Three biological replicates were conducted for the experiments. The concentration of cholesterol in these experiments (386 μg/ml media) is approximately double the concentration of the average cholesterol levels measured in the fecal samples of hamsters consuming the control diet (170 ± 111 μg/g) but still around 20 times lower than the fecal cholesterol levels present in SA-treated hamsters (4,303 ± 1,930 μg/g). Unfortunately, generation of micelles containing higher amounts of cholesterol, which would have better reflected the in vivo conditions, was not possible due to the formation of a precipitate.
Statistical analysis.
Results were expressed as means ± standard deviations (SD) unless otherwise stated. The impact of dietary treatments on the abundance of individual bacterial taxa was analyzed by one-way analyses of variance (ANOVA) and Tukey's post hoc tests. Correlations between bacterial groups and host physiological measurements were assessed with Pearson's correlations and nonlinear regressions. A nonlinear four-parameter sigmoidal inhibitory model that describes dose-response relationships between bacteria and inhibitory compounds was used to fit the data (27, 28). The model is represented by the equation Y = Yo + (Ymax × Xslope)/(EC50slope + Xslope), where X represents the metabolic parameter, Y the abundance of the bacterial taxon in the fecal sample, Yo and Ymax are the minimum and maximum effects, respectively, EC50 is the concentration where 50% of the maximum effect is measured, and the slope is the sigmoidicity coefficient.
The statistical analyses were performed using GraphPad Prism version 5.0 (GraphPad Software). P < 0.05 in the ANOVAs and correlation coefficients of r > 0.60 (in absolute values) were considered significant. In addition to the analyses described above, partial correlations were performed with SAS/STAT software to evaluate the associations between metabolic markers and abundance of bacterial populations when excluding the effect of diet using linear regressions. To decrease the number of variables in this analysis and increase the power of the test, only a few bacterial taxa (Coriobacteriaceae, Erysipelotrichaceae, unclassified Erysipelotrichaceae, and OTU1) and metabolic markers (whole-body cholesterol synthesis, fecal cholesterol excretion, fecal neutral sterols, and fecal bile acid) that were determined to be pertinent based on the previous analyses were selected to conduct the partial correlation statistics.
RESULTS
Dietary supplementation with 5% PSE induced substantial alterations of the fecal microbiota of hamsters.
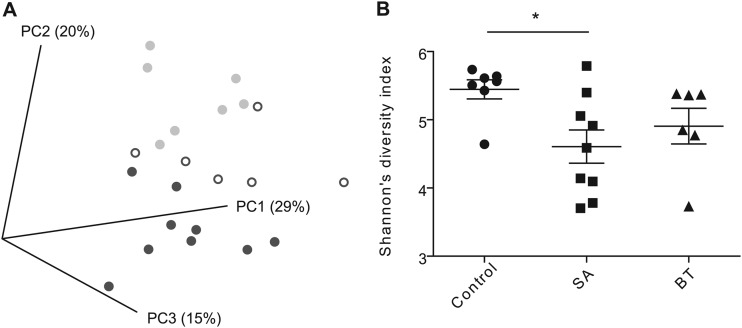
454 Pyrosequencing was used to characterize the fecal microbiota of hamsters fed a control diet or a diet supplemented with 5% plant sterols esterified with stearic acid (SA) or beef tallow (BT). The inclusion of PSE in the diet, in particular SA, had extensive effects on the fecal microbiota composition. UniFrac analysis revealed that the fecal microbial communities of hamsters fed the SA diet clustered separately from those of the control group (Fig. 1A). Analysis of Shannon diversity coefficients revealed that dietary addition of SA reduced the diversity of the fecal microbiota compared to that of the control (P < 0.05) (Fig. 1B).
Fig 1.
Alpha and beta diversity measurements of the fecal bacterial communities. (A) Principal coordinate analysis based on UniFrac distances, segregated fecal microbial communities of hamsters fed plant sterols esterified with stearic acid (dark gray closed circles), and beef tallow (open circles) from animals fed a control diet (light gray closed circles). (B) Shannon's diversity index. *, P < 0.05.
Dietary PSE significantly altered the gut microbiota at all taxonomic levels (Table 1). PSE had a dramatic effect on the abundance of the phylum Actinobacteria, and especially on the family Coriobacteriaceae, which declined 10- and 4-fold with SA or BT intake, respectively. Three OTUs belonging to this family were significantly reduced through PSE (Table 1; also see Fig. S1 in the supplemental material). SA induced a significant increase in the phylum Firmicutes which was, to a large degree, due to a bloom of a single OTU (OTU16) within the family Eubacteriaceae (Table 1). It is possible that this increase was due to the ability of this organism to utilize the additional cholesterol that became available as a growth substrate in the gut of PSE-treated hamsters. However, the presence of OTU16 is strictly linked to the absence of the OTUs whose abundance was reduced by PSE (data not shown), suggesting that the bloom was caused by OTU16 expanding into niches that became vacant. Although total Firmicutes levels showed an increase with SA, five OTUs belonging to the family Erysipelotrichaceae did not follow this general trend and instead showed a dramatic reduction (Table 1; also see Fig. S2).
Table 1.
Abundance of fecal bacterial taxa of hamsters fed a control diet or diets enriched in plant sterol estersa
| Taxon | Abundance level by diet (% of total sequences [mean ± SD])b |
P value (by ANOVA) | ||
|---|---|---|---|---|
| Control | SA | BT | ||
| Phylum | ||||
| Actinobacteria | 16.55 ± 7.27 | 3.24 ± 1.90*** | 8.37 ± 5.04* | 0.0002 |
| Firmicutes | 72.88 ± 5.50 | 85.07 ± 7.25**§ | 76.44 ± 4.90 | 0.0025 |
| Family | ||||
| Coriobacteriaceae | 11.63 ± 7.30 | 0.88 ± 0.49*** | 2.71 ± 2.32** | 0.0002 |
| Eubacteriaceae | 0.02 ± 0.05 | 10.85 ± 11.58*§ | 0.61 ± 1.32 | 0.0156 |
| Genus | ||||
| Unc. Coriobacteriaceae | 11.57 ± 7.30 | 0.87 ± 0.49*** | 2.66 ± 2.27** | 0.0002 |
| Unc. Erysipelotrichaceae | 1.08 ± 0.30* | 0.66 ± 0.24 | 0.76 ± 0.40 | 0.0409 |
| Unc. Eubacteriaceae | 0.02 ± 0.05 | 10.85 ± 11.58***§§§ | 0.61 ± 1.32 | <0.0001 |
| OTUs (family, genus and species, similarity to closest type strain) | ||||
| OTU1 (Coriobacteriaceae, Eggerthella lenta, 97%) | 6.98 ± 7.46 | 0.24 ± 0.31* | 0.04 ± 0.07* | 0.0078 |
| OTU2 (Coriobacteriaceae, Gordonibacter pamelaeae, 93%) | 2.05 ± 0.76 | 0.12 ± 0.10*** | 0.81 ± 1.19* | 0.0002 |
| OTU4 (Coriobacteriaceae, Slackia heliotrinireducens, 97%) | 1.22 ± 0.93 | 0.03 ± 0.10** | 0.24 ± 0.30* | 0.0011 |
| OTU8 (Erysipelotrichaceae, Allobaculum stercoricanis, 91%) | 6.56 ± 4.80 | 1.48 ± 2.24* | 2.71 ± 2.51 | 0.0205 |
| OTU9 (Erysipelotrichaceae, Allobaculum stercoricanis, 86%) | 3.30 ± 3.19 | 0.15 ± 0.14** | 0.31 ± 0.28* | 0.0051 |
| OTU10 (Erysipelotrichaceae, Eubacterium cylindroides, 87%) | 1.03 ± 1.00 | 0.08 ± 0.07* | 0.28 ± 0.23 | 0.0125 |
| OTU12 (Erysipelotrichaceae, Allobaculum stercoricanis, 94%) | 3.14 ± 1.55 | 0.71 ± 0.86** | 1.43 ± 1.08* | 0.0021 |
| OTU13 (Erysipelotrichaceae, Eubacterium biforme, 87%) | 0.52 ± 0.25 | 0.09 ± 0.09** | 0.22 ± 0.40 | 0.0100 |
| OTU15 (Eubacteriaceae, Clostridium sufflavum, 90%) | 0.91 ± 0.67 | 0.20 ± 0.29* | 0.29 ± 0.21* | 0.0105 |
| OTU16 (Eubacteriaceae, Eubacterium limosum, 93%) | 0.04 ± 0.06 | 8.55 ± 12.71 | 0.62 ± 1.49 | 0.0972 |
The taxa presented were significantly affected by plant sterols esterified with stearic acid (SA) or beef tallow (BT) or were determined to be associated with host metabolic markers of the lipid metabolism. Unc., unclassified.
*, P < 0.05; **, P < 0.01; *** P < 0.001 compared to the control group. §, P < 0.05; §§, P < 0.01; §§§, P < 0.001 compared to the BT group.
Correlation analysis showed an extensive link between the microbiota and host lipid metabolism.
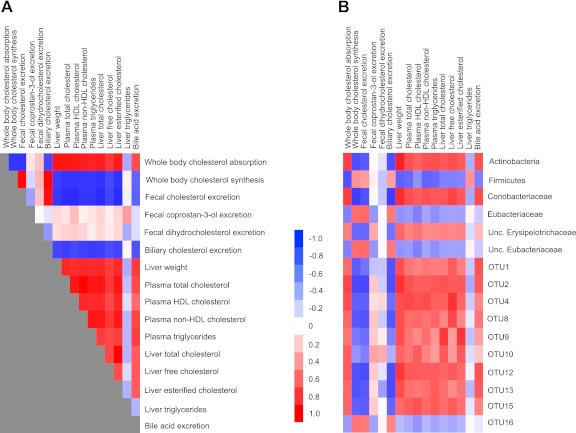
The feeding of PSE induced extensive changes in the lipid and cholesterol metabolism of hamsters (19), and many of the individual metabolites were highly interrelated (Fig. 2A). One of the goals of the present study was to investigate whether the lipidemic effects were associated with shifts in the gut microbiota. The correlation analysis revealed that host cholesterol metabolism was extensively interlinked with the bacterial community (Fig. 2). The tightest associations were observed within the phylum Actinobacteria and especially the family Coriobacteriaceae, which showed remarkable associations with cholesterol absorption (r = 0.75, P < 0.0001), whole-body cholesterol synthesis (r = −0.75, P < 0.0001), fecal biliary cholesterol excretion (r = −0.75, P < 0.0001), liver free cholesterol (r = 0.73, P < 0.0001), plasma non-HDL cholesterol (r = 0.68, P = 0.0005), and liver weight (r = 0.82, P < 0.0001), among others. Similar high correlations were observed for the OTUs belonging to the Coriobacteriaceae and Erysipelotrichaceae families that were affected by PSE administration. Total Firmicutes and unclassified Erysipelotrichaceae also displayed significant associations with host lipidemic markers, although they were less significant than associations detected for Coriobacteriaceae and the individual OTUs (Fig. 2).
Fig 2.
Associations among host markers of the lipid metabolism and with fecal bacterial populations. Heat maps display correlation coefficients among markers of the host lipid metabolism (A) and between host lipid profile and abundance of fecal bacterial populations (B). Unc., unclassified.
Improvements in the host lipid metabolism appear to be independent from metabolic modifications of intestinal cholesterol by the gut microbiota.
One mechanism through which gut bacteria could lower host cholesterol levels is the transformation of cholesterol to coprostan-3-one, coprostan-3-ol, and dihydrocholesterol, which are, to a large degree, excreted (29). However, fecal concentrations of these cholesterol derivatives were not affected by PSE intake, and their fecal concentrations did not correlate with any of the host lipid metabolism markers tested (Fig. 2A). In addition, the correlation analysis did not provide evidence for an association between coprostan-3-one, coprostan-3-ol, and dihydrocholesterol and specific bacterial members of the gut microbiota (Fig. 2). These findings suggest that PSE-induced shifts of the gut microbiota did not impact bacterial cholesterol metabolites in the gastrointestinal lumen, and the bacterial action on the cholesterol pool was not responsible for the improvements observed in host lipid metabolism.
Cholesterol excreted by the host via bile appears to inhibit specific bacterial taxa.
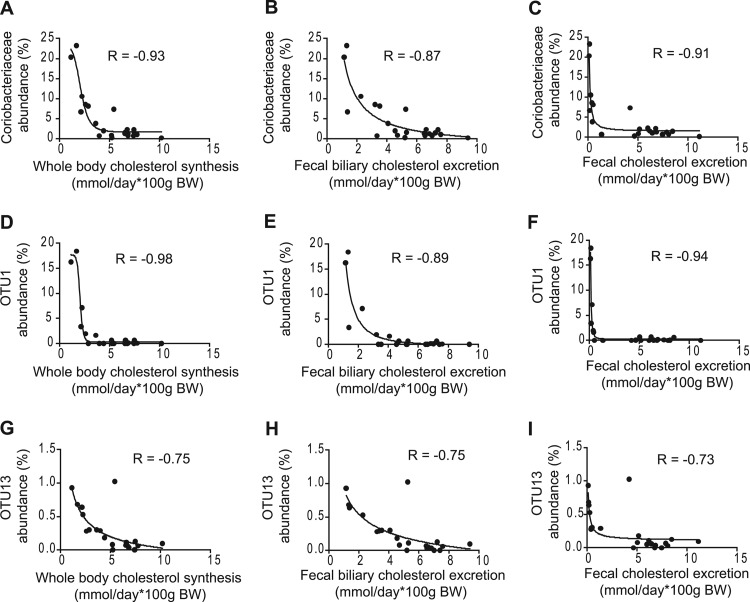
Upon visual inspection of the correlation plots, the associations of bacterial taxa with host cholesterol synthesis and excretion appeared to follow an exponential relationship. In addition, these associations appeared to resemble dose-response curves observed in microbial antibiotic assays. We therefore tested how bacterial proportions and cholesterol synthesis/excretion fitted a four-parameter sigmoidal model for bacterial inhibition used to describe dose-response relationships between bacteria and inhibitory compounds (27, 28). This analysis revealed that the relationships between cholesterol synthesis, fecal biliary cholesterol excretion, and fecal cholesterol with several bacterial taxa showed an almost perfect fit to the inhibitory model (Fig. 3). For example, the associations between Coriobacteriaceae and host cholesterol synthesis, fecal biliary cholesterol excretion, and fecal cholesterol excretion fitted the model with regression coefficients of −0.93, −0.87, and −0.91, respectively. The model was an even better fit for the data obtained for OTU1, which is related to Eggerthella lenta. Interestingly, the fecal proportions of OTU13, which was classified as a member of the Erysipelotrichaceae family, also showed a tight association with cholesterol synthesis and excretion with an excellent fit to the model. In total, four OTUs, two belonging to the Coriobacteriaceae (OTUs 1 and 4) and two belonging to the Erysipelotrichaceae (OTUs 13 and 15), displayed a significant fit to the four-parameter sigmoidal model (r = −0.94, −0.71, −0.73, and −0.68, respectively). The fact that the abundance of taxonomically distinct bacterial taxa displayed associations with fecal cholesterol concentrations that fit an inhibition model suggests an antibacterial role of the excreted cholesterol. In contrast, bile acid concentrations showed no negative associations with the fecal proportions of Coriobacteriaceae- or Erysipelotrichaceae-related phylotypes that were affected by the feeding of PSE (see Fig. S3 in the supplemental material).
Fig 3.
Associations between fecal bacterial populations and markers of host cholesterol metabolism. Four-parameter sigmoidal inhibitory regressions between fecal abundance of Coriobacteriaceae (A, B, and C), OTU1 (D, E, and F), and OTU13 (G, H, and I) with whole-body cholesterol synthesis (A, D, and G), fecal biliary cholesterol excretion (B, E, and H), and fecal cholesterol excretion (C, F, and I). BW, body weight.
Partial correlation analysis revealed that the negative associations between the abundance of Erysipelotrichaceae with whole-body cholesterol synthesis and concentrations of fecal neutral sterols (P = 0.033 and 0.034, respectively) were independent of dietary PSE. This analysis suggested that the links between fecal proportions of Erysipelotrichaceae and Coriobacteriaceae with fecal cholesterol also are independent of PSE intake (P = 0.076 and 0.069, respectively). Importantly, the abundance of the family Coriobacteriaceae (P = 0.0726, r = −0.7122) and OTUs 1 (P = 0.0244, r = −0.82) and 13 (P = 0.0032, r = −0.92) also showed significant negative associations with fecal cholesterol excretion in hamsters on the control diet alone (see Fig. S4 in the supplemental material). These findings suggest that the associations detected between host cholesterol metabolism and the microbiota are not solely caused by PSE but also by a direct association of the host phenotype with the bacterial taxa.
Cholesteryl-linoleate but not plant sterols inhibits growth of gut bacteria in vitro.
To gain insight into the functional interactions between bacteria and cholesterol metabolism, we determined the capability of cholesterol, cholesteryl-linoleate, and the plant sterols stigmastanol and β-sitosterol to inhibit bacterial growth when included in model bile mixed micelles containing a bile salt and lecithin, which are natural components of micelles in the gut (35). We used plant sterols for these experiments as PSE get hydrolyzed in the gut, and the unesterified sterol is the compound that is incorporated into intestinal micelles (20). We tested for the inhibition of seven strains of gut bacteria, including strains belonging to the Coriobacteriaceae, as members of this bacterial family were most affected in our animal experiments. No statistically significant levels of inhibition were detected for any of the treatments. However, out of the seven strains tested, Lactobacillus reuteri, Clostridium histolyticum, and Collinsella aerofaciens showed a modest degree of inhibition. All three showed reduced growth in the presence of micelles containing cholesteryl-linoleate compared to control micelles containing only lecithin and bile acid (Fig. 4). Growth of Lactobacillus reuteri also appeared to be inhibited by micelles containing cholesterol.
Fig 4.
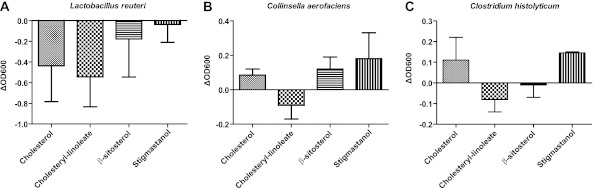
In vitro inhibition of fecal bacterial isolates by sterols. Shown are optical density differences between micelles containing cholesterol, cholesteryl-linoleate, β-sitosterol, and stigmastanol compared to control micelles without sterols for the gut bacterial isolates Lactobacillus reuteri (A), Collinsella aerofaciens (B), and Clostridium histolyticum (C). ΔOD600, change in optical density at 600 nm.
DISCUSSION
In this study, we have characterized the interplay between the gut microbiota and cholesterol metabolism in hamsters treated with PSE. The study revealed that PSE-induced alterations of cholesterol metabolism were tightly associated with specific compositional shifts of the gut microbiota. The strongest associations were identified between the families Coriobacteriaceae and Erysipelotrichaceae, and several OTUs within these families, and host cholesterol concentrations in plasma, the liver, and fecal samples. Although it is difficult to determine cause-effect relationships among these associations because of the impossibility of raising hamsters germ free, this study provided several lines of evidence that indicate that the bacterial shifts induced by PSE are a consequence of changes in host cholesterol metabolism.
First, the capability of PSE to decrease cholesterol absorption and ultimately modify the host cholesterol metabolism is based on physicochemical interactions independent of bacterial action. Second, levels of cholesterol derivatives (coprostan-3-one, coprostan-3-ol, and dihydrocholesterol) that are considered to contribute to the cholesterol-lowering activity of the gut microbiota were not affected by PSE and showed no association with host cholesterol phenotypes. Third, PSE have not been described to be antibacterial and therefore are unlikely to directly cause the dramatic shifts in the microbiota when added to the diet. In contrast, cholesterol derivatives, and especially cholesterol-linoleate, have been shown to be antibacterial (36, 37), and associations between several bacterial taxa affected by dietary PSE with fecal and biliary cholesterol excretion showed an excellent fit to a sigmoidal inhibitory nonlinear model of dose-response relationships between bacteria and inhibitory compounds (Fig. 3). Fourth, fecal cholesterol excretion was negatively associated with bacterial taxa when only hamsters in the control treatment were considered.
The data obtained in this study therefore suggest that changes in host cholesterol metabolism induced through dietary PSE were the main drivers of the modulation in gut microbiota composition. A schematic summary illustrating the physiological processes that are likely to have caused the PSE-induced associations between host cholesterol metabolism and the gut microbiota is shown in Fig. 5. Intake of PSE decreased plasma and liver cholesterol levels through an inhibition of both dietary and biliary cholesterol absorption in the small intestine, with a consequent increase in fecal cholesterol excretion (Fig. 5A). In order to maintain cholesterol homeostasis, the host compensated for the decrease in the total cholesterol pool by increasing cholesterol synthesis (Fig. 5B), which resulted in a further increase of bile-excreted cholesterol. The combination of higher biliary cholesterol excretion and decreased cholesterol absorption resulted in increased concentrations of free and esterified cholesterol in the gastrointestinal tract. These cholesterol derivatives exert an antibacterial effect on specific members of the gastrointestinal microbiota, causing alterations in the microbial community. Since cholesterol excretion strongly correlated with plasma cholesterol levels, the antimicrobial effect of cholesterol is ultimately causing detectable correlations between specific bacterial taxa and host plasma and liver cholesterol levels (Fig. 5B).
Fig 5.
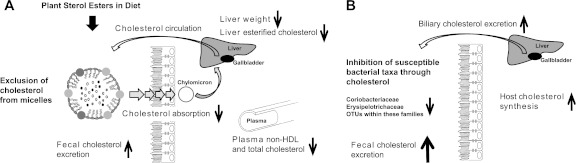
Model depicting the impact of plant sterol esters on the host lipid metabolism and on the gut microbiota. The inclusion of dietary plant sterol esters (PSE) precludes the incorporation of cholesterol into the micelles in the small intestine, lowering cholesterol absorption and increasing cholesterol excretion. (A) The removal of cholesterol results in reductions in plasma non-HDL, liver weight, and hepatic liver esterified cholesterol. The decreased cholesterol pools are compensated for by increased hepatic cholesterol synthesis. (B) The higher concentrations of fecal cholesterol coupled with the inhibitory capacity of cholesterol result in the decreased abundance of bacterial populations of Coriobacteriaceae and Erysipelotrichaceae in the gastrointestinal tract.
The findings obtained in this study are relevant to our understanding of the gut microbiota-host lipid metabolism interplay. The linkages between the gut microbiota and cholesterol metabolism found in PSE-treated hamsters recapitulated previous findings for hamsters fed grain sorghum lipid extracts (GSL) (15). In addition, negative correlations between Coriobacteriaceae and Erysipelotrichaceae with fecal cholesterol excretion were also detected by our group in an independent experiment with hamsters that were fed whole sorghum kernels (I. Martínez, T. P. Carr, C. L. Weller, and J. Walter, unpublished observations). Bacterial taxa within the families Coriobacteriaceae and Erysipelotrichaceae have been recurrently associated with host dyslipidemic phenotypes in mice and humans in the context of obesity, metabolic syndrome, and hypercholesterolemia (8, 9, 16). If microbiome alterations contributed to lipidmic aberrancies, they could constitute pharmaceutical targets to improve host metabolic functions. In fact, we have previously suggested Coriobacteriaceae as therapeutic targets to improve host cholesterol metabolism (15). However, the data obtained with PSE-treated hamsters suggest that the strong associations between Coriobacteriaceae and Erysipelotrichaceae and host cholesterol metabolism are caused by the host phenotype affecting the bacteria. Analogous interactions could exist in mice and humans, especially in relation to metabolic disorders that are associated with an altered cholesterol metabolism, such as obesity and the metabolic syndrome. The findings also indicate that diet can modulate gut microbiota composition through an effect on host metabolism, which has also been demonstrated for dietary fat-induced changes in host bile acid composition (38).
The findings obtained from PSE-treated hamsters provided the first evidence for a role of cholesterol as a relevant host factor that modulates the gut microbiota. The in vitro assays performed in this study confirmed the antibacterial effect of cholesteryl-linoleate on selected strains of gut bacteria. Although the antibacterial effect detected in the in vitro experiments was modest, even small levels of inhibition could be relevant under the competitive conditions in the gastrointestinal tract, where even a minor reduction in growth rate could translate into a significant ecological disadvantage. It is important to point out that the approximate concentrations of cholesterol in the in vitro experiments, due to experimental limitations, were around 20 times lower than those present in the gut of hamsters during PSE treatment. Interestingly, cholesterol and its derivatives have antibacterial activity in the nose and eye epithelial linings (36, 37). In addition, the findings of Do and coworkers suggest that the antibacterial effect of cholesterol-esters in nasal fluid acts in synergism with that of the α-defensin HNP-2 (36). Enteric defensins play a significant role in regulating the gut microbiota (39), and the strong inhibitory effects of cholesterol detected in hamsters therefore might be caused by a synergistic effect of the two compounds. Clearly, the in vitro experiments on the antibacterial effects of cholesterol included in this study are only preliminary, and further research should be targeted towards elucidating the role of cholesterol as a host factor that modulates the gut microbiota.
Bile acids, which are synthesized from cholesterol, also have antimicrobial activity (40, 41). Bile acids have been demonstrated to modulate gut microbiota composition (38, 42), and since the concentration and composition of excreted bile are influenced by dietary fat, bile acids have been suggested to be one cause for the dysbiosis that is associated with obesity-related pathologies (14). However, bile acids did not appear to be a contributing factor in the population shifts observed in PSE-treated hamsters, as fecal bile acid excretion was reduced by PSE and showed significant positive correlations with the bacteria of the Coriobacteriaceae and Erysipelotrichaceae families that were affected by PSE. Although the reduction of bile excretion in PSE-treated hamsters was likely caused by the smaller cholesterol pool, only fecal cholesterol showed highly significant correlations with both the improvements in host lipid metabolism and the abundance of bacterial taxa.
Although this study revealed an example of how a host metabolic factor influences the gut microbiota composition, research with germ-free animals has clearly shown that gut microbes impact host metabolism (including cholesterol metabolism), and it is likely that some alterations of the gut microbiome associated with host metabolism have functional consequences for the host (12, 43–45). Specific bacterial taxa have been determined to improve lipid markers in the host. Bifidobacteria, which showed positive associations with plasma HDL cholesterol in our previous study (15), have been identified to alleviate dyslipidemia and high-fat-induced insulin resistance when administered as probiotics (46–50). In addition, some changes in gut microbiota composition induced through host factors might still have pathological consequences, as shown recently for a fat-induced pathobiont expansion caused by changes in the bile acid pool (38). Given that the gut microbiota reduces liver and plasma cholesterol levels (11, 12), we cannot exclude that the dramatic changes in Coriobacteriaceae and Erysipelotrichaceae contribute to the cholesterol-lowering effects of PSEs. However, the findings obtained in this study provide evidence that interactions between the gut microbiota and host metabolism are bidirectional, and some patterns of dysbiosis associated with metabolic dysfunctions might be a consequence rather than a cause of the host phenotype.
Supplementary Material
ACKNOWLEDGMENTS
Susan Hammons was supported by the UCARE Program of the University of Nebraska.
We thank John H. Rupnow for organizing and supporting Diahann J. Perdicaro internship at the University of Nebraska.
Footnotes
Published ahead of print 2 November 2012
Supplemental material for this article may be found at 10.1128/AEM.03046-12.
REFERENCES
- 1. Allen CA, Torres AG. 2008. Host-microbe communication within the GI tract. Adv. Exp. Med. Biol. 635:93–101 [DOI] [PubMed] [Google Scholar]
- 2. Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, de Waziers I, Cloarec O, Richards SE, Wang Y, Dumas ME, Ross A, Rezzi S, Kochhar S, Van Bladeren P, Lindon JC, Holmes E, Nicholson JK. 2011. Colonization-induced host-gut microbial metabolic interaction. mBio 2:e00271–10 doi:10.1128/mBio.00271-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tremaroli V, Backhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249 [DOI] [PubMed] [Google Scholar]
- 4. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920 [DOI] [PubMed] [Google Scholar]
- 5. Cani PD, Delzenne NM. 2009. Interplay between obesity and associated metabolic disorders: new insights into the gut microbiota. Curr. Opin. Pharmacol. 9:737–743 [DOI] [PubMed] [Google Scholar]
- 6. Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. 2010. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5:e9085 doi:10.1371/journal.pone.0009085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102:11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. 2011. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 140:976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, Krajmalnik-Brown R. 2009. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U. S. A. 106:2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 101:15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Danielsson H, Gustafsson B. 1959. On serum-cholesterol levels and neutral fecal sterols in germ-free rats; bile acids and steroids 59. Arch. Biochem. Biophys. 83:482–485 [DOI] [PubMed] [Google Scholar]
- 12. Wostmann BS. 1973. Intestinal bile acids and cholesterol absorption in the germfree rat. J. Nutr. 103:982–990 [DOI] [PubMed] [Google Scholar]
- 13. de Vos WM, de Vos EA. 2012. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr. Rev. 70(Suppl. 1):S45–S56 [DOI] [PubMed] [Google Scholar]
- 14. Yokota A, Fukiya S, Ooka T, Ogura Y, Hayashi T, Ishizuka S. 2012. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes 3:455–459 [DOI] [PubMed] [Google Scholar]
- 15. Martínez I, Wallace G, Zhang C, Legge R, Benson AK, Carr TP, Moriyama EN, Walter J. 2009. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl. Environ. Microbiol. 75:4175–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, Mao Y, Zhang X, Pang X, Wei C, Zhao G, Chen Y, Zhao L. 2010. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 4:232–241 [DOI] [PubMed] [Google Scholar]
- 17. Mitchell PL, McLeod RS. 2008. Conjugated linoleic acid and atherosclerosis: studies in animal models. Biochem. Cell Biol. 86:293–301 [DOI] [PubMed] [Google Scholar]
- 18. Angulo AF, Spaans J, Zemmouchi L, van der Waaij D. 1978. Selective decontamination of the digestive tract of Syrian hamsters. Lab. Anim. 12:157–158 [DOI] [PubMed] [Google Scholar]
- 19. Rasmussen HE, Guderian DM, Jr, Wray CA, Dussault PH, Schlegel VL, Carr TP. 2006. Reduction in cholesterol absorption is enhanced by stearate-enriched plant sterol esters in hamsters. J. Nutr. 136:2722–2727 [DOI] [PubMed] [Google Scholar]
- 20. Brown AW, Hang J, Dussault PH, Carr TP. 2010. Phytosterol ester constituents affect micellar cholesterol solubility in model bile. Lipids 45:855–862 [DOI] [PubMed] [Google Scholar]
- 21. Brown AW, Hang J, Dussault PH, Carr TP. 2010. Plant sterol and stanol substrate specificity of pancreatic cholesterol esterase. J. Nutr. Biochem. 21:736–740 [DOI] [PubMed] [Google Scholar]
- 22. Heinemann T, Kullak-Ublick GA, Pietruck B, von Bergmann K. 1991. Mechanisms of action of plant sterols on inhibition of cholesterol absorption. Comparison of sitosterol and sitostanol. Eur. J. Clin. Pharmacol. 40(Suppl. 1):S59–S63 [PubMed] [Google Scholar]
- 23. Normen L, Dutta P, Lia A, Andersson H. 2000. Soy sterol esters and beta-sitostanol ester as inhibitors of cholesterol absorption in human small bowel. Am. J. Clin. Nutr. 71:908–913 [DOI] [PubMed] [Google Scholar]
- 24. Ntanios FY, Jones PJ. 1999. Dietary sitostanol reciprocally influences cholesterol absorption and biosynthesis in hamsters and rabbits. Atherosclerosis 143:341–351 [DOI] [PubMed] [Google Scholar]
- 25. Roberts PJ, Riley GP, Morgan K, Miller R, Hunter JO, Middleton SJ. 2001. The physiological expression of inducible nitric oxide synthase (iNOS) in the human colon. J. Clin. Pathol. 54:293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trautwein EA, Duchateau GSMJE, Lin Y, Mel'nikov SM, Molhuzien HOF, Ntanios FY. 2003. Proposed mechanisms of cholesterol-lowering action of plant sterols. Eur. J. Lipid Sci. Technol. 105:171–185 [Google Scholar]
- 27. Macdougall J. 2006. Analysis of dose-response studies–Emax model. In Ting N. (ed), Dose finding in drug development. Springer, New York, NY [Google Scholar]
- 28. Mouton JW, Dudley MN, Cars O, Derendorf H, Drusano GL. 2005. Standardization of pharmacokinetic/pharmacodynamic (PK/PD) terminology for anti-infective drugs: an update. J. Antimicrob. Chemother. 55:601–607 [DOI] [PubMed] [Google Scholar]
- 29. Schneider CL, Cowles RL, Stuefer-Powell CL, Carr TP. 2000. Dietary stearic acid reduces cholesterol absorption and increases endogenous cholesterol excretion in hamsters fed cereal-based diets. J. Nutr. 130:1232–1238 [DOI] [PubMed] [Google Scholar]
- 30. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martínez I, Kim J, Duffy PR, Schlegel VL, Walter J. 2010. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 5:e15046 doi:10.1371/journal.pone.0015046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 34. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 35. Mukhopadhyay S, Maitra U. 2004. Chemistry and biology of bile acids. Curr. Sci. 87:1666–1682 [Google Scholar]
- 36. Do TQ, Moshkani S, Castillo P, Anunta S, Pogosyan A, Cheung A, Marbois B, Faull KF, Ernst W, Chiang SM, Fujii G, Clarke CF, Foster K, Porter E. 2008. Lipids including cholesteryl linoleate and cholesteryl arachidonate contribute to the inherent antibacterial activity of human nasal fluid. J. Immunol. 181:4177–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marquart ME, Monds KS, McCormick CC, Dixon SN, Sanders ME, Reed JM, McDaniel LS, Caballero AR, O'Callaghan RJ. 2007. Cholesterol as treatment for pneumococcal keratitis: cholesterol-specific inhibition of pneumolysin in the cornea. Investig. Ophthalmol. Vis. Sci. 48:2661–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. 2012. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487:104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. 2010. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11:76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Floch MH, Gershengoren W, Elliott S, Spiro HM. 1971. Bile acid inhibition of the intestinal microflora–a function for simple bile acids? Gastroenterology 61:228–233 [PubMed] [Google Scholar]
- 41. Stacey M, Webb M. 1947. Studies on the antibacterial properties of the bile acids and some compounds derived from cholanic acid. Proc. R. Soc. Med. 134:523–537 [DOI] [PubMed] [Google Scholar]
- 42. Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. 2011. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141:1773–1781 [DOI] [PubMed] [Google Scholar]
- 43. Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. 2007. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U. S. A. 104:979–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. 2008. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57:1470–1481 [DOI] [PubMed] [Google Scholar]
- 45. Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. 2010. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328:228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen JJ, Wang R, Li XF, Wang RL. 2011. Bifidobacterium longum supplementation improved high-fat-fed-induced metabolic syndrome and promoted intestinal Reg I gene expression. Exp. Biol. Med. 236:823–831 [DOI] [PubMed] [Google Scholar]
- 47. Kiessling G, Schneider J, Jahreis G. 2002. Long-term consumption of fermented dairy products over 6 months increases HDL cholesterol. Eur. J. Clin. Nutr. 56:843–849 [DOI] [PubMed] [Google Scholar]
- 48. Kondo S, Xiao JZ, Satoh T, Odamaki T, Takahashi S, Sugahara H, Yaeshima T, Iwatsuki K, Kamei A, Abe K. 2010. Antiobesity effects of Bifidobacterium breve strain B-3 supplementation in a mouse model with high-fat diet-induced obesity. Biosci. Biotechnol. Biochem. 74:1656–1661 [DOI] [PubMed] [Google Scholar]
- 49. Sadrzadeh-Yeganeh H, Elmadfa I, Djazayery A, Jalali M, Heshmat R, Chamary M. 2010. The effects of probiotic and conventional yoghurt on lipid profile in women. Br. J. Nutr. 103:1778–1783 [DOI] [PubMed] [Google Scholar]
- 50. Yin YN, Yu QF, Fu N, Liu XW, Lu FG. 2010. Effects of four bifidobacteria on obesity in high-fat diet induced rats. World J. Gastroenterol. 16:3394–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.