Abstract
In this study, we propose the use of the marine green alga Ostreococcus tauri, the smallest free-living eukaryotic cell known to date, as a new luminescent biosensor for toxicity testing in the environment. Diuron and Irgarol 1051, two antifouling biocides commonly encountered in coastal waters, were chosen to test this new biosensor along with two degradation products of diuron. The effects of various concentrations of the antifoulants on four genetic constructs of O. tauri (based on genes involved in photosynthesis, cell cycle, and circadian clock) were compared using 96-well culture microplates and a luminometer to automatically measure luminescence over 3 days. This was compared to growth inhibition of O. tauri wild type under the same conditions. Luminescence appeared to be more sensitive than growth inhibition as an indicator of toxicity. Cyclin-dependent kinase (CDKA), a protein involved in the cell cycle, fused to luciferase (CDKA-Luc) was found to be the most sensitive of the biosensors, allowing an accurate determination of the 50% effective concentration (EC50) after only 2 days (diuron, 5.65 ± 0.44 μg/liter; Irgarol 1015, 0.76 ± 0.10 μg/liter). The effects of the antifoulants on the CDKA-Luc biosensor were then compared to growth inhibition in natural marine phytoplankton. The effective concentrations of diuron and Irgarol 1051 were found to be similar, indicating that this biosensor would be suitable as a reliable ecotoxicological test. The advantage of this biosensor over cell growth inhibition testing is that the process can be easily automated and could provide a high-throughput laboratory approach to perform short-term toxicity tests. The ability to genetically transform and culture recombinant O. tauri gives it huge potential for screening many other toxic compounds.
INTRODUCTION
The presence of pollutants in the natural environment is one of the main causes of ecosystem degradation. Increased anthropogenic activity results in a global degradation of marine and estuary environments (1). Marine organisms are exposed to a suite of pollutants, including heavy metals, polycyclic aromatic hydrocarbons, pesticides, and antifouling biocides. There is an urgent need to develop new sensitive, rapid, automatic, and inexpensive toxicity tests to analyze the effects of the increasing numbers of chemicals released into the marine environment and also the combined effect of these pollutants, which may exhibit synergistic toxicity (2, 3).
Microalgae are well suited for toxicity bioassays because they are sensitive to a wide variety of both organic and inorganic pollutants (4). There are several ways to determine the response of marine phytoplankton species to toxic substances. Growth inhibition can be tested by measuring cell density (e.g., by measuring optical density or chlorophyll a) (5). It can also be measured by cell counting using an optical microscope (6) or a flow cytometer (7). The toxic effect of chemicals on phytoplankton can also be assessed by measuring the effective photosystem II (PSII) quantum yield with a pulse-amplified modulation fluorometer (5, 7). Naturally bioluminescent marine microorganisms can also be useful indicators of toxicity. The best-known test, Microtox, is based on the naturally luminescent marine bacteria Vibrio fischeri and has been internationally adopted as a rapid screening test (8, 9). This test, however, is not suitable for all pollutants, particularly those targeting photosynthetic species. To complete the Microtox test, the use of bioluminescent marine dinoflagellates, including Ceratocorys horrida, Pyrocystis noctiluca, Pyrocystis lunula, Pyrocystis fusiformis, and Pyrophacus stienii, has been developed in the QwikLite test (10), which has yielded results similar to those of classical ecotoxicological tests based on enumeration of phytoplankton cells (11). An alternative strategy is to develop genetic engineering tools to express recombinant luciferase (Luc) in fast-growing microorganisms expressing recombinant luciferase. Recombinant microorganisms, such as Escherichia coli encoding the thermostable lux luciferase from Photorhabdus luminescens (12) and Synechocystis sp., a freshwater cyanobacterium, encoding the thermostable luciferase from the firefly Photinus pyralis (13) have been used for both industrial effluent (14, 15) and soil environment (16, 17) monitoring. To the best of our knowledge, marine microorganism recombinant biosensors have never been developed for ecotoxicological testing.
Recently, following the development of genetic engineering tools (18), the green unicellular alga Ostreococcus tauri (Mamiellophyceae class, Mamelliales order), has emerged as new model organism. O. tauri, the smallest free-living eukaryotic cell known to date (19, 20), was isolated from the Thau lagoon (Mediterranean coastal lagoon, France), a biotope particularly exposed to anthropic pollution. This fast-growing picoalga does not produce natural bioluminescence. Firefly luciferase transcriptional and translational reporter lines have been produced to monitor the expression of genes/proteins involved in diverse biological functions such as cell division (21), circadian clock (22), photoperception (22, 23), or photosynthesis (18). Furthermore, an automated process has been developed to continuously monitor luminescence of reporter lines grown in 96-well microplates.
In this study, we determine the feasibility of using these O. tauri luminescent recombinant lines in toxicity testing using two antifouling biocides, diuron and Irgarol 1051, as toxic models. These are two of the most popular biocide boosters used to replace organotins in antifouling paints for preventing ship hulls from biofouling (24). Diuron and Irgarol 1051 are two photosynthesis inhibitors which prevent oxygen production by blocking electron transfer to the plastoquinone B (QB) of the photosystem II (25, 26). They are therefore highly phytotoxic, and this toxicity and that of their degradation products for nontarget species has been described for various aquatic organisms (3, 27–29). Initially we monitored the effect of the selected antifoulants on the kinetics of luminescence of several O. tauri reporter lines to identify the best suited for ecotoxicological studies. Biosensor responses were compared with growth inhibition of wild-type (WT) O. tauri. Finally, we compared the sensitivity of the O. tauri luminescent biosensor with that of a natural marine phytoplankton community when exposed to diuron and Irgarol 1051.
MATERIALS AND METHODS
Preparation of pesticides.
Four analytical-grade standards of pesticides were selected for the study. We used the diuron Pestanal ([3-(3,4-dichlorophenyl)-1,1 dimethyl-urea]) (99% purity; Fluka), Irgarol 1051 (2-methythiol-4-tert-butylamino-6-cyclopropylamino-s-triazine; Fluka), DCPMU [3-(3,4-dichlorophenyl)-1-methylurea; Sigma], and DCPU [3-(3,4-dichlorophenyl)-urea; Sigma]. Diuron and Irgarol 1051 are two herbicides, belonging to the urea and triazine families, respectively, and are known to induce photosynthesis inhibition by blocking the photosystem II electron-transporting chain. DCPMU and DCPU are two degradation products of diuron. The different pesticides were diluted in acetone in order to obtain the same final concentration of 0.1% (vol/vol) acetone carrier in the different treatments.
Ostreococcus tauri alga and culture conditions.
Wild-type O. tauri strain OTTH0595 and genetically modified lines with a luciferase gene reporter system (these strains have been described previously [18]) were used for the growth and luminescence assays, respectively. Wild-type and genetically modified lines were both grown in Keller medium in aerated flasks (Sarstedt) for several days and then transferred into white 96-well microplates (Nunc; Perkin-Elmer) for growth over 3 days under constant illumination (at 16 μmol · quanta/cm2 · s−1). The cells were then refreshed into new 96-well microplates at final densities of 5 × 106 cells/ml for the wild-type and 10 × 106 cells/ml for the genetically modified lines. Luciferin (10 μM final concentration) was added to the microplates containing the genetically modified lines. For both luminescence and cytometry approaches, the pesticides were tested at different concentrations: 0.5, 1, 2, 5, and 10 μg/liter for diuron; 0.05, 0.1, 0.2, 0.5, and 1 μg/liter for Irgarol 1015; 6.25, 12.5, 25, 50, 100, 200, and 400 μg/liter for DCPMU and DCPU. Each concentration was tested in triplicate using independent wells. Control samples containing 0.1% (vol/vol) acetone were carried out in triplicate for both assays. Luminescence linked to the expression of TOC1 (time of chlorophyll a binding protein [CAB] expression), pCAB (promoter of chlorophyll a binding protein gene), cyclin A, and cyclin-dependent kinase (CDKA) reporter lines was measured using a Mithras LG 940 (Berthold Technologies) luminometer every 2 h. For the cell growth approach, a subsampling of 30 μl was taken from each well of the plate after 48 h and 72 h and then diluted in 120 μl of Keller medium with glutaraldehyde (0.25%, vol/vol, final concentration). Samples were stored at −80°C before the cells were counted by flow cytometry (Cell Lab Quanta MLP; Beckman Coulter).
Determination of EC50s.
The acute toxicity of the biocides after 48 h or 72 h exposure was expressed as the concentration of pesticide inducing a reduction of 50% in growth or luminescence relative to the control (50% effective concentration [EC50]). Luminescence curves or cell concentration curves of the treated samples were normalized with those of the control samples. The percentages were then transformed in Probit units (30) and plotted against the logarithm of the pesticide concentrations to obtain a linear regression. The equation of the regression was used to determine the EC50 (i.e., pesticide concentration corresponding to a Probit unit equal to 5).
Ecotoxicological test using a natural marine phytoplankton community.
Seawater was sampled with a Niskin bottle at 3-m depth in April 2010 from a coastal station (42°29′N, 03°08′E; maximum depth, ∼26 m) in the northwestern Mediterranean Sea (∼500 m offshore Banyuls-sur-Mer, France). The water sample was filtered through 100-μm-pore-size nylon mesh and enriched with NO3 and PO4 (3.2 μM and 0.2 μM final concentrations, respectively) to avoid nutrient limitation during incubation. Twenty-four 500-ml transparent bottles were filled, and the assays were done in triplicate. Diuron or Irgarol 1051 was added at three different concentrations: 0.25, 2.5, and 25 μg/liter for diuron and 0.1, 1, and 10 μg/liter for Irgarol 1051. Two different control samples were used, one with and the other without the addition of acetone at a final concentration of 0.1% (vol/vol) corresponding to the value present in each of the pesticide samples tested. Bottles were incubated for 5 days outdoors in a water bath in which the temperature was controlled by continuously circulating surface seawater from the harbor (temperature, ∼15°C).
After 2 days, samples (3 ml) from each bottle were fixed with formaldehyde (2%, vol/vol, final concentration) and then stored at −80°C. They were analyzed with a flow cytometer (FACScan; Becton, Dickinson) to determine the abundance of Synechococcus spp. and photosynthetic pico- and nanoeukaryotes. Synechococcus cells were identified on the basis of cytograms of scatter versus phycoerythrin fluorescence (31). For the two groups of photosynthetic eukaryotes, cytograms of scatter versus chlorophyll fluorescence were used. After 5 days, duplicate 200-ml chlorophyll a samples were collected on GF/F filters (Millipore), extracted in 90% acetone, and quantified fluorometrically using a Turner-Designs 10-AU fluorometer (32).
RESULTS
Responses of different luminescent Ostreococcus tauri biosensors and comparison with the growth inhibition approach.
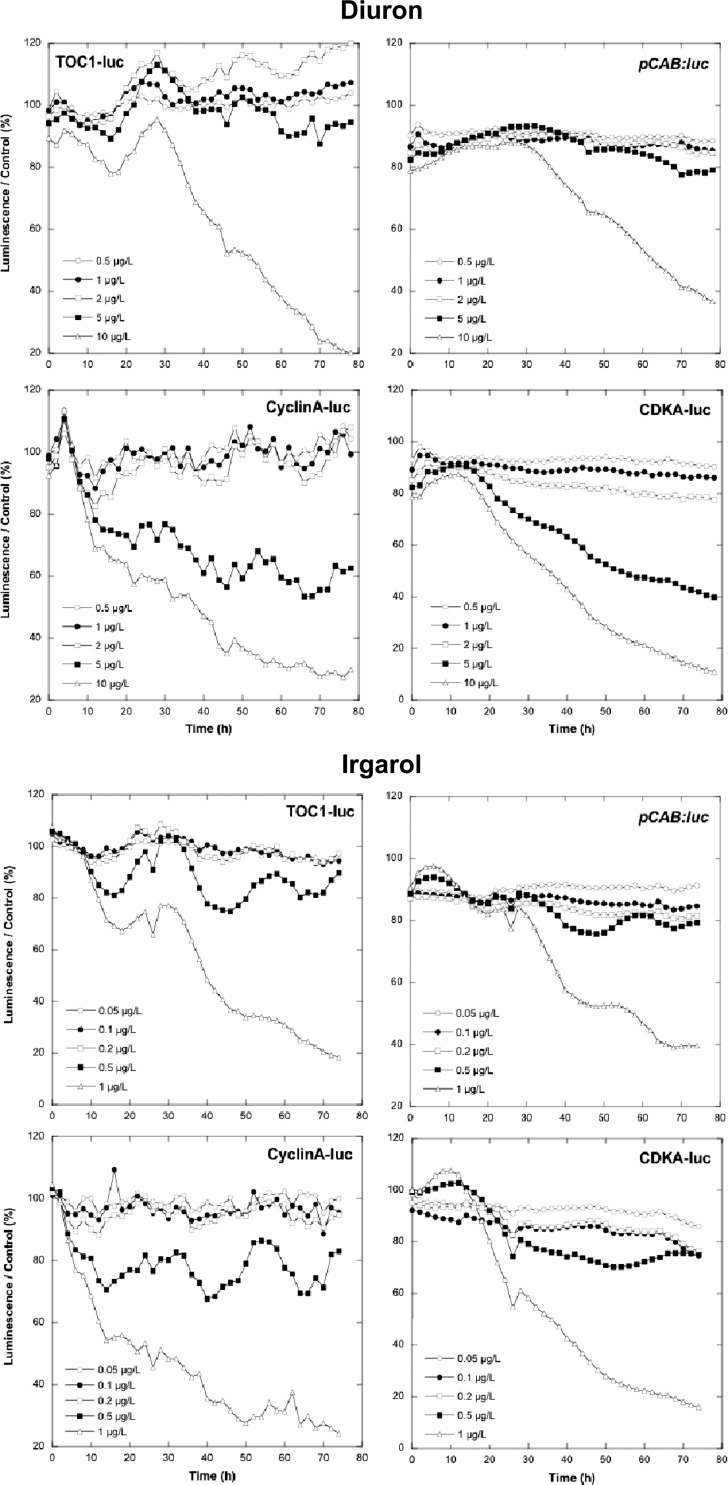
Luminescence curves were obtained after exposure to different concentrations of diuron and Irgarol 1051 for each of the different reporter lines tested (i.e., TOC1-Luc, pCAB::luc, cyclinA-Luc, and CDKA-Luc). The genetically transformed lines were characterized by different luminescence levels at the start of the experiment: the cyclin A-Luc line had the weakest luminescence (2,000 relative luminescence units [RLU]), a value only 10-fold higher than background, with TOC1-Luc at 10,000 RLU, CDKA-Luc at 60,000 RLU, and pCAB::luc at 220,000 RLU (data not shown). The luminescence values measured with the different concentrations of diuron and Irgarol 1051 over 72 h were normalized to the luminescence measured in the controls (Fig. 1). A clear decrease in luminescence was observed in the case of cyclin A-Luc and CDKA-Luc at 5 and 10 μg/liter of diuron, whereas TOC1-Luc and pCAB::luc lines responded only at the highest concentration. The toxicity of diuron increased over time for the different luminescent lines but at different rates. The sharpest decrease in luminescence was observed for CDKA-Luc (40% after 24 h). For Irgarol 1051, luminescence of all transformed lines was significantly inhibited at 1 μg/liter, demonstrating the higher toxicity of this antifouling biocide (Fig. 1). If we consider that a decrease of 20% in luminescence is sufficient to define a toxic effect, then toxicity was observed for Irgarol 1051 at 0.5 μg/liter using cyclin A-Luc and CDKA-Luc lines. With respect to diuron, the luminescence decrease showed different kinetics depending on the reporter line being used. After 24 h, the strongest inhibition was seen in the cyclin A-Luc line (50%), with inhibition of 35% for CDKA-Luc and 15% for TOC1-Luc and nonsignificant inhibition with and pCAB::luc. EC50s were calculated for each luminescent line and for both antifoulants after 24, 48, and 72 h. The EC50s found at 24 h exhibited marked variance between the replicated assays for each reporter and therefore were not considered. The EC50s calculated at 48 and 72 h, however, were more reproducible for each of the tested lines (Table 1). The CDKA-Luc line appeared to be the most sensitive O. tauri line for detecting the toxicity of both antifoulants, with the EC50 decreasing by 40% between 48 h and 72 h (P < 0.05, Student's t test) (Table 1). Given its high level of luminescence and its high sensitivity to the two antifoulants tested, the CDKA-Luc line was therefore used for subsequent experiments and comparisons.
Fig 1.
Comparison of the different O. tauri luminescent lines, as indicated, after exposure to diuron and Irgarol 1051. Graphs represent normalized data compared to the control lines (without antifoulant). Values correspond to the means of triplicate wells on the microplate.
Table 1.
Comparison of the EC50s of different O. tauri luminescent lines and differences of sensibility of the two methods tested
| Pesticide | Cell line (assay)a | EC50 (μg/liter) at the indicated timeb |
|
|---|---|---|---|
| 48 h | 72 h | ||
| Diuron | TOC1-Luc (LI) | 20.29 ± 1.78 | 11.19 ± 4.03 |
| pCAB::luc line (LI) | 57.36 ± 34.21 | 13.91 ± 2.53 | |
| Cyclin A-Luc (LI) | 9.19 ± 1.21 | 6.64 ± 0.95 | |
| CDKA-Luc (LI) | 5.65 ± 0.44 | 3.51 ± 0.09 | |
| Wild type (GI) | 26.37 ± 3.69 | 10.22 ± 4.48 | |
| Irgarol 1051 | TOC1-Luc (LI) | 1.05 ± 0.28 | 1.04 ± 0.56 |
| pCAB::luc line (LI) | 1.96 ± 0.48 | 1.27 ± 0.50 | |
| Cyclin A-Luc (LI) | 0.89 ± 0.03 | 0.92 ± 0.27 | |
| CDKA-Luc (LI) | 0.76 ± 0.10 | 0.42 ± 0.04 | |
| Wild type (GI) | 3.63 ± 0.97 | 2.20 ± 0.58 | |
| DCPMU | CDKA-Luc (LI) | 14.45 ± 1.22 | 14.26 ± 0.63 |
| Wild type (GI) | 50.81 ± 1.91 | ND | |
| DCPU | CDKA-Luc (LI) | >400 | > 400 |
| Wild type (GI) | >400 | ND | |
GI, growth inhibition test; LI, luminescence inhibition test.
Data represent the means ± standard deviations of three replicates. ND, not determined.
The luminescence response of CDKA-Luc was compared to the measurement of growth inhibition of O. tauri WT cells (Table 1). For both antifoulants the EC50 was significantly lower (P < 0.05, Student's t test) for the luminescence assay than for the growth experiment after 48 and 72 h (Table 1). After 48 h, the ratio between the EC50 estimated by growth inhibition and the EC50 estimated by CDKA-Luc luminescence was 4.7 for diuron and 4.8 for Irgarol 1051 (Table 1). After 72 h, this ratio was still in favor of the luminescent biosensor (2.9 for diuron and 5.2 for Irgarol 1051) (Table 1), underlining the greater sensitivity of the luminescent biosensor approach for assessing the toxicity of these antifoulants. To complete this comparison, we also tested the toxicity of two degradation products of diuron: DCPU and DCPMU. We did not measure any significant toxicity of DCPU with either approach (up to 400 μg/liter). After 48 h, toxicity of DCPMU was detected by the CDKA-Luc biosensor and growth inhibition (EC50 of 14.45 ± 1.22 g/liter and 50.81 ± 1.91 g/liter, respectively) but was lower than that of its parent molecule (EC50 of 5.65 ± 0.44 g/liter and 26.37 ± 3.69 g/liter, respectively) (Table 1).
Relevance of the response measured with CDKA-Luc compared to a natural phytoplankton community.
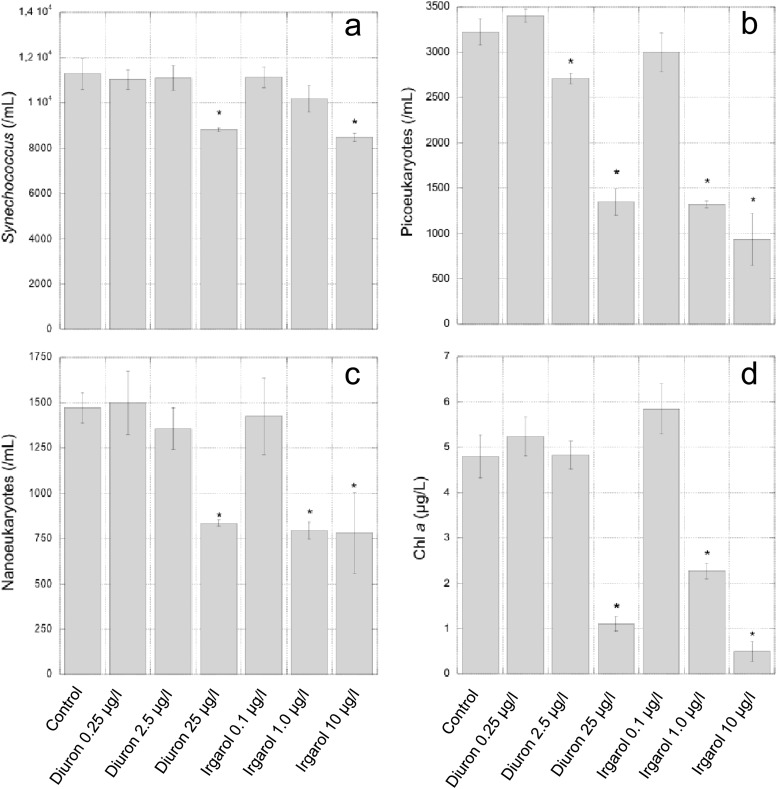
The next experiment was designed to determine the effect of diuron and Irgarol 1051 on natural communities at concentrations in the range of those tested by the luminescence assay. We did not measure any significant effect on the concentration of phytoplankton cells after 2 days or on the chlorophyll a content after 5 days in the control samples with and without acetone, suggesting that acetone at a concentration of 0.1% is harmless for phytoplankton. After 2 days of incubation, flow cytometry analysis revealed significant differences in Synechococcus abundance between the controls at the highest concentrations of diuron and Irgarol 1051 (22% and 25% inhibition at 25 μg/liter and 10 μg/liter, respectively) (Fig. 2). At the same concentrations, photosynthetic nanoeukaryotes appeared more sensitive than Synechococcus cells to these two antifoulants, with inhibition of 43% and 47% for diuron and Irgarol 1051, respectively. Photosynthetic picoeukaryotes were the most sensitive of the three phytoplanktonic groups analyzed by flow cytometry, showing a significant inhibition at both 2.5 and 25 μg/liter of diuron and 1.0 and 10 μg/liter of Irgarol 1051. For the highest concentrations tested for each antifoulant, the inhibition was 58% and 71% for diuron and Irgarol 1051, respectively. After 5 days of incubation, the chlorophyll a concentrations were significantly reduced in the presence of diuron at 25 μg/liter (by a factor 4) and Irgarol 1051 at 1.0 and 10 μg/liter (by factors of 2 and 10, respectively) (Fig. 2).
Fig 2.
Effect of diuron and Irgarol 1051 on Synechococcus, picoeukaryote, and nanoeukayote concentrations after 2 days of incubation, and on chlorophyll a (Chl a) concentration after 5 days of incubation. Data represent the means ± standard deviations of three replicates. Asterisks indicate treatments that were significantly different from the control based on Student's t test (P < 0.05).
DISCUSSION
There is an ongoing need to develop sensitive, rapid, automatic, and inexpensive toxicity tests to monitor the effect of the increasing number of chemicals released into the aquatic environment. Such ecotoxicological tests need to consider the complex mixture of pollutants and other stressors (e.g., temperature and UV light) (33). Ostreococcus sp. is a prasinophyte widely distributed in the marine environment from coastal lagoon to open waters (34). In this study, we propose the use of the marine green alga O. tauri, the smallest free-living eukaryotic cell known to date, as a new environmental luminescent biosensor for toxicity testing.
CDKA-Luc as the most effective O. tauri luminescent biosensor.
The differences observed in the luminescence raw data (RLU) for each reporter line can be explained by the different levels of gene expression. CDKA-Luc, cyclin A-Luc, and TOC1-Luc are translational reporter proteins fused in frame with luciferase. In these reporter lines the luminescence is a result of both luciferase decay upon oxidation of luciferin and turnover/degradation of the fused protein. In contrast, the pCAB::luc transcriptional reporter, which produces only luciferase, displays higher levels of luminescence. Considering the normalized results compared to controls, all luminescent biosensors were affected by the action of the antifoulants. Of the different O. tauri luminescent lines, CDKA-Luc was the most sensitive to diuron and Irgarol 1051. A possible explanation is that cell division cycle regulators like CDKA are very quickly downregulated when cells are exposed to toxic compounds. Surprisingly, cyclin A, which is also a cell cycle regulator, was less sensitive than CDKA to these antifoulants. This result needs to be carefully interpreted, however, since the average expression level of cyclin A-Luc was lower than that of CDKA-Luc, making it difficult to detect small changes in luminescence. A major advantage of the O. tauri luminescent assay over the conventional cytometry test and other phytoplankton growth inhibition tests is the ability to fully automate the process, allowing high throughput of samples at low cost. Because growth inhibition tests are cumbersome, only a few measurements can be reasonably recorded, e.g., at 48 h or 72 h, whereas with the luminescence method, a high throughput can be automatically achieved using a laboratory work station.
Comparison with other phytoplankton growth inhibition and whole-biosensor approaches.
Comparison of our results of the CDKA-Luc biosensor to literature reports of growth inhibition assays using phytoplankton strains shows that the luminescence assay is within a reasonable range of EC50s recorded for both diuron and Irgarol 1051 (Tables 2 and 3). According to our evidence, the O. tauri luminescence assay is the most sensitive test of all the whole-cell luminescence biosensors (Tables 2 and 3). The CDKA-Luc reporter particularly showed a much higher sensitivity to both antifoulants than the frequently used Vibrio fischeri Microtox luminescence test, with a difference of three to four orders of magnitude (i.e., from μg/liter to mg/ml concentrations) (Tables 2 and 3).
Table 2.
Comparison of different approaches to measure diuron toxicity using phytoplankton growth inhibition tests and different whole-cell biosensors
| Test or indicator | Organism (phylum/division, test, or indicator) | EC50 (μg/liter) | Time (h) | Reference |
|---|---|---|---|---|
| Phytoplankton growth inhibitiona | Synechococcus sp (Cyanobacteria) | 0.55 | 72 | 35 |
| Emiliania huxleyi (Haptophyta) | 2.3 | 72 | 35 | |
| Chlorella vulgaris (Chlorophyta) | 4.3 | 96 | 36 | |
| Chaetoceros gracilis (Bacillariophyta) | 4.9 | 96 | 37 | |
| Dunaliella tertiolecta (Chlorophyta) | 5.9 | 96 | 38 | |
| Ostreococcus tauri (Chlorophyta) | 26 | 48 | This study | |
| Navicula forcipata (Bacillariophyta) | 27 | 96 | 38 | |
| Chaetoceros gracilis (Bacillariophyta) | 36 | 96 | 38 | |
| Raphidocelis subcapitata (Chlorophyta) | 45 | 72 | 35 | |
| Whole-cell biosensor | Ostreococcus tauri, luminescent biosensor (CDKA-Luc) | 5.7 | 48 | This study |
| 3.5 | 72 | This study | ||
| Vibrio fischeri (Microtox) | 58,000 | 0.5 | 26 | |
| Pyrocystis lunula (QwickLite) | 19,000 | 24 | 11 | |
| Synechocystis (luc) | 29,700 | 24 | 13 | |
| Escherichia coli (lux) | >20,000 | 2 | 39 | |
| Saccharomyces cerevisiae (ATP synthesis) | 11,600 | 0.5 | 40 |
All organisms used for the phytoplankton growth inhibition test were from marine water except Chlorella vulgaris and Raphidocelis subcapitata, which were from freshwater.
Table 3.
Comparison of different approaches to measure Irgarol 1051 toxicity using phytoplankton growth inhibition tests and different whole-cell biosensors
| Test and/or sample type or indicator | Organism name (phylum or division) | EC50 (μg/liter) | Time (h) | Reference |
|---|---|---|---|---|
| Phytoplankton growth inhibition | ||||
| Marine water | Tetraselmis sp. (Chlorophyta) | 0.12 | 72 | 7 |
| Synechococcus sp. (Cyanobacteria) | 0.16 | 72 | 35 | |
| Emiliania huxleyi (Haptophyta) | 0.25 | 72 | 35 | |
| Thalassiosira weisflogii (Bacillariophyta) | 0.30 | 72 | 7 | |
| Emiliania huxleyi (Haptophyta) | 0.41 | 72 | 7 | |
| Chaetoceros gracilis (Bacillariophyta) | 0.49 | 96 | 37 | |
| Fibrocapsa japonica (Raphidophyta) | 0.62 | 72 | 7 | |
| Dunaliella tertiolecta (Chlorophyta) | 1.17 | 96 | 38 | |
| Ostreococcus tauri (Chlorophyta) | 3.63 | 48 | This study | |
| Freshwater | Navicula accomoda (Bacillariophyta) | 0.5 | 96 | 38 |
| Chlamydomonas intermedia (Chlorophyta) | 0.5 | 144 | 41 | |
| Nitszchia sp. (Bacillariophyta) | 0.8 | 96 | 41 | |
| Chlorella vulgaris (Chlorophyta) | 1.5 | 96 | 41 | |
| Staurastrum sebaldii (Zygnematophyceae) | 2.5 | 144 | 41 | |
| Pseudokirchneriella subcapitata (Chlorophyta) | 3.3 | 96 | 41 | |
| Scenedesmus acutus (Chlorophyta) | 5.1 | 96 | 41 | |
| Selenastrum capricornutum (Chlorophyta) | 10.8 | 72 | 3 | |
| Raphidocelis subcapitata (Chlorophyta) | 11 | 72 | 35 | |
| Asterionella formasa (Bacillariophyta) | >253 | 96 | 41 | |
| Whole-cell biosensor | ||||
| CDKA-Luc | Ostreococcus tauria | 0.76 | 48 | This study |
| 0.42 | 72 | This study | ||
| Microtox | Vibrio fischeri | 50,800 | 0.25 | 3 |
Luminescent biosensor.
Our study revealed that O. tauri was less sensitive to DCPMU (and not sensitive at all to DCPU) than the diuron parent molecule. This has also been observed for the microalgae Dunaliella tertiolecta (38), Chaetoceros gracilis (37), and Selenastrum capricornotum (3). In contrast, higher toxicity has been observed for DCPMU and DCPU than for diuron in the two marine ciliates Tetrahymena pyriformis and Spirostomum teres and using the Microtox test (3, 26, 38). The toxicity of degradation products of Irgarol 1051 has been studied less than that of the diuron degradation products. Biological and chemical degradation of Irgarol 1051 can lead to the formation of the stable degradation product 2-methylthio-4-tert-butylamino-6-amino-s-triazine (M1) (29). This compound has been shown to be less toxic than its parent compound to S. capricornotum but more toxic for higher plants (3, 29). It will be interesting to test the toxicity of this degradation product using the O. tauri CDKA-Luc biosensor in future study.
Comparison with the response of a natural phytoplankton community.
In our study, the EC50s of diuron and Irgarol 1051 using the CDKA-Luc reporter (5.65 ± 0.44 and 0.76 ± 0.10 μg/liter, respectively) were similar to those measured for natural phytoplankton communities (∼2.5 and 1 μg/liter, respectively). Moreover, our results show that among the phytoplankton community studied, photosynthetic picoeukaryotes were the most sensitive population to both antifoulants, reinforcing the suitability of the O. tauri CDKA-Luc reporter as an ecotoxicological biosensor.
Overall, our measurements of the effects of diuron and Irgarol 1051 (demonstrated both by growth inhibition of natural marine phytoplankton and by the O. tauri monoculture approach) yielded consistent results, with Irgarol 1051 showing higher toxicity than diuron. This is consistent with other studies which have shown the toxic effect of diuron and Irgarol 1051 at low concentrations on a natural phytoplankton community in freshwater ecosystems (42, 43). The effective concentrations of diuron and Irgarol 1051 measured on both O. tauri and the natural phytoplankton community are close to the concentrations measured in some marinas and coastal areas. For instance, in Japanese coastal areas, diuron and Irgarol 1051 concentrations have been measured up to 3.05 μg/liter and 0.262 μg/liter, respectively (44). Levels of up to 1.7 μg/liter and 0.64 μg/liter were observed in the French Mediterranean Sea (45, 46), while in United Kingdom waters, they have reached even higher values, up to 6.74 μg/liter and 1.42 μg/liter, respectively (47). These results provide evidence that current environmental concentrations of diuron and Irgarol 1051 could impair the activity and composition of the phytoplankton community, with possible consequences for higher trophic levels and the global functioning of aquatic ecosystems.
Concluding remarks.
We conclude that the O. tauri microplate-based recombinant biosensor constitutes a reliable high-throughput laboratory approach to performing short-term toxicity tests in application to antifouling biocides and could potentially be used for screening other toxic compounds in the marine environment. This assay offers many advantages over phytoplankton growth inhibition assays and is much more sensitive than other existing whole-cell biosensors. More than 20 luciferase transcriptional/translational reporters are currently available in O. tauri, and the number is likely to rapidly expand in the near future (18, 21–23; F.-Y. Bouget, personal communication). These reporters cover diverse biological processes, including cell division, circadian clock, nutriments assimilation, and photosynthesis, that are potentially specific targets of biocides. In the future more sensitive and pollutant-specific biosensors could be developed by using specific target genes fused to luciferase. Such target genes could also be defined using next-generation sequencing of RNA to identify genes that are specifically and rapidly induced or repressed in response to low doses of specific molecules (48–50). Automated monitoring of these luminescent reporters should allow the development of new platforms for high-throughput screening of the effect of pollutants on marine eukaryotic picophytoplankton.
ACKNOWLEDGMENTS
This work was supported by the CNRS-INSU through the program EC2CO (Écosphère Continentale et Côtière).
We thank L. Oriol for chlorophyll determination, L. Bariat for flow cytometry analysis of natural samples, and C. Jacquet, F. Sanchez, and P. Schatt for the maintenance of Ostreococcus tauri cultures. We are deeply grateful to Joan and Eamon Groves for extensive editing and proofreading the manuscript.
Footnotes
Published ahead of print 9 November 2012
REFERENCES
- 1. Edgar G, Barrett N, Graddon D, Last P. 2000. The conservation significance of estuaries: a classification of Tasmanian estuaries using ecological, physical and demographic attributes as a case study. Biol. Conserv. 92:383–397 [Google Scholar]
- 2. Faust M, Altenburger R, Backhaus T, Blanck H, Boedeker W, Gramatica P, Hamer V, Scholze M, Vighi M, Grimme LH. 2001. Predicting the joint algal toxicity of multi-component s-triazine mixtures at low-effect concentrations of individual toxicants. Aquat. Toxicol. 56:13–32 [DOI] [PubMed] [Google Scholar]
- 3. Fernández-Alba AR, Hernando MD, Piedra L, Chisti Y. 2002. Toxicity evaluation of single and mixed antifouling biocides measured with acute toxicity bioassays. Anal. Chim. Acta 456:303–312 [Google Scholar]
- 4. Klaine SJ, Lewis MA. 1995. Algal and plant toxicity testing, p 163–184 In Hoffman DJ, Rattner BA, Burton GA, Jr, Cairns J., Jr (ed), Handbook of ecotoxicology. Lewis Publishers, Boca Raton, FL [Google Scholar]
- 5. Chung MK, Wong MH, Cheung KC. 2007. Comparative toxicity of hydrophobic contaminants to microalgae and higher plants. Ecotoxicology 16:393–402 [DOI] [PubMed] [Google Scholar]
- 6. DeLorenzo M, Serrano L. 2003. Individual and mixture toxicity of three pesticides; atrazine, chlorpyrifos, and chlorothalonil to the marine phytoplankton species Dunaliella tertiolecta. J. Environ. Sci. Health B 38:529–538 [DOI] [PubMed] [Google Scholar]
- 7. Buma AGJ, Sjollema SB, van de Poll WH, Klamer HJC, Bakker JF. 2009. Impact of the antifouling agent Irgarol 1051 on marine phytoplankton species. J. Sea Res. 61:133–139 [Google Scholar]
- 8. Isenberg DL. 1993. The Microtox toxicity test: a developer's commentary, p 3–15 In Richardson M. (ed), Ecotoxicology monitoring. VCH, Weinheim, Germany [Google Scholar]
- 9. Steinberg SM, Poziomek EJ, Englemann WH, Rogers KR. 1995. A review of environmental applications of bioluminescence measurements. Chemosphere 30:2155–2197 [Google Scholar]
- 10. Lapota D, Robayo Osorio A, Liao C, Bjorndal B. 2007. The use of bioluminescent dinoflagellates as an environmental risk assessment tool. Mar. Pollut. Bull. 54:1857–1867 [DOI] [PubMed] [Google Scholar]
- 11. Stauber JL, Binet MT, Bao VWW, Boge J, Zhang AQ, Leung KMY, Adams MS. 2008. Comparison of the QwikLite algal bioluminescence test with marine algal growth rate inhibition bioassays. Environ. Toxicol. 23:617–625 [DOI] [PubMed] [Google Scholar]
- 12. Kurvet I, Ivask A, Bondarenko O, Sihtmäe M, Kahru A. 2011. LuxCDABE-transformed constitutively bioluminescent Escherichia coli for toxicity screening: comparison with naturally luminous Vibrio fischeri. Sensors 11:7865–7878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shao CY, Howe CJ, Porter AJR, Glover LA. 2002. Novel cyanobacterial biosensor for detection of herbicides. Appl. Environ. Microbiol. 68:5026–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paton GI, Palmer G, Kindness A, Campbell CD, Glover LA, Killham K. 1995. Use of luminescence marked bacteria to assess copper bioavailability in malt whisky distillery effluent. Chemosphere 31:3217–3224 [Google Scholar]
- 15. Brown JS, Rattray EAS, Paton GI, Reid G, Caffoor I, Killham K. 1996. Comparative assessment of the toxicity of a papermill effluent by respirometry and a luminescence-based bacterial assay. Chemosphere 32:1553–1561 [Google Scholar]
- 16. Paton GI, Rattray EAS, Campbell CD, Cresser MS, Glover LA, Meeussen JCL, Killham K. 1996. Use of genetically modified microbial biosensor for soil ecotoxicity testing, p 394–418 In Pankhurst CF, Doube BM, Gupta VVSR. (ed), Biological indicators of soil health. CAB International Press, Oxon, United Kingdom [Google Scholar]
- 17. McGrath SP, Knight B, Killham K, Preston S, Paton GI. 1999. Assessment of the toxicity of metals in soils amended with sewage sludge using a chemical speciation technique and a lux-based biosensor. Environ. Toxicol. Chem. 18:659–663 [Google Scholar]
- 18. Corellou F, Schwartz C, Motta Djouani-Tahri J-PEB, Sanchez F, Bouget F-Y. 2009. Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote Ostreococcus. Plant Cell 21:3436–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Courties C, Vaquer A, Troussellier M, Lautier J, Chrétiennot-Dinet MJ, Neveux J, Machado C, Claustre H. 1994. Smallest eukaryotic organism. Nature 370:255 doi:10.1038/370255a0 [Google Scholar]
- 20. Chrétiennot-Dinet M-J, Courties C, Vaquer A, Neveux J, Claustre H, Lautier J, Machado M-C. 1995. A new marine picoeucaryote: Ostreococcus tauri gen. et sp. nov. (Chlorophyta, Prasinophyceae). Phycologia 34:285–292 [Google Scholar]
- 21. Moulager M, Corellou F, Vergé V, Escande M-L, Bouget F-Y. 2010. Integration of light signals by the retinoblastoma pathway in the control of S phase entry in the picophytoplanktonic cell Ostreococcus. PLoS Genet. 6:e1000957 doi:10.1371/journal.pgen.1000957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Djouani-Tahri E, Christie JM, Sanchez-Ferandin S, Sanchez F, Bouget FY, Corellou F. 2011. A eukaryotic LOV-histidine kinase regulates circadian clock function in the picoalga Ostreococcus. Plant J. 65:578–588 [DOI] [PubMed] [Google Scholar]
- 23. Heijde M, Zabulon G, Corellou F, Ishikawa T, Brazard J, Usman A, Sanchez F, Plaza P, Martin M, Falciatore A, Todo T, Bouget Bowler F-YC. 2010. Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant Cell Environ. 33:1614–1626 [DOI] [PubMed] [Google Scholar]
- 24. Konstantinou IK, Albanis TA. 2004. Worldwide occurrence and effects of antifouling paint booster biocides in the aquatic environment: a review. Environ. Int. 30:235–248 [DOI] [PubMed] [Google Scholar]
- 25. Eullaffroy P, Vernet G. 2003. The F684/F740 chlorophyll fluorescence ratio: a tool for a quick detection and determination of phytotoxicity of herbicides in algae. Water Res. 37:1983–1990 [DOI] [PubMed] [Google Scholar]
- 26. Bonnet J-L, Bonnemoy F, Dusser M, Bohatier J. 2007. Assessment of the potential toxicity of herbicides and their degradation products to nontarget cells using two microorganisms, the bacteria Vibrio fischeri and the ciliate Tetrahymena pyriformis. Environ. Toxicol. 22:78–91 [DOI] [PubMed] [Google Scholar]
- 27. Chesworth JC, Donkin ME, Brown MT. 2004. The interactive effects of the antifouling herbicides Irgarol 1051 and Diuron on the seagrass Zostera marina (L.). Aquat. Toxicol. 66:293–305 [DOI] [PubMed] [Google Scholar]
- 28. Manzo S, Buono S, Cremisini C. 2006. Toxic effects of Irgarol and diuron on sea urchin Paracentrotus lividus early development, fertilization, and offspring quality. Arch. Environ. Contam. Toxicol. 51:61–68 [DOI] [PubMed] [Google Scholar]
- 29. Okamura H, Aoyama I, Liu D, Maguire RJ, Pacepavicius GJ, Lau YL. 2000. Fate and ecotoxicity of the new antifouling compounds Irgarol 1051 in the aquatic environment. Water Res. 34:3523–3530 [Google Scholar]
- 30. Finney DJ. 1989. Probit analysis, 3rd ed Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 31. Joux F, Servais P, Naudin Lebaron J-JP, Oriol L, Courties C. 2005. Distribution of picophytoplankton and bacterioplankton along a river plume gradient in the Mediterranean Sea. Vie Milieu 55:197–208 [Google Scholar]
- 32. Lorenzen C. 1967. Determination of chlorophyll and pheopigments: spectrometric equations. Limnol. Oceanogr. 12:343–346 [Google Scholar]
- 33. Eggen RIL, Behra R, Burkhardt-Holm P, Escher BI, Schweigert N. 2004. Challenges in ecotoxicology. Environ. Sci. Technol. 38:58A–64A [DOI] [PubMed] [Google Scholar]
- 34. Worden A. 2006. Picoeukaryote diversity in coastal waters of the Pacific Ocean. Aquat. Microb. Ecol. 43:165–175 [Google Scholar]
- 35. Devilla RA, Brown MT, Donkin M, Tarran GA. 2005. Impact of antifouling booster biocides on single microalgal species and on a natural marine phytoplankton community. Mar. Ecol. Progr. Ser. 286:1–12 [Google Scholar]
- 36. Ma J, Xu L, Wang S, Zheng R, Jin S, Huang S, Huang Y. 2002. Toxicity of 40 herbicides to the green alga Chlorella vulgaris. Ecotoxicol. Environ. Saf. 51:128–132 [DOI] [PubMed] [Google Scholar]
- 37. Sauren S, Arzul G, Durand G, Hureau H. 2005. Toxic effects of the antifoulants Diuron and Irgarol 1051 on the diatom Chaetoceros gracilis, poster. Abstr. Soc. Environ. Toxicol. Chem. (SETAC) Europe 15th Annu. Meet., Lille, France Society of Environmental Toxicology and Chemistry, Brussels, Belgium: (In French.) http://wwz.ifremer.fr/pollution/content/download/31847/437618/file/poster_GFP_05_Champs_sur_marne.pdf [Google Scholar]
- 38. Gatidou G, Thomaidis N. 2007. Evaluation of single and joint toxic effects of two antifouling biocides, their main metabolites and copper using phytoplankton bioassays. Aquat. Toxicol. 85:184–191 [DOI] [PubMed] [Google Scholar]
- 39. Strachan G, Preston S, Maciel H, Porter AJR, Paton G. 2001. Use of bacterial biosensors to interpret the toxicity and mixture toxicity of erbicides in freshwater. Water Res. 35:3490–3495 [DOI] [PubMed] [Google Scholar]
- 40. Estève K, Popot C, Dabert P, Mietton-Peuchot M, Milisic V. 2009. A Saccharomyces cerevisiae-based bioassay for assessing pesticide toxicity. J. Ind. Microbiol. Biotechnol. 36:1529–1534 [DOI] [PubMed] [Google Scholar]
- 41. Bérard A, Dorigo U, Mercier I, Becker-van Slooten K, Grandjean D, Leboulanger C. 2003. Comparison of the ecotoxicological impact of the triazines Irgarol 1051 and atrazine on microalgal cultures and natural microalgal communities in Lake Geneva. Chemosphere 53:935–944 [DOI] [PubMed] [Google Scholar]
- 42. Backhaus T, Faust M, Scholze M, Gramatica P, Vighi M, Grimme LH. 2004. Joint algal toxicity of phenylurea herbicides is equally predictable by concentration addition and independent action. Environ. Toxicol. Chem. 23:258–264 [DOI] [PubMed] [Google Scholar]
- 43. Mohr S, Schöder H, Feibicke M, Berghahn R, Arp W, Nicklisch A. 2008. Long-term effects of the antifouling booster biocide Irgarol 1051 on periphyton, plankton and ecosystem function in freshwater pond mesocosms. Aquat. Toxicol. 90:109–120 [DOI] [PubMed] [Google Scholar]
- 44. Okamura H, Aoyama II, Ono Y, Nishida T. 2003. Antifouling herbicides in the coastal waters of western Japan. Mar. Pollut. Bull. 47:59–67 [DOI] [PubMed] [Google Scholar]
- 45. Readman JW, Kwong LLW, Grondin D, Bartocci J, Villeneuve JP, Mee LD. 1993. Coastal water contamination from a triazine herbicide used in antifouling paints. Environ. Sci. Technol. 27:1940–1942 [Google Scholar]
- 46. Tolosa IJ, Readman W, Blaevoet A, Ghilini S, Bartocci J, Horvat M. 1996. Contamination of Mediterranean (Cote d'Azur) coastal waters by organotins and Irgarol 1051 used in antifouling paints. Mar. Pollut. Bull. 32:335–341 [Google Scholar]
- 47. Thomas K, Fileman T, Readman J, Waldock M. 2001. Antifouling paint booster biocides in the UK coastal environment and potential risks of biological effects. Mar. Pollut. Bull. 42:677–688 [DOI] [PubMed] [Google Scholar]
- 48. Schirmer K, Fischer BB, Madureira DJ, Pillai S. 2010. Transcriptomics in ecotoxicology. Anal. Bioanal. Chem. 397:917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ekblom R, Galindo J. 2011. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 107:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Manikandan J. 2012. Integration of next-generation sequencing based multi-omics approaches in toxicogenomics. Front. Genet. 3:88 doi:10.3389/fgene.2012.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]