Abstract
Pseudomonas syringae pv. tomato DC3000 contains genes for 15 sigma factors. The majority are members of the extracytoplasmic function class of sigma factors, including five that belong to the iron starvation subgroup. In this study, we identified the genes controlled by three iron starvation sigma factors. Their regulons are composed of a small number of genes likely to be involved in iron uptake.
TEXT
Pseudomonas syringae is a globally dispersed bacterial pathogen that is well known for its ability to cause disease in plants (1, 2). Plant-pathogenic bacteria tend to possess functions that enable them to adapt and thrive under diverse environmental conditions. One feature that may contribute to survival in different habitats is the abundance of TonB-dependent signal transduction systems (3). These systems often activate gene expression through a signal transduction cascade that activates a cytoplasmic iron starvation (IS) sigma factor in response to siderophore binding (4–6). Sigma factors are exchangeable subunits of RNA polymerase that function in promoter recognition and transcription initiation (7, 8).
P. syringae pv. tomato DC3000 contains five IS sigma factor genes (9, 10). We previously determined the regulons of two IS sigma factors, PvdS and PSPTO_1203, which control genes for the production/uptake of pyoverdine and expression of genes in response to hydroxamate siderophores, respectively (11, 12). All five IS sigma factors are positioned downstream of Fur binding sites (13), and expression of pvdS, PSPTO_1209 and PSPTO_1286 sigma factor genes is controlled by iron concentration in the growth medium (14). Here we determined the regulons of three IS sigma factors encoded by PSPTO_0444, PSPTO_1209, and PSPTO_1286.
Chromatin immunoprecipitation sequencing (ChIP-seq) was used to screen the P. syringae DC3000 genome for sites bound by PSPTO_0444, PSPTO_1209, and PSPTO_1286 as described by Markel et al. (11). In these experiments, C-terminally Flag-tagged derivatives of each sigma factor were constitutively expressed from plasmids (see Table S1 in the supplemental material for a complete list and description of plasmids used in this study) in P. syringae DC3000 cells grown to an optical density at 600 nm (OD600) of 0.5 in a low-iron minimal medium (11). After formaldehyde cross-linking, sigma factor-DNA complexes were immunoprecipitated via the Flag epitope, the DNA was analyzed by high-throughput sequencing, and sequence reads were aligned with the P. syringae DC3000 genome. Enrichment was evaluated by comparing the number of sequence reads at each locus in experimental samples to the number of reads from controls prepared from cells containing the empty vector.
Four loci were enriched in the PSPTO_1209 ChIP-seq, and two loci were enriched in the PSPTO_1286 and PSPTO_0444 ChIP-seq (Table 1). As expected, each sigma factor captured regions upstream of genes encoding TonB-dependent receptors. The genes encoding these types of TonB-dependent receptors are typically colocated with the sigma factor and anti-sigma factor genes that control their expression (3). The TonB-dependent receptor genes bound by PSPTO_1286 and PSPTO_1209 exhibited this typical genomic arrangement. However, the TonB-dependent siderophore receptor gene PSPTO_4128, which was immediately downstream of a genomic site bound by PSPTO_0444, was located more than 2.2 Mb from the sigma factor gene itself.
Table 1.
Genomic loci enriched in sigma factor ChIP-seq experiments
| ChIP-seq yielding enrichment | Peak | Peak coordinatesa | Flanking gene | Product | Expression ratiob |
|---|---|---|---|---|---|
| PSPTO_0444 | 1 | 490523–490864 | PSPTO_0446 | Uncharacterized membrane protein | 70.8 (1.3) |
| 2 | 4653465–4653851 | PSPTO_4128 | TonB-dependent siderophore receptor | 477 (1.5) | |
| PSPTO_1209 | 1 | 1277234–1277588 | PSPTO_1164 | OmpA-family protein | 0.47 |
| 2 | 1323698–1323965 | PSPTO_1207 | TonB-dependent siderophore receptor | 34.4 (2.2) | |
| 3 | 1826690–1827094 | PSPTO_1660 | Helicase/SNF2 family domain protein | 0.67 | |
| 4 | 6178066–6178398 | PSPTO_5432 | Type VI protein secretion system | 0.57 | |
| PSPTO_1286 | 1 | 1410711–1410926 | PSPTO_1283 | Heme oxygenase | 84.04 (0.9) |
| 2 | 1413374–1413707 | PSPTO_1284 | TonB-dependent heme receptor | 84.11 (11.7) |
Peak coordinates refer to positions in the P. syringae pv. tomato DC3000 genome (9).
Expression ratios show the difference in expression (obtained by qRT-PCR) of genes flanking ChIP-seq peaks in cells containing the sigma factor expression vector relative to cells containing the empty vector control plasmid, pBS60 (12). Expression ratios are the averages of three biological replicates (standard deviations are in parentheses). Data without standard deviations are representative results, and genes are not considered differentially expressed by the corresponding sigma factor under these conditions. Flag-tagged and native sigma factor expression vectors are described in Table S1 in the supplemental material.
Genes downstream of loci identified in the ChIP-seq experiments were tested to determine whether their transcription was dependent on the associated sigma factor. Using quantitative reverse transcription-PCR (qRT-PCR), we compared the relative abundance of selected transcripts in cells constitutively expressing each of the three sigma factors versus with the abundance of those containing the empty vector, as described in reference 11.
This analysis confirmed that each sigma factor regulates the expression of a TonB-dependent receptor gene and showed that PSPTO_0444 and PSPTO_1286 each regulate another gene (Table 1). PSPTO_0444 controls the expression of PSPTO_0446, which is predicted to encode a 377-amino-acid protein annotated as an uncharacterized iron-regulated membrane protein similar to PiuB in the Conserved Domain database (E value = 3.93e-51). PiuB is a transmembrane permease/iron uptake ABC transporter, first characterized in the Gram-positive bacterium Streptococcus pneumoniae (15, 16). PSPTO_1286 regulates PSPTO_1283, predicted to encode a heme oxygenase. Proteins of this class are often involved in the degradation of chelators to extract the iron (17). Three additional locations were also enriched in the PSPTO_1209 ChIP-seq experiments, but we were unable to detect regulation of the genes downstream of them. These genes may be regulated by PSPTO_1209 under different conditions, or binding at these locations may be an artifact of constitutive expression of the sigma factor.
The 5′ ends of transcripts regulated by each sigma factor were determined using 5′ random amplification of cDNA ends (RACE) (11) and RNA collected from P. syringae DC3000 expressing each of the respective sigma factors (Fig. 1). Cells used in this experiment were grown under the same conditions as those used in the ChIP-seq experiments. This confirmed that promoters regulated by these sigma factors are located in regions enriched by ChIP and defines the relationship between the transcription start sites and the annotated initiation codon of the downstream gene. Approximate locations of the −10 and −35 promoter elements were assigned based on their distance from the 5′ end (Fig. 1). These sequence elements are used by sigma factors to locate RNA polymerase on the DNA when transcription is being initiated. There are few clearly defined promoter elements for this sigma factor class because the IS sigma factor regulons are generally small, making it difficult to identify conserved sequences (11, 12, 18). An amalgamated IS sigma-regulated promoter motif presented by Staron et al. (19) suggests that a T-rich −35 region and a conserved TGT in the −10 region are common features of these types of promoters. There is some evidence of a T-rich −35 element upstream of PSPTO_1284, but the −10 elements are quite different from the generalized motif. This is likely due to specialization of the promoters recognized by IS-class sigma factors, a feature that is presumably necessary to avoid cross regulation in organisms that carry genes encoding multiple IS sigma factors.
Fig 1.
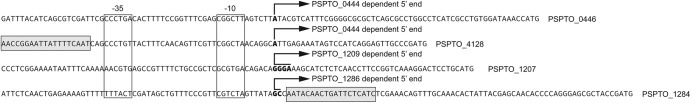
Transcription start sites for PSPTO_0444-, PSPTO_1209-, and PSPTO_1286-regulated genes. Transcript 5′ ends were mapped on the P. syringae DC3000 genome sequence using 5′ RACE as described in reference 11. Potential start sites are indicated with bold type. Multiple boldface nucleotides shown for PSPTO_1207 and PSPTO_1286 indicate uncertainty regarding the position of the 5′ end due technical limitations that prevent mapping the start site to a single nucleotide. The annotated ATG initiation codon is at the 3′ end of each sequence, followed by the gene name. The locations of Fur boxes from reference 13 are indicated in gray shading. PSPTO_0444, PSPTO_1209, and PSPTO_1286 were expressed from pBS161, pBS164, and pBS165, respectively.
Fur box operator sites were found upstream of the TonB-dependent receptor genes regulated by PSPTO_0444 and PSPTO_1286 (Fig. 1) (13). Fur binding at these locations has been observed using ChIP-seq (13), and earlier microarray experiments demonstrated that PSPTO_1284 but not PSPTO_4128 is controlled by iron status (14). The discrepancy in iron-dependent regulation at these locations may be due to the positioning of the Fur box relative to the promoter sequences. Together, these data suggest that the regulatory circuit controlling PSPTO_1284 expression involves Fur-mediated control at two levels: expression of both the sigma factor and the target TonB-dependent receptor gene. It has been proposed that Fur-dependent regulation of TonB-dependent receptor genes is an ancestral regulatory state that has been replaced by IS sigma factor-dependent regulation (20). Accordingly, direct Fur regulation of both sigma factor genes and their downstream regulon targets may represent an intermediate state (20). Alternatively, there may be selective pressures that maintain this more complex arrangement for certain receptors.
Supplementary Material
ACKNOWLEDGMENTS
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
We thank Deepti Thete for pDT7.
Footnotes
Published ahead of print 2 November 2012
Supplemental material for this article may be found at 10.1128/AEM.02801-12.
REFERENCES
- 1. Lindeberg M, Cunnac S, Collmer A. 2009. The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol. Plant Pathol. 10:767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffiere A, Yan S, Dominguez H, Thompson BM. 2008. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2:321–334 [DOI] [PubMed] [Google Scholar]
- 3. Koebnik R. 2005. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13:343–347 [DOI] [PubMed] [Google Scholar]
- 4. Braun V, Mahren S, Sauter A. 2006. Gene regulation by transmembrane signaling. Biometals 19:103–113 [DOI] [PubMed] [Google Scholar]
- 5. Draper RC, Martin LW, Beare PA, Lamont IL. 2011. Differential proteolysis of sigma regulators controls cell-surface signalling in Pseudomonas aeruginosa. Mol. Microbiol. 82:1444–1453 [DOI] [PubMed] [Google Scholar]
- 6. Mettrick KA, Lamont IL. 2009. Different roles for anti-sigma factors in siderophore signalling pathways of Pseudomonas aeruginosa. Mol. Microbiol. 74:1257–1271 [DOI] [PubMed] [Google Scholar]
- 7. Helmann JD. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47–110 [DOI] [PubMed] [Google Scholar]
- 8. Paget MS, Helmann JD. 2003. The σ70 family of sigma factors. Genome Biol. 4:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Daugherty S, Brinkac L, Beanan MJ, Haft DH, Nelson WC, Davidsen T, Zafar N, Zhou LW, Liu J, Yuan QP, Khouri H, Fedorova N, Tran B, Russell D, Berry K, Utterback T, Van Aken SE, Feldblyum TV, D'Ascenzo M, Deng WL, Ramos AR, Alfano JR, Cartinhour S, Chatterjee AK, Delaney TP, Lazarowitz SG, Martin GB, Schneider DJ, Tang XY, Bender CL, White O, Fraser CM, Collmer A. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 100:10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oguiza JA, Kiil K, Ussery DW. 2005. Extracytoplasmic function sigma factors in Pseudomonas syringae. Trends Microbiol. 13:565–568 [DOI] [PubMed] [Google Scholar]
- 11. Markel E, Maciak C, Butcher BG, Myers CR, Stodghill P, Bao Z, Cartinhour S, Swingle B. 2011. An ECF sigma factor mediated cell surface signaling system in Pseudomonas syringae pv. tomato DC3000 regulates gene expression in response to heterologous siderophores. J. Bacteriol. 193:5775–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swingle B, Thete D, Moll M, Myers CR, Schneider DJ, Cartinhour S. 2008. Characterization of the PvdS-regulated promoter motif in Pseudomonas syringae pv. tomato DC3000 reveals regulon members and insights regarding PvdS function in other pseudomonads. Mol. Microbiol. 68:871–889 [DOI] [PubMed] [Google Scholar]
- 13. Butcher BG, Bronstein PA, Myers CR, Stodghill PV, Bolton JJ, Markel EJ, Filiatrault MJ, Swingle B, Gaballa A, Helmann JD, Schneider DJ, Cartinhour SW. 2011. Characterization of the Fur regulon in Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 193:4598–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bronstein PA, Filiatrault MJ, Myers CR, Rutzke M, Schneider DJ, Cartinhour SW. 2008. Global transcriptional responses of Pseudomonas syringae DC3000 to changes in iron bioavailability in vitro. BMC Microbiol. 8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown JS, Gilliland SM, Holden DW. 2001. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol. Microbiol. 40:572–585 [DOI] [PubMed] [Google Scholar]
- 16. Brown JS, Gilliland SM, Ruiz-Albert J, Holden DW. 2002. Characterization of pit, a Streptococcus pneumoniae iron uptake ABC transporter. Infect. Immun. 70:4389–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li C, Stocker R. 2009. Heme oxygenase and iron: from bacteria to humans. Redox Rep. 14:95–101 [DOI] [PubMed] [Google Scholar]
- 18. Enz S, Mahren S, Menzel C, Braun V. 2003. Analysis of the ferric citrate transport gene promoter of Escherichia coli. J. Bacteriol. 185:2387–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Staron A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 74:557–581 [DOI] [PubMed] [Google Scholar]
- 20. Cornelis P, Matthijs S, Van Oeffelen L. 2009. Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22:15–22 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


