Abstract
α-Amylase-binding streptococci (ABS) are a heterogeneous group of commensal oral bacterial species that comprise a significant proportion of dental plaque microfloras. Salivary α-amylase, one of the most abundant proteins in human saliva, binds to the surface of these bacteria via specific surface-exposed α-amylase-binding proteins. The functional significance of α-amylase-binding proteins in oral colonization by streptococci is important for understanding how salivary components influence oral biofilm formation by these important dental plaque species. This review summarizes the results of an extensive series of studies that have sought to define the molecular basis for α-amylase binding to the surface of the bacterium as well as the biological significance of this phenomenon in dental plaque biofilm formation.
INTRODUCTION
It is well established that tooth-borne bacterial biofilm (also known as dental plaque) is the cause of the two most common oral diseases, dental caries and periodontal disease (1). Essential to oral biofilm development is the initial colonization of the oral cavity by the commensal (also known as “normal”) microflora. Humans and their commensal partners have evolved together through history. While most commensal organisms benefit human health, some can also serve as pathogens. For example, following the ingestion of dietary carbohydrate, a shift to an acidogenic, aciduric flora that fosters dental caries results. Further, the administration of broad-spectrum antibiotics that select for antibiotic-resistant bacteria can result in imbalances in the commensal flora that foster opportunistic infections. In many cases, while potential disease-causing bacteria are present, the commensal bacteria, which are typically more abundant, prevent potential pathogens from increasing in numbers that can tip the ecological balance toward a pathogenic state. It is thus imperative to learn details of the biology of commensal bacteria like the oral streptococci to understand the microbial ecologic factors that support a healthy flora and thereby prevent the transition to a pathogenic flora (2).
Oral streptococci are pioneer colonizers of the oral cavity and are abundant in dental plaque (3–5). They are among the first species to colonize the newly erupted tooth and to attach to a salivary pellicle that forms on recently cleaned teeth (6). These bacteria likely facilitate the colonization of the dental biofilm by other species (3). There are many oral streptococcal commensal species, including the viridans group streptococci (Streptococcus mitis, salivarius, and anginosus groups), which are generally benign, and the cariogenic mutans group streptococci (7–9). Many studies have provided considerable evidence that prevalent viridans group streptococci, primarily the mitis and salivarius groups (Table 1), are associated with oral health, while the mutans group streptococci are associated with dental caries (8, 10).
Table 1.
α-Amylase binding in oral streptococcia
| Oral streptococcus group | Streptococcus species | α-Amylase-binding component in supernatant (size[s] in kDa) | α-Amylase binding to bacterial surface | Bacterial binding to surface-bound α-amylase | Reference(s) |
|---|---|---|---|---|---|
| Mitis | S. mitis | + (20, 26, 30, 31, 36, 87) | + | U | 31, 14, 30, 15, 33 |
| S. peroris | U | U | U | ||
| S. oralis | U | U | + | 53 | |
| S. infantis | U/A | U | U | This study | |
| S. gordonii | + (20, 82) | + | + | 31, 14, 30, 15, 32 | |
| S. parasanguinis | + (20, 21, 25, 26, 27, 82, 87) | + | U | 31, 30 | |
| S. sinensis | U | U | U | ||
| S. australis | U/A | U | U | This study | |
| S. sanguinis | U | + | + | 47 | |
| S. cristatus | + (30, 32, 82) | + | U | 31, 14, 30 | |
| Anginosus (Milleri) | S. anginosus | U | + | U | 31 |
| S. constellatus | U | U | U | ||
| S. intermedius | U | U | U | ||
| Salivarius | S. salivarius | + (29) | + | U | 30, 15 |
| S. vestibularis | + (27, 33) | U | U | 30 | |
| Mutans | S. sobrinus | − | U | U | 53 |
| S. mutans | + (65) | U | U | 48 |
+, presence of α-amylase-binding ability in at least some streptococcal strains of the species; −, species not known to bind α-amylase; U, unknown; A, AbpA-like protein sequence is present in the NCBI databases.
Dental plaque formation is a complex process that involves the participation of many salivary components (11). Such interactions mediate adhesion of microbes to surfaces, aggregation of microbes to hinder adhesion, and antimicrobial effects. One such interaction is that between oral streptococci and the abundant salivary protein α-amylase (12, 13). α-Amylase binds to Streptococcus gordonii, Streptococcus mitis, Streptococcus cristatus, Streptococcus parasanguinis, Streptococcus salivarius, and several other species (12–15), collectively referred to as the α-amylase-binding streptococci (ABS) (Table 1). ABS have been found to colonize only animals that are able to secrete α-amylase in their saliva (16). Thus, it is likely that the α-amylase-streptococcus interaction evolved with the host to play an important role in the ability of these bacteria to colonize the oral cavity. A series of studies has characterized α-amylase binding to human ABS, such as S. gordonii, S. parasanguinis, and S. mitis. This review will summarize the existing knowledge of the α-amylase-streptococcus interaction and suggest additional questions to guide directions for future research in this area. It will discuss the characteristics of the interaction of α-amylase with S. gordonii and other streptococcal species, the potential benefits of this interaction for the bacteria, and the effects of α-amylase binding on bacterial gene expression, colonization, biofilm formation, and host adaptation.
SALIVARY α-AMYLASE
α-Amylase is an abundant enzyme produced primarily by serous cells of the parotid gland but also by sublingual, submaxillary, and minor glands. The concentration of α-amylase in human saliva ranges from 0.04 to 0.4 mg/ml and can comprise up to 5% of the total salivary protein (17). The α-amylase concentration in saliva typically increases during food consumption and with increased stress levels (18). Salivary α-amylase is an enzyme of the α-1,4-glucan-4-glucanohydrolase family, catalyzing the hydrolysis of α-1,4-glucosidic bonds of starch, glycogen, and other polysaccharides (19). The initial enzymatic digestion of dietary starch begins in the oral cavity with the release of maltose and maltodextrin, providing an abundant source of carbohydrate for oral bacterial nutrition. Besides its hydrolytic activity, α-amylase adsorbs to tooth enamel (20, 21) and selectively binds to prominent pioneer bacterial colonizers of the oral cavity (16). Thus, considering the abundance of α-amylase in saliva and dental pellicle (22, 23), it has been postulated that this α-amylase-bacterium interaction contributes to the establishment of tooth biofilm.
The three-dimensional structure of salivary α-amylase has been determined (24). α-Amylase consists of 496 amino acids and is present in saliva as both glycosylated (62-kDa) and nonglycosylated (56-kDa) forms. While α-amylase in general is involved in the hydrolysis of long-chain maltooligosaccharides to maltose as the end product, glycosylated salivary α-amylase has been reported to have a higher capacity for converting maltotriose into maltose and glucose (25). However, both glycosylated and nonglycosylated forms of the enzyme bind to ABS (13). Salivary α-amylase is present both as a monomeric protein (26) and as a dimer (27), with calcium and chloride ions essential for its enzymatic function. Binding of α-amylase to bacteria is calcium independent, suggesting a mechanism distinct from enzymatic hydrolysis.
The active site of α-amylase mediates saccharide binding and hydrolysis of starch, with multiple secondary oligosaccharide-binding sites thought to enhance α-amylase affinity to starch granules (26). These secondary oligosaccharide-binding sites have also been shown to play a role in bacterial binding (26). Mutation of aromatic residues in the secondary oligosaccharide-binding sites decreased the affinity of α-amylase binding to S. gordonii (26). Because α-amylase bound to bacteria retains enzymatic activity (26, 28), the bacterium-binding site must be distinct from that involved in enzymatic activity. Mutations in aromatic residues of secondary oligosaccharide-binding sites did not reduce the binding of α-amylase to hydroxylapatite, suggesting other yet-to-be-identified sites mediating this interaction (26).
Some studies suggest that α-amylase dimerizes in saliva and interacts with the glycosylated form of salivary carbonic anhydrase (27). Though colocalization of both enzymes has been reported in human enamel pellicle, the biological significance of this interaction has not been determined (29). Notably, salivary α-amylase bound to the bacterial surface retains ∼60% of its hydrolytic activity, suggesting that the bound enzyme may be dimerized (28).
Salivary α-amylase thus participates in at least three important functions in the oral cavity affecting biofilm: (i) hydrolysis of dietary starch, (ii) binding to the tooth surface, and (iii) binding to oral streptococci. An understanding of the role of this enzyme in oral bacterium-host interactions is required for an understanding of oral biofilm formation.
α-AMYLASE-BINDING PROTEINS (ABPs)
ABS express several proteins that specifically bind α-amylase. In most ABS species, multiple ABPs are found as both low-molecular-weight and high-molecular-weight proteins, ranging from 20 to 87 kDa. However, there is heterogeneity in the size of these components between species (30). In some species, such as Streptococcus vestibularis and S. salivarius, only one lower-molecular-weight ABP is observed. In contrast, S. parasanguinis strain 2156 displays four ABP (30, 31). The sizes of the lower-molecular-weight ABPs between streptococcal species range from 20 kDa to 36 kDa, while the high-molecular-weight ABPs range from 82 kDa to 87 kDa. In a preliminary study, we determined the N-terminal sequences of ABPs from several oral streptococci in order to identify the full-length genes in available databases (see Table S1 in the supplemental material). The N-terminal sequence of each ABP matched the species sequence available in the NCBI database. Only the N-terminal sequence of the 82-kDa protein of S. cristatus strain CR311 did not match any available proteins in the S. cristatus database.
The most-studied ABPs are AbpA (20 kDa) and AbpB (82 kDa) from S. gordonii (30, 32). α-Amylase-binding protein C (AbpC), isolated from S. mitis strain NS51 (36 kDa), also interacts with salivary α-amylase and has been cloned and sequenced (33). AbpC shows no sequence homology or secondary-structure similarity with other ABPs. As genome databases continue to expand, additional ABPs with homology to known ABP have been identified, allowing for further study of the similarity among these proteins and their evolutionary origin. AbpA, AbpB, and AbpC share neither amino acid sequence homology nor similarity in secondary structures, as predicted by protein modeling (see Fig. S1 in the supplemental material).
Interestingly, although AbpA and AbpB of S. gordonii Challis are the predominant and best-studied ABPs, SDS-PAGE analysis of α-amylase precipitates indicates the presence of other as-yet-uncharacterized components that also appear to interact with α-amylase (34). Whether these are unique proteins or proteolytic breakdown products of known proteins remains to be determined. Ultimately, identification of the complete ABP sequences will allow detailed comparison of the proteins, identification of conserved domains, and prediction of secondary structure and may suggest a potential mechanism of their interaction with salivary α-amylase. Further studies are required to identify uncharacterized ABPs, which may provide clues to the functions and mechanisms of bacterium-host interactions of oral streptococci.
α-AMYLASE-BINDING PROTEIN A
The most-studied ABP of S. gordonii CH1, AbpA (20 kDa), is an extracellular cell wall-associated surface protein that is maximally expressed during mid-log phase of bacterial growth (32). AbpA appears to be an essential receptor for α-amylase binding to the bacterial surface. Inactivation of AbpA entirely eliminates α-amylase binding to the bacterium (35). Immunogold electron microscopy studies localized α-amylase binding to the cell division septum and poles on the streptococcal surface (32). Cells in the exponential phase of growth bind more α-amylase than cells in stationary phase. Thus, it appears that AbpA is localized to the nascent cell wall, and as the cell matures, it is shed into the supernatant. Abundant amounts of AbpA protein are present in the supernatants of bacterial cultures. α-Amylase-treated bacteria do not appear to display morphological changes, as observed by electron microscopy (32), which indicates that α-amylase binding does not cause overall perturbation to the bacterial cell surface.
Identification of S. gordonii biofilm determinant genes was conducted by screening biofilm-deficient mutants generated following Tn916 mutagenesis (36). These studies suggested abpA as a potential biofilm determinant gene. The mutants selected in this study were found to have similar growth characteristics. Thus, impairment of biofilm formation was not likely due to growth defects (36). Another study from our laboratory found that an abpA-deficient mutant showed impaired biofilm formation in a standard microtiter biofilm assay as well as in a flow model system containing 25% saliva in distilled water (35). These findings suggest that the AbpA protein modulates adhesion and biofilm formation of oral streptococci in vitro. Not only did mutation of abpA reduce biofilm formation, it also reduced growth of S. gordonii in human saliva (35).
Mutation of abpA also impaired the ability of S. gordonii to grow with starch as the predominant carbon source (35). S. gordonii does not produce extracellular α-amylase (35). Thus, the ability to bind salivary α-amylase to the bacterial surface via AbpA appears critical for bacterial growth from starch (35). It is possible that S. gordonii and other ABS contribute to oral microbial colonization by metabolizing dietary starch and providing nutrition for non-ABS species within the biofilm. In contrast, this interaction would make ABS better competitors against pathogenic species. This, however, has yet to be established and requires further studies using multispecies biofilm models.
In vivo studies of the AbpA mutant strain conducted on pathogen-free Osborne-Mendel rats showed quite contradictory results to those of the in vitro studies described above. Curiously, AbpA mutant strains sometimes colonized teeth to a better extent than did the wild type, especially when the rats were fed a sucrose-rich diet and, to some degree, when they were fed a starch diet (37). Interestingly, the activity of glucosyltransferase G (GtfG), an enzyme thought to promote biofilm formation (38), was greater in the AbpA mutant strains, which may have been responsible for the slightly augmented cariogenicity of the mutant strain (37). Further studies of AbpA confirmed its interaction with GtfG of S. gordonii, in which AbpA was found to form complexes with GtfG and salivary α-amylase (34). However, this interaction was suggested to increase GtfG enzymatic activity (34).
The AbpA-α-amylase complex is also known to increase enzymatic activity of GtfB of S. mutans in vitro (34). To explain this, it was suggested that a conformational change occurs in GtfG following interaction with the α-amylase-AbpA complex. This may result in a change in the structure of the synthesized glucan which has been shown to occur in the presence of starch hydrolysates (34, 39). These findings suggest that the α-amylase-AbpA complex represents an interaction that involves other yet-to-be-determined factors, all of which may modulate bacterial adhesion and colonization.
It is also possible that the α-amylase-streptococcus interaction functions in ways other than in nutrition or adhesion. In order to further investigate the mechanism and significance of α-amylase binding to the bacterial surface via AbpA, studies of gene expression of S. gordonii following the binding of salivary α-amylase were conducted using microarray analysis (40). When the bacterium was cultured in a minimal medium, a total of 33 genes were differentially expressed following exposure to purified salivary α-amylase. The greatest change in expression was observed for genes involved in fatty acid synthesis. Several of these genes were highly upregulated in response to α-amylase binding compared to the control exposure, heat-denatured salivary α-amylase. Not only was gene expression altered when the bacteria were exposed to native α-amylase, there were also changes in bacterial phenotype, as observed by increased bacterial growth, increased resistance to low pH, and increased resistance to the detergent triclosan. Mutation of abpA, which abolishes the binding of α-amylase to the bacterial surface, eliminated the gene responses and phenotype changes, suggesting a role for the AbpA protein in this response. These findings suggest that α-amylase binding to AbpA initiates a signal resulting in differential gene expression. This may serve as an environmental sensing mechanism specific for the oral environment. Identification of other proteins that interact with AbpA will be crucial for understanding this mechanism.
Analysis of the AbpA protein sequence to predict the secondary structure identified several sites that may potentially participate in AbpA-protein interaction and signaling. Two predicted N-myristoylation sites at residues 83 to 88 and 147 to 152 may be involved in protein-protein interaction. A tyrosine phosphorylation site (residues 32 to 39) and an ATP/ADP-binding site (residues 121 to 128) may be involved in phosphorylation and signal transduction. These findings support a possible role for AbpA in environmental surveillance through α-amylase binding and interaction with other components of a putative signaling system to modulate gene expression and adaptation to the host environment.
Furthermore, multiple sequence alignment of amino acid sequences of AbpA-like proteins indicates several highly conserved areas that may mediate α-amylase binding and the interaction with other bacterial proteins (see Fig. S2 in the supplemental material). However, though there is evident homology in AbpA-like proteins, the α-amylase-binding activity of these hypothetical proteins has yet to be experimentally established. In fact, that AbpA-like proteins are conserved among some streptococcal species but not present in all the strains among the same species suggests that AbpA-like proteins likely have a common origin and perhaps were acquired by horizontal gene transfer.
Regulation of AbpA expression in S. gordonii appears to involve at least two mechanisms: catabolite repression in response to glucose and substrate induction via maltose/maltodextrin transport (41, 42). The promoter region of the abpA gene possesses the catabolite responsive element (cre), which is the binding site for LacI/GalR transcriptional regulators. It has been determined that the RegG protein, a LacI/GalR transcriptional regulator and homolog of catabolite control protein A (CcpA), is responsible for the catabolite repressive effect on abpA (42). Mutation of regG in S. gordonii eliminated repression of abpA transcription in the presence of glucose, suggesting that RegG is a transcriptional regulator responsible for repression.
A recent study found that that the products of starch hydrolysis produced from the action of salivary α-amylase, particularly maltose and maltotriose, upregulate AbpA expression in S. gordonii (41). While RegG represses transcription of abpA in the presence of glucose, the identity of the transcriptional regulator that activates the expression of abpA in the presence of maltose/maltodextrin is presently unknown. Interestingly, previous studies showed that maltotriose enhanced the binding of S. gordonii to α-amylase-coated hydroxylapatite (43). Thus, the products of starch hydrolysis by α-amylase on the surface of the streptococcus may increase the expression of AbpA to maximize binding of the host enzyme to the bacterial surface for better utilization of a significant dietary nutrient source or to increase adhesion of the bacterial cell to a host surface. Because starch and salivary α-amylase are abundant in the oral cavity, it seems that AbpA expression would be beneficial for bacterial colonization and proliferation. Multispecies biofilm models of oral colonization would be useful to determine the effect of increased expression of AbpA on overall bacterial survival, bacterial competence, and effects on other oral bacterial species.
α-AMYLASE-BINDING PROTEIN B
The 82-kDa AbpB protein coprecipitates from S. gordonii culture supernatants together with α-amylase, AbpA, and GtfG. Though this protein has affinity for salivary α-amylase, as observed on a Far Western blot (30, 44), it does not share sequence homology with either AbpA or AbpC. Comparative amino acid sequence analysis reveals that AbpB shares homology with other bacterial dipeptidases (44). This protein has been shown to have hydrolytic activity for Ala-Pro, Gly-Pro, and Arg-Pro peptides, a finding which suggests restriction of its enzymatic activity to proline-containing residues (44). This is in accordance with evidence that AbpB cleaves two human proline-rich proteins, namely, type IV collagen and fibrinogen. AbpB does not cleave human salivary α-amylase, which is low in proline. Overall, enzymatic activity of the purified protein was modest, possibly due to the limitations of enzyme assays, absence of yet-unknown cofactors, or structural modification of AbpB (44). Purification of AbpB from S. gordonii revealed additional peptides of 52, 36, and 26 kDa to interact with anti-AbpB antibody. It has not yet been established if these peptides are products of AbpB self-hydrolysis or if they are cross-reactive peptides encoded by other abpB-like genes (44). As with AbpA and α-amylase, AbpB coprecipitated with GtfG of S. gordonii and enhanced the transferase and sucrase activity of GtfG (44). Also, as demonstrated by an enzyme-linked immunosorbent assay (ELISA), the affinity of AbpB and AbpA toward GtfG appeared greater than that between α-amylase and GtfG (34, 44).
AbpB is an extracellular secreted protein, and it is not responsible for α-amylase binding to the bacterial surface (44, 45). Mutation of abpB did not abolish α-amylase binding to the S. gordonii surface, whereas mutation of abpA resulted in no α-amylase binding to S. gordonii (45). In vivo studies showed that abpB mutants of S. gordonii colonize the oral cavity of pathogen-free rats fed by starch-supplemented diet less well. However, colonization of the abpB mutant was rescued by a sucrose-supplemented diet (37). These results suggest that AbpB appears important to bacterial colonization of the oral cavity, perhaps as part of nutrient acquisition pathways. It is possible that salivary proline-rich proteins, which are abundant in dental plaque, may serve as substrates for AbpB peptidase activity. AbpB also shares homology with streptococcal lipoproteins, suggesting its potential to be involved in activities characteristic of bacterial lipoproteins, including sensory signaling, cell growth and division, and nutrient uptake.
α-AMYLASE-BINDING PROTEIN C
A 36-kDa α-amylase-binding protein isolated from culture supernatants of S. mitis NS51 was sequenced, cloned, and designated AbpC (31, 33). A similar protein was also found in S. mitis K208 but not in S. mitis OP51, OS51, and NCTC 10712 (30). While the molecular weight of this component suggests that it is related to AbpA of S. gordonii, the amino acid sequence of AbpC does not share homology with either AbpA or AbpB, suggesting that it has a unique origin. Dot plot alignment of AbpA and AbpC (see Fig. S3 in the supplemental material) shows some similarity in amino acid sequence between these components and other ABPs in the middle region of the proteins, which may account for their α-amylase-binding ability. Interestingly, neither AbpA nor AbpB was reported in S. mitis strains that encode AbpC (30, 31, 33, 46). The C terminus of the AbpC protein has two conserved domains: one is a pfam01473 choline-containing cell wall-binding domain (CW repeats) and the other is a COG5263 glucan-binding domain (YG repeat). This suggests that AbpC may be associated with the bacterial cell wall. However, the role of AbpC as an α-amylase receptor has not yet been definitively established. The presence of two conserved domains in AbpC demonstrates homology with other streptococcal choline-binding proteins and glycosyltransferases.
OTHER α-AMYLASE-STREPTOCOCCUS INTERACTIONS
A recent study of S. sanguinis SK36 demonstrated a pilus-like structure, similar to those found on the pathogenic streptococci Streptococcus pyogenes (group A), Streptococcus agalactiae (group B), and Streptococcus pneumoniae, that binds to surface-adsorbed salivary α-amylase to facilitate biofilm formation on saliva-coated surfaces (47). A pilus-deficient mutant of this strain failed to form multilayered biofilm structures when grown on noncoated and saliva-coated surfaces (47). Out of the three structural proteins of streptococcal pili, PilC (36 kDa) and PilB (36 kDa) had the highest α-amylase-binding capacity while PilA binding was low. The C terminus of PilC, thought to contain a collagen-binding domain, interacted with α-amylase, although binding of collagen was not detected. While mutation of all three pili proteins significantly reduced binding to immobilized α-amylase, this reduction was limited and α-amylase was still able to interact with the bacteria to some degree. This suggests that other yet-unidentified bacterial components in addition to pili that are responsible for α-amylase binding to the bacterial surface are present on the surface of S. sanguinis. The interaction of pili proteins with salivary α-amylase must be structurally distinct from that of conventional ABPs, since the ABP-binding site in α-amylase partially overlaps with its sugar-binding site and the secondary structure of the enzyme is important for this interaction. In the case of PilC, disruption of the secondary structure of the α-amylase enzyme by heat and SDS did not abolish the binding of biotinylated PilC to α-amylase in a ligand blot analysis. Furthermore, PilC and PilB also do not share homology with AbpA or AbpB. These findings suggest that the mechanism of PilC binding to surface-immobilized salivary α-amylase is distinct from that of ABP.
Another interaction was described between α-amylase and S. mutans strain A32-2 fimbria-associated surface proteins (48). A purified 65-kDa fimbrial protein was shown to bind to saliva-coated surfaces, with α-amylase as the major receptor. Although it is possible that S. mutans binds salivary α-amylase via its surface fimbriae, this has not been established experimentally. Interestingly, previous studies (12, 15, 31) have not identified α-amylase-binding mutans group streptococci. In our previous work (16, 49), in which isolates from dental plaque were screened for soluble α-amylase binding to bacterial surfaces, none of the α-amylase-binding strains were identified as mutans group streptococci. This may be explained by the possibility that S. mutans can bind surface-absorbed α-amylase but not soluble α-amylase and can use protein segments that are unordered or present in the coiled regions of the structure.
BIOLOGICAL SIGNIFICANCE OF α-AMYLASE BINDING TO ORAL STREPTOCOCCI
Several assumptions can be made regarding the function of α-amylase-streptococcus interaction on the basis of the available evidence. It is likely that host salivary α-amylase plays a multifaceted role in the physiology and colonization process of oral streptococci. Some functions may include, but are not limited to, adhesion of streptococci to the salivary pellicle, interbacterial adhesion and interaction within the dental plaque, nutrition of oral streptococci and other coinhabitants of dental plaque, and/or clearance of bacteria from the oral cavity by inhibition of adhesion and/or by interaction of the bacteria and dietary starch granules that are swallowed.
α-AMYLASE BINDING AND ABS ADHESION TO TEETH
Because salivary α-amylase is an abundant constituent of the dental pellicle, α-amylase adsorbed to the tooth surface may provide a receptor for ABS adhesion. The structure of this receptor differs from that of the site influencing adsorption of the enzyme to hydroxylapatite (26, 28). Several in vitro studies found that adhesion of ABS to hydroxylapatite-bound α-amylase contributed to biofilm formation and that mutation of AbpA resulted in impaired biofilm formation by S. gordonii (35, 36). Although there is evidence for the role of α-amylase-streptococcus interaction in biofilm formation in vitro, the results of in vivo studies did not support this idea (37). However, the α-amylase-streptococcus interaction is likely one of many redundant bacterial host interactions that together influence bacterial colonization.
An interesting observation that also requires further study is the unique localization of AbpA on the bacterial surface and its shedding into the culture medium. What is the function of AbpA localization to regions of cellular division and polar zones? What controls its localization on the cell wall, and what triggers its shedding into the environment? Is AbpA shed when grown exclusively in saliva? Perhaps the timing of AbpA expression on the surface at mid-log phase (32) increases α-amylase binding in the signal-enhanced fatty acid synthesis, which, in turn, fosters increased membrane synthesis (41). It is also possible that the increased expression of AbpA in the presence of starch and α-amylase is important for S. gordonii biofilm establishment. The presence of maltose and maltooligosaccharides increases expression of AbpA in S. gordonii (41), which may help explain the observation that the presence of maltotriose increased the adhesion of S. gordonii to α-amylase-coated hydroxylapatite (43). However, it is not yet clear what this effect would have on the S. gordonii overall oral colonization.
α-AMYLASE BINDING IN INTERBACTERIAL INTERACTIONS WITHIN ORAL BIOFILMS
The α-amylase-streptococcus interaction may foster the adhesion of different bacterial species to each other. It is possible for dimerized α-amylase in saliva (27) to provide two bacterial interaction sites and thus provide a bridge between two bacterial surfaces. It is possible for α-amylase to mediate such interactions via the interaction of α-amylase with multiple bacterial proteins, for example, AbpA, AbpB, and GtfG. Such protein complexes may act as interbacterial bridges connecting bacterial cells to each other. The presence in these complexes of α-amylase and GtfG in the presence of starch may provide a substrate for synthesis of biofilm matrix exopolysaccharides.
α-AMYLASE IN NUTRITION OF S. GORDONII AND OTHER ORAL BACTERIAL SPECIES
ABS likely benefit from capturing α-amylase activity to enable utilization of carbohydrates from the human diet as a consequence of starch hydrolysis. Considering the fact that most oral streptococci do not secrete α-amylase into the milieu and are not able to hydrolyze extracellular starch, binding of host salivary α-amylase allows the bacteria to access a rich nutrient source from the human diet. Furthermore, ABS would likely have a growth advantage over organisms that lack the ability to bind α-amylase in a mixed-species environment containing starch (35). It is also possible that maltose resulting from starch hydrolysis on the ABS surface can “cross-feed” other non-ABS species in close proximity to ABS. In addition, the lactic acid metabolite produced by S. gordonii may, in turn, be used as a nutrient for other dental plaque species, such as Aggregatibacter actinomycetemcomitans (50).
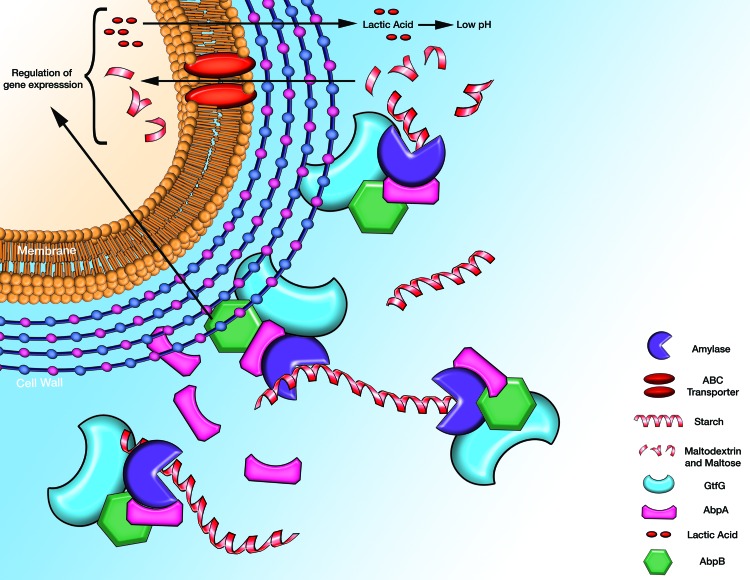
Another potential mechanism by which α-amylase may modulate bacterial nutrition is through interaction between α-amylase AbpA, GtfG, and AbpB (Fig. 1). These proteins together may play a role in nutrient acquisition whereby the dipeptidase activity of AbpB can assist in the peptide hydrolysis of glycoproteins and proline-rich proteins and GtfG can assist in carbohydrate utilization.
Fig 1.
Schematic model of the localization and function of the α-amylase-binding proteins in S. gordonii. AbpA is bound to the cell wall and then released. It interacts with α-amylase, AbpB, and GtfG to form a complex. α-Amylase remains enzymatically active within the complex to allow hydrolysis of dietary starch to maltose and maltooligosaccharides, which are transported into the cell via an ABC transporter. These carbohydrates are ultimately metabolized for energy production, with lactic acid as an end product. Gene expression is influenced by metabolic end products as well as by α-amylase itself through its interaction with AbpA on the bacterial surface.
α-AMYLASE BINDING IN BACTERIAL CLEARANCE
Another possible interesting function of the α-amylase-streptococcus interaction is to mediate the attachment of the bacteria to dietary starch granules. This may provide protection from the host response for the bacteria within the starch granule structure (51, 52). Such a phenomenon has been observed in the case of probiotic bacteria that enter the intestines from the oral cavity and manage to avoid destruction by the highly acidic gastric environment through “hiding” in starch granules until they transit to a less-severe environment (52). Because α-amylase is specific for α-1,4-glucosidic bonds present in starch, glycogen, and other polysaccharides (26), such structures present in the oral environment may provide a substrate for attachment of α-amylase-binding streptococci. Thus, the oral bacteria may have evolved the mechanism to “hitchhike” along with the human enzyme in order to localize within the most nutrient-rich environment.
ENVIRONMENTAL SIGNALING THROUGH α-AMYLASE BINDING AND PHENOTYPE ADJUSTMENT
α-Amylase and AbpA may play a role in signaling that results in differential gene expression after binding of α-amylase (40). Though it is still unclear how this is achieved and to what extent other proteins are involved, the observation of differential gene regulation following α-amylase binding implies that the bacteria may exploit this interaction. Thus, the genes responsible for initiation of fatty acid synthesis are upregulated following α-amylase binding, which may help to explain the stimulation of bacterial growth and proliferation (Fig. 1). Interestingly, upon binding of salivary α-amylase to bacteria, the cells appear to adjust their phenotype to become more resistant to an acidic environment. This adjustment may anticipate the increase in metabolites from carbohydrate utilization provided by the action of α-amylase or prepare the bacterium to survive the highly acidic gastric environment following attachment to starch granules (52).
CONCLUSION AND FURTHER QUESTIONS
The binding of α-amylase to the surface of oral streptococci is an example of a bacterium-host interaction that likely influences commensal bacterial colonization. This notion is supported by the fact that ABS colonize only the oral cavity of species with active salivary α-amylase. It is likely that the acquisition of α-amylase binding by oral streptococci was selected through evolutionary mechanisms to contribute to bacterial survival in the host. Further questions remain. What role does α-amylase binding to AbpA play in bacterial gene expression? How does the effect of amylase on S. gordonii gene expression compare with the effects induced by other salivary proteins? Does AbpA serve as a component of a unique signal transduction system? How are the unique sequences of ABP from different streptococcal species explained? Do ABP from different species share an amylase-binding motif? Answers to these questions will improve understanding of such bacterium-host interactions. This information may one day be applied to manipulate bacterial colonization in an attempt to prevent and control infectious disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank James Malone of the Department of Oral Biology, University at Buffalo, for assistance with protein modeling.
This work was supported in part by grants from the National Institute of Dental and Craniofacial Research (DE09838, DE022673, and DE007034).
Biographies

Anna Nikitkova received her D.D.S. from Pavlov's State Medical University, St. Petersburg, Russia, and her Ph.D. in oral biology from the University at Buffalo. Her research interests focus on pioneer oral colonizers, including molecular mechanisms of bacterium-host interactions and the role of salivary components in metabolic gene regulation of oral bacteria.

Elaine Haase received her B.S. and M.S. in medical technology from the University at Buffalo, SUNY, and worked for several years as a medical technologist at Mt. St. Mary's Hospital in Lewiston, NY. She returned to the University at Buffalo and earned a Ph.D. in microbiology working in the laboratory of Tim Murphy on the P2 protein of Haemophilus influenzae. Her postdoctoral training with Frank Scannapieco began with studies of the amylase-binding protein A of Streptococcus gordonii. She has also identified and characterized components of the flp fimbrial operon of Aggregatibacter actinomycetemcomitans. Since then, as Research Associate Professor, she has performed studies on the molecular epidemiology of respiratory pathogens from the oral cavity, biofilms, and transcriptional regulation of the flp operon and the role of small RNAs (sRNAs) in A. actinomycetemcomitans pathogenesis.

Frank Scannapieco is Professor and Chair of the Department of Oral Biology, School of Dental Medicine, State University of New York at Buffalo. He received a B.S. in biology from the University of Connecticut, an M.S. in biology from Northeastern University, a D.M.D. from the University of Connecticut, and a Certificate in Periodontology and Ph.D. in oral biology, both from the University at Buffalo. His research focuses on oral microbial ecology, including molecular mechanisms of oral bacterial colonization, the role of saliva in oral microbial colonization, and the interface between oral and systemic health, especially the role of oral conditions in the process of respiratory infection.
Footnotes
Published ahead of print 9 November 2012
Supplemental material for this article may be found at 10.1128/AEM.02581-12.
REFERENCES
- 1. Loesche W. 2007. Dental caries and periodontitis: contrasting two infections that have medical implications. Infect. Dis. Clin. North Am. 21:471–502 [DOI] [PubMed] [Google Scholar]
- 2. Avila M, Ojcius DM, Yilmaz O. 2009. The oral microbiota: living with a permanent guest. DNA Cell Biol. 28:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413–437 [DOI] [PubMed] [Google Scholar]
- 4. Nyvad B, Kilian M. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369–380 [DOI] [PubMed] [Google Scholar]
- 5. Nyvad B, Kilian M. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24:267–272 [DOI] [PubMed] [Google Scholar]
- 6. Rosan B, Lamont RJ. 2000. Dental plaque formation. Microbes Infect. 2:1599–1607 [DOI] [PubMed] [Google Scholar]
- 7. Banas JA. 2004. Virulence properties of Streptococcus mutans. Front. Biosci. 9:1267–1277 [DOI] [PubMed] [Google Scholar]
- 8. Kuramitsu HK. 2003. Molecular genetic analysis of the virulence of oral bacterial pathogens: an historical perspective. Crit. Rev. Oral Biol. Med. 14:331–344 [DOI] [PubMed] [Google Scholar]
- 9. Whitmore SE, Lamont RJ. 2011. The pathogenic persona of community-associated oral streptococci. Mol. Microbiol. 81:305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanzer JM, Livingston J, Thompson AM. 2001. The microbiology of primary dental caries in humans. J. Dent. Educ. 65:1028–1037 [PubMed] [Google Scholar]
- 11. Scannapieco FA. 1994. Saliva-bacterium interactions in oral microbial ecology. Crit. Rev. Oral Biol. Med. 5:203–248 [DOI] [PubMed] [Google Scholar]
- 12. Douglas CW. 1983. The binding of human salivary α-amylase by oral strains of streptococcal bacteria. Arch. Oral Biol. 28:567–573 [DOI] [PubMed] [Google Scholar]
- 13. Scannapieco FA, Bergey EJ, Reddy MS, Levine MJ. 1989. Characterization of salivary α-amylase binding to Streptococcus sanguis. Infect. Immun. 57:2853–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Douglas CW, Pease AA, Whiley RA. 1990. Amylase-binding as a discriminator among oral streptococci. FEMS Microbiol. Lett. 54:193–197 [DOI] [PubMed] [Google Scholar]
- 15. Kilian M, Nyvad B. 1990. Ability to bind salivary α-amylase discriminates certain viridans group streptococcal species. J. Clin. Microbiol. 28:2576–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scannapieco FA, Solomon L, Wadenya RO. 1994. Emergence in human dental plaque and host distribution of amylase-binding streptococci. J. Dent. Res. 73:1627–1635 [DOI] [PubMed] [Google Scholar]
- 17. Jacobsen N, Melvaer KL, Hensten-Pettersen A. 1972. Some properties of salivary amylase: a survey of the literature and some observations. J. Dent. Res. 51:381–388 [DOI] [PubMed] [Google Scholar]
- 18. Rohleder N, Wolf JM, Maldonado EF, Kirschbaum C. 2006. The psychosocial stress-induced increase in salivary α-amylase is independent of saliva flow rate. Psychophysiology 43:645–652 [DOI] [PubMed] [Google Scholar]
- 19. Scannapieco FA, Torres G, Levine MJ. 1993. Salivary α-amylase: role in dental plaque and caries formation. Crit. Rev. Oral Biol. Med. 4:301–307 [DOI] [PubMed] [Google Scholar]
- 20. Al-Hashimi I, Levine MJ. 1989. Characterization of in vivo salivary-derived enamel pellicle. Arch. Oral Biol. 34:289–295 [DOI] [PubMed] [Google Scholar]
- 21. Orstavik D, Kraus FW. 1973. The acquired pellicle: immunofluorescent demonstration of specific proteins. J. Oral Path. 2:68–76 [DOI] [PubMed] [Google Scholar]
- 22. Jensen JL, Lamkin MS, Oppenheim FG. 1992. Adsorption of human salivary proteins to hydroxyapatite: a comparison between whole saliva and glandular salivary secretions. J. Dent. Res. 71:1569–1576 [DOI] [PubMed] [Google Scholar]
- 23. Yao Y, Grogan J, Zehnder M, Lendenmann U, Nam B, Wu Z, Costello CE, Oppenheim FG. 2001. Compositional analysis of human acquired enamel pellicle by mass spectrometry. Arch. Oral Biol. 46:293–303 [DOI] [PubMed] [Google Scholar]
- 24. Ramasubbu N, Paloth V, Luo Y, Brayer GD, Levine MJ. 1996. The structure of human salivary α-amylase at 1.6Å resolution: implications for its role in the oral cavity. Acta Crystallogr. D Biol. Crystallogr. 52:435–446 [DOI] [PubMed] [Google Scholar]
- 25. Koyama I, Komine S, Yakushijin M, Hokari S, Komoda T. 2000. Glycosylated salivary α-amylases are capable of maltotriose hydrolysis and glucose formation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 126:553–560 [DOI] [PubMed] [Google Scholar]
- 26. Ragunath C, Manuel SG, Venkataraman V, Sait HB, Kasinathan C, Ramasubbu N. 2008. Probing the role of aromatic residues at the secondary saccharide-binding sites of human salivary α-amylase in substrate hydrolysis and bacterial binding. J. Mol. Biol. 384:1232–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fisher SZ, Govindasamy L, Tu C, Agbandje-McKenna M, Silverman DN, Rajaniemi HJ, McKenna R. 2006. Structure of human salivary α-amylase crystallized in a C-centered monoclinic space group. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62:88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scannapieco FA, Bhandary K, Ramasubbu N, Levine MJ. 1990. Structural relationship between the enzymatic and streptococcal binding sites of human salivary α-amylase. Biochem. Biophys. Res. Commun. 173:1109–1115 [DOI] [PubMed] [Google Scholar]
- 29. Li J, Helmerhorst EJ, Troxler RF, Oppenheim FG. 2004. Identification of in vivo pellicle constituents by analysis of serum immune responses. J. Dent. Res. 83:60–64 [DOI] [PubMed] [Google Scholar]
- 30. Gwynn JP, Douglas CW. 1994. Comparison of amylase-binding proteins in oral streptococci. FEMS Microbiol. Lett. 124:373–379 [DOI] [PubMed] [Google Scholar]
- 31. Brown AE, Rogers JD, Haase EM, Zelasko PM, Scannapieco FA. 1999. Prevalence of the amylase-binding protein A gene (abpA) in oral streptococci. J. Clin. Microbiol. 37:4081–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scannapieco FA, Haraszthy GG, Cho MI, Levine MJ. 1992. Characterization of an amylase-binding component of Streptococcus gordonii G9B. Infect. Immun. 60:4726–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vorrasi J, Chaudhuri B, Haase EM, Scannapieco FA. 2010. Identification and characterization of amylase-binding protein C from Streptococcus mitis NS51. Mol. Oral Microbiol. 25:150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chaudhuri B, Rojek J, Vickerman MM, Tanzer JM, Scannapieco FA. 2007. Interaction of salivary α-amylase and amylase-binding-protein A (AbpA) of Streptococcus gordonii with glucosyltransferase of S. gordonii and Streptococcus mutans. BMC Microbiol. 7:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rogers JD, Palmer RJ, Jr, Kolenbrander PE, Scannapieco FA. 2001. Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation. Infect. Immun. 69:7046–7056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Loo CY, Corliss DA, Ganeshkumar N. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanzer JM, Grant L, Thompson A, Li L, Rogers JD, Haase EM, Scannapieco FA. 2003. Amylase-binding proteins A (AbpA) and B (AbpB) differentially affect colonization of rats' teeth by Streptococcus gordonii. Microbiology 149:2653–2660 [DOI] [PubMed] [Google Scholar]
- 38. Vickerman MM, Clewell DB, Jones GW. 1991. Sucrose-promoted accumulation of growing glucosyltransferase variants of Streptococcus gordonii on hydroxyapatite surfaces. Infect. Immun. 59:3523–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vacca-Smith AM, Venkitaraman AR, Quivey RG, Jr, Bowen WH. 1996. Interactions of streptococcal glucosyltransferases with α-amylase and starch on the surface of saliva-coated hydroxyapatite. Arch. Oral Biol. 41:291–298 [DOI] [PubMed] [Google Scholar]
- 40. Nikitkova AE, Haase EM, Vickerman MM, Gill SR, Scannapieco FA. 2012. Response of fatty acid synthesis genes to the binding of human salivary amylase by Streptococcus gordonii. Appl. Environ. Microbiol. 78:1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nikitkova AE, Haase EM, Scannapieco FA. 2012. Effect of starch and amylase on the expression of amylase-binding protein A in Streptococcus gordonii. Mol. Oral Microbiol. 27:284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rogers JD, Scannapieco FA. 2001. Catabolite repression and regulation of the amylase-binding protein gene of Streptococcus gordonii. J. Bacteriol. 183:3521–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scannapieco FA, Torres GI, Levine MJ. 1995. Salivary amylase promotes adhesion of oral streptococci to hydroxyapatite. J. Dent. Res. 74:1360–1366 [DOI] [PubMed] [Google Scholar]
- 44. Chaudhuri B, Paju S, Haase EM, Vickerman MM, Tanzer JM, Scannapieco FA. 2008. Amylase-binding protein B of Streptococcus gordonii is an extracellular dipeptidyl-peptidase. Infect. Immun. 76:4530–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li L, Tanzer JM, Scannapieco FA. 2002. Identification and analysis of the amylase-binding protein B (AbpB) and gene (abpB) from Streptococcus gordonii. FEMS Microbiol. Lett. 212:151–157 [DOI] [PubMed] [Google Scholar]
- 46. Douglas CW. 1990. Characterization of the α-amylase receptor of Streptococcus gordonii NCTC 7868. J. Dent. Res. 69:1746–1752 [DOI] [PubMed] [Google Scholar]
- 47. Okahashi N, Nakata M, Terao Y, Isoda R, Sakurai A, Sumitomo T, Yamaguchi M, Kimura RK, Oiki E, Kawabata S, Ooshima T. 2011. Pili of oral Streptococcus sanguinis bind to salivary amylase and promote the biofilm formation. Microb. Pathog. 50:148–154 [DOI] [PubMed] [Google Scholar]
- 48. Ray CA, Gfell LE, Buller TL, Gregory RL. 1999. Interactions of Streptococcus mutans fimbria-associated surface proteins with salivary components. Clin. Diagn. Lab. Immunol. 6:400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tseng CC, Scannapieco FA, Levine MJ. 1992. Use of a replica-plate assay for the rapid assessment of salivary protein-bacteria interactions. Oral Microbiol. Immunol. 7:53–56 [DOI] [PubMed] [Google Scholar]
- 50. Ramsey MM, Rumbaugh KP, Whiteley M. 2011. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 7:e1002012 doi:10.1371/journal.ppat.1002012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Topping DL, Clifton PM. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81:1031–1064 [DOI] [PubMed] [Google Scholar]
- 52. Topping DL, Fukushima M, Bird AR. 2003. Resistant starch as a prebiotic and synbiotic: state of the art. Proc. Nutr. Soc. 62:171–176 [DOI] [PubMed] [Google Scholar]
- 53. Murray PA, Prakobphol A, Lee T, Hoover CI, Fisher SJ. 1992. Adherence of oral streptococci to salivary glycoproteins. Infect. Immun. 60:31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.