Abstract
The ability of Pseudomonas syringae pv. syringae to use nitrate as a nitrogen source in culture and on leaves was assessed. Substantial amounts of leaf surface nitrate were detected directly and by use of a bioreporter of nitrate on bean plants grown with a variety of nitrogen sources. While a nitrate reductase mutant, P. syringae ΔnasB, exhibited greatly reduced growth in culture with nitrate as the sole nitrogen source, it exhibited population sizes similar to those of the wild-type strain on leaves. However, the growth of the ΔnasB mutant was much less than that of the wild-type strain when cultured in bean leaf washings supplemented with glucose, suggesting that P. syringae experiences primarily carbon-limited and only secondarily nitrogen-limited growth on bean leaves. Only a small proportion of the cells of a green fluorescent protein (GFP)-based P. syringae nitrate reductase bioreporter, LK2(pOTNas4), exhibited fluorescence on leaves. This suggests that only a subset of cells experience high nitrate levels or that nitrate assimilation is repressed by the presence of ammonium or other nitrogenous compounds in many leaf locations. While only a subpopulation of P. syringae consumes nitrate at a given time on the leaves, the ability of those cells to consume this resource would be strongly beneficial to those cells, especially in environments in which nitrate is the most abundant form of nitrogen.
INTRODUCTION
Nitrate assimilation is a central metabolic process that contributes to bacterial growth in a variety of habitats such as the ocean (1), rhizosphere (2), and soils (3). While there are numerous studies of bacterial nitrate assimilation in such locations, very little is known about the importance of bacterial nitrate assimilation in other habitats such as the aerial portions of plants known as the phyllosphere. The phyllosphere is considered to be a relatively stressful habitat due to nutrient limitation, desiccation stress, low and variable water availability, and high fluxes of UV radiation; nonetheless, it is colonized by specialized microbes (4). The fitness of epiphytic bacteria is associated with their ability to tolerate or avoid these various stresses and to utilize the limited nutrients available to them on leaves.
Various studies have suggested that the phyllosphere is generally a nutrient-limited habitat. A wide variety of both inorganic and organic compounds are found on a given plant species. Amino acids, organic acids, and carbohydrates were detected in all of the species tested, although the quantity and abundance of a given compound varied from species to species (5, 6). Inorganic nitrogen is also found on the surface of plants, but its form has generally not been determined. Many factors, such as leaf age and the hydrophobic properties of the leaf, affect the amount and composition of leaf leachates. Younger leaves are typically more hydrophobic than older leaves, and their thicker cuticle restricts nutrient flux. Leaves that are more wettable, such as those of beans (Phaseolus vulgaris), experience more leaching (5–7). The leaching of nutrients onto the leaf surface enables the growth of bacterial epiphytes, which grow on the leaf surface and then compete for consumption of these limited resources in this habitat (8).
Despite the fact that a variety of metabolites are known to leach onto the leaf surface, overall carbon and nitrogen availability limits bacterial multiplication. The bacterial plant pathogen Pseudomonas syringae exhibits carbon-limited growth on potato plants grown under both greenhouse and field conditions (9), since growth of this species was stimulated only when exogenous glucose, and not ammonium ions, was added to leaves. Nitrogen appeared to be a secondarily limiting resource on the leaf surface, since more growth of P. syringae was seen when both carbon and nitrogen were added to leaves than when only carbon compounds were added. It is likely that the carbon-to-nitrogen ratio of leaf surface exudates varies between plant species and is especially influenced by plant growth conditions. The carbon or nitrogen limitation that epiphytes might experience is thus probably very context dependent. Indeed, Salmonella enterica and Escherichia coli encounter nitrogen-limited and not carbon-limited growth on lettuce (10). Thus, a better understanding of the processes by which bacteria acquire needed nitrogen on leaves is of importance in understanding the population dynamics of these important microbes.
Much of our understanding of phyllosphere microbiology has come from the study of P. syringae. This plant-pathogenic bacterium is nearly ubiquitous on plants and is also found in other habitats such as many aqueous environments (11, 12). There has been considerable interest in understanding the adaptations that make this species such a successful epiphyte. In one such approach to identifying fitness factors, Marco et al. identified promoters in P. syringae that were induced in cells on leaf surfaces that were presumed to contribute to the growth of the bacteria on the plant (18). One such promoter, PIG 17.1, was localized to a region that was annotated to be putatively involved in nitrate assimilation, suggesting that nitrate utilization was operative on leaf surfaces and that nitrate utilization contributes to the fitness of this epiphyte. Here we explore the ability of P. syringae to assimilate nitrate and address both the levels of nitrate found on leaves and the role of the assimilatory nitrate reductase of P. syringae in determining the population size that it can attain on leaves. We provide evidence that at the population level, nitrogen compounds are only secondarily limiting on bean leaves under our growth conditions but that nitrate reductase is expressed in a subset of the cells on leaves, suggesting that the relative amounts of nitrate and other nitrogenous compounds are spatially heterogeneous but that nitrate is locally a substantial form of nitrogen available to epiphytes at the sites where the nitrate reductase genes are reported to be induced. We conclude that while the majority of the cells in the P. syringae population on bean leaves consume preferred nitrogen forms such as amino acids and ammonium, there is a subpopulation that assimilates nitrate, and that nitrate is a significant source of nitrogen to at least some cells in the population.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Pseudomonas syringae pv. syringae B728a is a spontaneous rifampin-resistant mutant (13) isolated from a bean leaf in Wisconsin. LK2 is a derivative of B728a containing an insertion of the kanamycin gene into the gene conferring ice nucleation, inaB (14). Enterobacter cloacae EcCT501R is a spontaneous rifampin-resistant mutant of strain EcCT501 (15). Unless stated otherwise, all P. syringae strains were grown in King's B (KB) broth overnight or M9 minimal medium containing 0.4% glucose and 0.1% NH4Cl for 2 days at 28°C. All E. coli and E. cloacae strains were grown in Luria-Bertani (LB) broth at 37°C. Antibiotics and fungicides were used at the following concentrations (μg/ml): rifampin, 100; kanamycin, 50; tetracycline, 15; spectinomycin, 20; streptomycin, 20; and natamycin, 21.6.
Construction of P. syringae ΔnasB.
B728a chromosomal DNA was isolated using the DNeasy blood and tissue kit (Qiagen). The putative nasB gene, locus tag Psyr_2099, was PCR amplified with primers crt2099KO_F and 2099KO_R using Pfu DNA polymerase (Agilent Technologies) according to the manufacturer's instructions (Tables 1 and 2). The 978-bp fragment was subcloned into pENTR/D-TOPO (Invitrogen) and transformed into TOP10 cells, and the transformants were screened for the insert by PCR analysis. pENTR/D-TOPO contains recombination sequences flanking the cloned insert that allow the quick and efficient integration of the insert into the destination vector, pLVC-D, which contains the Gateway Cassette (Invitrogen). A Gateway LR Clonase II reaction (Invitrogen) was performed on pENTR-D+nasB and pLVC-D, and the reaction was transformed into TOP10 cells. The transformants were screened by PCR to confirm the proper structure. The resulting positive transformant was used in a triparental mating with B278a and the helper strain pRK2013 (Clonetech).
Table 1.
Strains and plasmids used
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| Pseudomonas syringae pv. syringae | ||
| B728a | Spontaneous Rifr mutant | |
| B728a ΔnasB | B728a with a nasB gene (psyr_2099) disrupted by the insertion of pLVC-D; Rifr Tetr | This study |
| LK2 | B728a containing a disruption of inaB (ΔinaB); Rifr Kanr | 14 |
| LK2(pPROBE-SK) | ΔinaB harboring the pPROBE-SK plasmid; Rifr Kanr | |
| LK2(pSKNas1) | ΔinaB harboring the pSKNas1 plasmid; Rifr Kanr | This study |
| LK2(pSKNas4) | ΔinaB harboring the pSKNas4 plasmid; Rifr Kanr | This study |
| B728a(pOTNas4) | B728a with pOTNas4; Rifr Stm/Spcr | This study |
| Other species | ||
| EcCT501R(pNICE) | Spontaneous Rifr mutant of Enterobacter cloacae harboring the pNICE plasmid; Rifr Kanr | 2 |
| HB101(pRK2013) | Escherichia coli helper strain harboring the plasmid pRK2013; Kanr | Clontech |
| Plasmids | ||
| pENTR-D | Subcloning vector; Kanr | Invitrogen |
| pENTR-D+nasB | pENTR-D plus 978-bp fragment of nasB; Kanr | This study |
| pLVC-D | Suicide vector containing the Gateway Cassette; Tetr | 16 |
| pLVC-D+nasB | pLVC-D plus 978-bp fragment of nasB; Tetr | This study |
| pRK2013 | Triparental helper plasmid; Kanr | Clontech |
| pPROBE-SK | Broad-host-range plasmid with a transcriptional fusion cassette containing a promoterless inaZ reporter gene; Sp/Stmr | 16 |
| pCR-Blunt II-TOPO | Subcloning vector; Kanr | Invitrogen |
| pBLUNT+Nas1 | pCR-Blunt II-TOPO plus region 1 of the nitrate/nitrite reductase promoter Kanr | This study |
| pBLUNT+Nas4 | pCR-Blunt II-TOPO plus region 4 of the nitrate/nitrite reductase promoter Kanr | This study |
| pSKNas1 | pPROBE-SK plus region 1 of the nitrate/nitrite reductase promoter; Sp/Stmr | This study |
| pSKNas4 | pPROBE-SK plus region 4 of the nitrate/nitrite reductase promoter; Sp/Stmr | This study |
| pPROBE-OT | Broad-host-range plasmid with a transcriptional fusion cassette containing a promoterless GFP reporter gene; Sp/Stmr | 17 |
| pOTNas4 | pPROBE-OT plus region 4 of the nitrate/nitrite reductase promoter; Sp/Stmr | This study |
Table 2.
Oligonucleotides used
| Oligonucleotide | Sequence |
|---|---|
| NasSK1aF | ATGGGATCCGTCTCTTGCAGATAATTCATAACA |
| NasSK1-2aR | GCGGGATCCTCACTTTGTATTACCTGCTGAC |
| NasSK4aF | CGCGGATCCGATATCAAACCGGAAAACCT |
| NasSK3-4aR | CCGGAATTCATCATCACCAGTTTGAGTTTG |
| crt2099KO_F | CACCCTGACCGAAGACTATTACG |
| 2099KO_R | AGTTTCGGTGGTTTTGAAC |
Construction of LK2(pSKNas1) and LK2(pSKNas4).
The regions termed 1 and 4 near the putative nitrite reductase gene, nasA (Fig. 1), were amplified from the chromosomal DNA of strain B728a using Pfu polymerase according to the manufacturer's instructions. The primers NasSK1aF and NasSK1-2aR and NasSK4aF and NasSK3-4aR were used to amplify regions 1 and 4, respectively (Table 2). Each primer set contains a BamHI and EcoRI restriction site at the 5′ end of the forward or reverse primers, respectively. The resulting PCR products were subcloned into pCR-Blunt II-TOPO (Invitrogen), resulting in the plasmids pBLUNT+Nas1 and pBLUNT+Nas4, respectively. The inserts were then digested from the resulting plasmids using BamHI and EcoRI (New England BioLabs, Inc. [NEB]) and ligated into the multiple cloning site of the transcriptional reporter plasmid pPROBE-SK (18), upstream of the promoterless ice nucleation reporter gene inaZ. The ligation reactions were transformed into TOP10 cells, and the transformants were screened for the presence of the insert. The resulting plasmid DNAs, pSKNas1 and pSKNas4, were harvested using the Qiaquick Miniprep kit (Qiagen) and introduced into P. syringae strain LK2 by electroporation.
Fig 1.
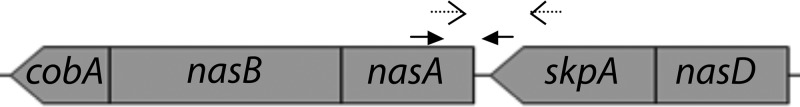
The nitrate assimilation operon of Pseudomonas syringae B728a. Genes (numbers in parentheses are locus tags): cobA (psyr_2098), uroporphyrinogen decarboxylase biosynthesis; nasB (psyr_2099), nitrate reductase (psyr_2100); nasA, nitrite reductase; skpA (psyr_2101), serine/threonine kinase/phosphatase; nasD (psyr_2102), nitrate/nitrite transporter. The filled arrows represent primers used to amplify region 1, while the open-ended arrows correspond to the primers used to amplify region 4.
Construction of B728a(pOTNas4).
pBLUNT+Nas4 was digested with BamHI and EcoRI (NEB), and the fragment was cloned upstream of the promoterless green fluorescent protein (GFP) gene in pPROBE-OT (19) that was also digested with BamHI and EcoRI. The products were ligated with T4 DNA ligase (NEB) and transformed into TOP10 cells. The transformants were screened for the insert by PCR, and the resulting plasmid, pOTNas4, was harvested using the Qiaquick Miniprep kit (Qiagen) and electroporated into strain B728a.
Plant propagation.
Bean plants (Phaseolus vulgaris L. cv. Bush Blue Lake 274) were grown in a soil mix in a greenhouse and fertilized daily with Hoagland's solution (20). Some plants were grown in vermiculate and bottom watered daily with the following nitrogen-containing solutions: nitrate only (5 mM), ammonium only (5 mM), and 50% nitrate (2.5 mM), as well as a solution with no nitrogen, which were derived from the NO3− and NH4+ nutrient solutions used by Van Beusichem et al. (21). While the sole NO3− and NH4+ treatments were essentially the same as that described by Van Beusichem et al., the 50% nitrate treatment differed from the nitrate-only solution in that half the amount of nitrate was used. The composition of the no-nitrogen treatment was the same as that of the ammonium-only treatment without the ammonium.
Bacterial inoculation of plants.
The bacterial inoculum was recovered by centrifugation from either KB or M9 medium, after which the bacteria were washed three times with 10 mM KPO4 buffer. Bean plants in which the second set of trifoliates had expanded but the third set had only recently emerged were spray inoculated with cells suspended in 1 mM KPO4 buffer to a final concentration of 105 to 108 cells/ml. The inoculated plants were enclosed in plastic bags to maintain 100% relative humidity and incubated at room temperature until sampled. Individual leaves were removed from the plant and submerged in 20 ml of washing buffer (0.1 M potassium phosphate buffer [pH 7.0] containing 0.1% peptone). Leaf surface bacteria were removed by sonication for 5 min (Branson Ultrasonic Cleaner 5510) followed by 30 s of vortexing. Bacteria were then enumerated by dilution plating onto KB containing rifampin or by their ice nucleus content determined as below.
Ice nucleation measurements.
Ice nucleation activity was measured using a droplet-freezing method as described previously at either −5°C or −9°C as appropriate (16). Ice nucleation activity was normalized for the number of bacterial cells recovered as estimated by dilution plating of samples onto appropriate antibiotic-containing media.
Leaf exudates.
Bean plants were misted with double-distilled water (ddH2O), bagged to keep the leaves moist, and placed at 4°C overnight to allow solubilization of any surface compounds while restricting bacterial growth. The primary leaves (30 leaves per sample) were carefully excised at the petiole and washed in beakers containing enough water to submerge the leaves without wetting the cut petiole and sonicated for 1 min in glass beakers in a Branson ultrasonic cleaner 5510. The leaves were then removed and dried in an oven for 2 days to obtain the dry weight. The leaf washes were dried under vacuum to a final volume of 5 ml using the Brinkman Rotovapor-R and then filtered through a 0.45-μm-pore-size filter and stored at −20°C. All glassware used to harvest and concentrate the leaf exudates was acid washed in 5% HCl.
Measurement of nitrate from leaf exudates.
Indirect measures of nitrate concentrations of exudates were determined using the nitrate biosensor EcCT501R(pNICE) (2), which was grown in M9 medium at 25°C for 2 days, after which 107 cells/ml were inoculated into 3-ml cultures of M9 containing 0.4% glucose (no nitrogen) or 1 ml of leaf exudate from bean leaves. The reporter was incubated in each sample at 25°C for 4 h before ice nucleation activity was measured by the droplet freezing assay. This same procedure was used to measure the nitrate reductase activity of the P. syringae bioreporter LK2(pSKNas4). Nitrate concentrations were also directly determined by ion chromatography (Dionex) based on the slope of the response obtained with different nitrate standards generated from the Dionex IC Combined Seven Anion Standard II.
Nitrate biosensor.
The nitrate biosensor strain, EcCT501R(pNICE) (14), was also used to measure nitrate availability on the surface of bean leaves. The plants were spray inoculated with a suspension of 108 cells/ml in water and incubated under moist conditions for 6 h before the primary bean leaves were harvested and bacterial population sizes and ice nucleation activity at −5°C were determined as described above.
Measuring GFP fluorescence.
B728a(pOTNas4) (this study) was grown in M9 medium and inoculated onto beans at a concentration of 107 cells/ml in 1 mM KPO4 buffer or in 0.1% KNO3 or 0.1% NH4Cl as positive or negative controls, respectively. Strain B728a(pOTNas4) grown overnight in M9 containing 0.4% glucose and 0.1% KNO3 also was used as an additional positive control. Plants were incubated at 100% humidity at room temperature to maintain leaf surface moisture, and two primary leaves per treatment were harvested after 24 h of incubation. Bacteria were removed from leaves by sonication as described before, cells were recovered by centrifugation at 2,390 × g for 10 min, and the cell pellet was treated with 2.5 mg/ml DAPI (4′,6-diamidino-2-phenylindole) for 5 min, snap chilled in a dry ice/ethanol bath, fixed with 4% paraformaldehyde overnight at 4°C, and washed once with 10 mM KPO4 buffer. Individual 10-μl droplets were then spotted onto ProbeOn Plus microscope slides (Fisherbrand), dried at 37°C for 30 min, and covered with Aqua Polymount anti-fade mounting reagent (Polyscience), and a coverslip was applied. The cells were visualized at ×1,000 magnification with a Hamamatsu digital camera attached to a Zeiss AxioImager 373 microscope, excited by a broad-spectrum mercury arc lamp, and visualized with both standard DAPI and EndowGFP filters. The intensity of GFP fluorescence in cells identified by DAPI fluorescence was analyzed by iVision software (ver. 4.5). A mask of the location of DAPI-stained cells was pasted onto the tandem GFP image, and the mean pixel intensity for each masked objected was quantified. The average background pixel intensity was subtracted from the GFP pixel intensities separately for each image.
Bacterial growth in leaf exudates.
Bacteria were grown in a defined medium composed of buffered leaf washings. The basal plant-derived medium consisted of 1 ml of 1× M9 medium without added nitrogen or glucose as well as 2 ml of bean leaf exudate (collected as above). Glucose- or nitrogen-containing compounds were then added to the medium as appropriate. P. syringae strains were initially grown in M9 medium for 2 days before transfer to M9 containing 0.4% glucose but with no added nitrogen and incubated overnight. The cells were then harvested, washed, transferred to the plant medium to a final concentration of 105 cells/ml, and incubated at 28°C for 48 h, and bacterial population size was enumerated by dilution plating.
RESULTS
Characterization of the assimilatory nitrate reduction operon.
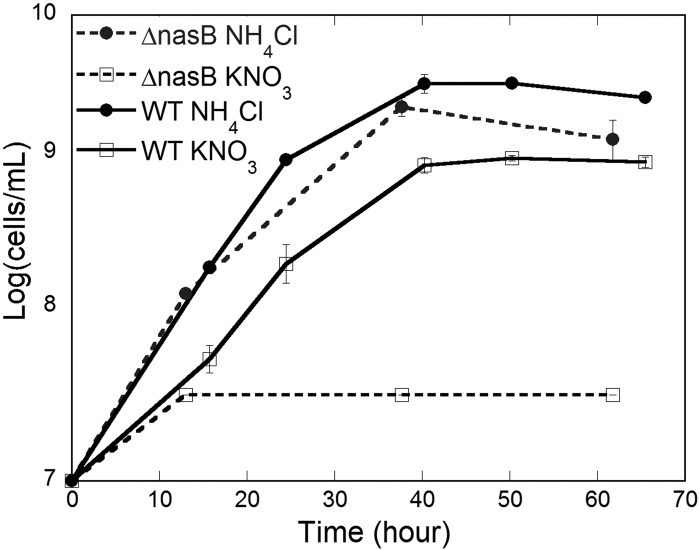
Nitrate assimilation has yet to be shown in Pseudomonas syringae but has been studied in a related member of the Pseudomonadales, Azotobacter vinelandii. These studies have identified the nitrate and nitrate reductase genes encoded in the same operon, nasB and nasA, respectively (22), while two additional regulatory proteins, NasS and NasT, have been identified upstream of this region (23). The localization of the nitrate assimilation operon in A. vinelandii is highly syntenous to the localization of Psyr_2099 to Psyr_2105 in P. syringae, where nasB and nasA are homologous to Psyr_2099 and Psyr_2100, respectively. Since nitrate assimilation has not been shown in P. syringae, we determined if genes Psyr_2099 to Psyr_2105, which were annotated as nitrate and nitrite reductases, a nitrate/nitrite transporter, and nitrate assimilation regulatory genes nasS and nasT, contributed to assimilatory nitrate reduction (Fig. 1). A mutant strain in which nasB, encoding a putative nitrate reductase, was disrupted showed growth deficiency on M9 minimal medium with nitrate as the sole nitrogen source (Fig. 2). A mutant in which the genes encoding a putative serine/threonine phosphatase/kinase and a nitrate/nitrite transporter, which we termed skpA and nasD, respectively (Fig. 1), were deleted also exhibited a growth deficiency in M9 medium containing only nitrate as a nitrogen source, confirming that this region is involved in nitrate assimilation in P. syringae (data not shown).
Fig 2.
Growth of the wild-type Pseudomonas syringae strain B728a and the ΔnasB mutant in M9 minimal medium containing 0.4% glucose and either 0.1% KNO3 or NH4Cl.
Construction of nitrate assimilation biosensors.
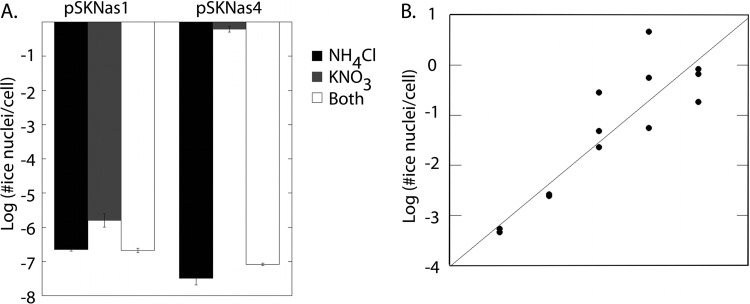
A bioreporter to study the expression of genes involved in nitrate reduction on the leaf surface was constructed. Two regions, labeled regions 1 and 4, upstream from the nitrite reductase (nasA) and putatively harboring the promoter for the operon containing both nasA and nasB (Fig. 1), were PCR amplified from the P. syringae genome and tested for their ability to confer nitrate-dependent transcription. Transcription was assessed by cloning these regions upstream of a promoterless inaZ reporter gene in the vector pPROBE-SK (19) and introducing the resulting plasmids, pSKNas1 and pSKNas4, into an ice nucleation-deficient strain of P. syringae (LK2). inaZ encodes an ice nucleation protein from P. syringae that is sufficient to catalyze ice formation at temperatures between −2°C and −5°C. The amount of ice nucleation activity is proportional to promoter activity. Only region 4 appeared to contain the nitrate-responsive promoter since pSKNas4 conferred much higher nitrate-dependent ice nucleation activity than pSKNas1 in cells grown in minimal medium containing nitrate (Fig. 3A). Cells harboring pSKNas4 or pOTNas4, a similar construct in which this promoter region was cloned into pPROBE-OT, a promoter-probe vector with a promoterless gfp reporter gene, were thus used as bioreporters in subsequent assays to assess nasB expression. In many other bacteria the expression of nitrate assimilation genes occurs only in the presence of nitrate and in the absence of other preferred nitrogen sources, such as ammonium or amino acids (24). We thus examined the expression of nasB in P. syringae under various conditions where alternative nitrogen sources were present. While the ice nucleation activity of LK2(pSKNas4) was about 160,000-fold higher when cells were grown in medium containing nitrate than in those grown in ammonium, little apparent induction of nasB was seen in media containing both nitrate and ammonium, indicating that nitrate assimilation is repressed in the presence of other nitrogen sources (Fig. 3A). The ice nucleation activity of LK2(pSKNas4) exhibited a log-linear relationship with increasing concentrations of nitrate in the culture medium (Fig. 3B). nasB expression is high even at very low concentrations of nitrate; the ice nucleation activity of cells grown in medium containing only 0.1 μM nitrate was 670,000-fold higher than those in ammonium-containing culture medium. Potentially, the reporter should be able to detect even lower nitrate concentrations than those tested.
Fig 3.
Ice nucleation activity reflecting the expression of the putative nitrate/nitrite reductase promoter in the presence of nitrate. (A) Ice nucleation activity of Pseudomonas syringae LK2(pSKNas1) and LK2(pSKNas4). The strains were incubated in M9 minimal medium containing ammonium (19 μM) (black), nitrate (10 μM) (gray), or both (white) for 4 h at 25°C before the ice nucleation activity was assessed at −5°C. (B) Relationship of ice nucleation activity of LK2(pSKNas4) and concentrations of nitrate in M9 minimal medium. The line drawn represents the linear regression: y = 0.81x + 2.56 (r2 = 0.81). The ice nucleation activity is expressed as the log of the number of ice nuclei present per cell.
Determining the levels of nitrate on leaf surfaces.
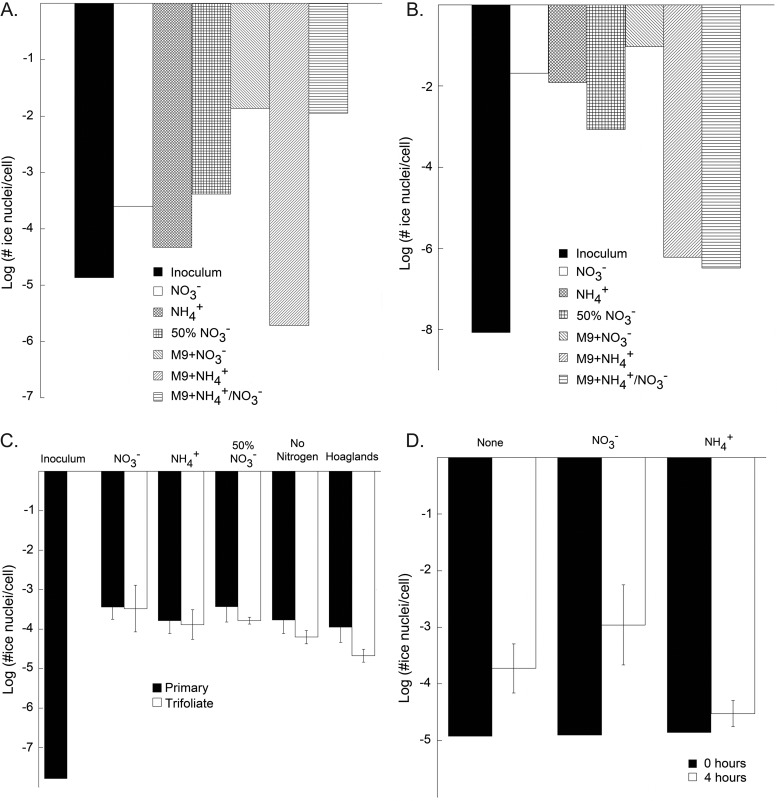
We examined the availability of nitrate on leaf surfaces to determine the potential for nitrate-dependent growth of P. syringae on plants. While several studies have examined nitrate concentrations in whole plants, leaves, stems, or roots (25, 26), it has yet to be determined whether nitrate is present on leaf surfaces. Two methods were used to estimate nitrate concentrations on the leaf surface. A nitrate bioreporter, Enterobacter cloacae EcCT501R(pNICE) (2), was used to determine the nitrate that is bioavailable to bacteria on leaves. This bioreporter contains the promoter from the gene encoding dissimilatory nitrate reductase, narG, of Escherichia coli MG1655 cloned upstream of a promoterless inaZ gene in pPROBE-KI. Regulation of dissimilatory nitrate reduction differs from that of assimilatory nitrate reduction in that the former is repressed by oxygen and not ammonium; hence, nitrate can be detected even in the presence of other nitrogen-containing compounds in this reporter that also harbors an altered transcriptional regulator, fnr, to allow for promoter activity in the presence of oxygen. To determine the range of nitrate levels that might be found on leaves, washings were obtained from the leaves of beans that were fertilized with various concentrations of nitrate and/or ammonium. EcCT501R(pNICE) was incubated in a minimal medium lacking nitrogen but containing 0.4% glucose and the various leaf washings (Fig. 4A). The ice nucleation activity of EcCT501R(pNICE) was induced from 3.5-fold to over 30-fold in the presence of various leaf washings, with the lowest induction seen from plants fertilized with ammonium alone (Fig. 4A). Thus, nitrate is seen on the leaves of all plants sampled, largely independently of the amount and type of nitrogen fertilizer applied to the plants. We note that there are differences in the ice nucleation activity of the inoculum, which was grown in M9 with ammonium, and the sample grown on fresh M9 medium with ammonium (M9+NH4+). These differences are primarily due to the fact that the inoculum consisted of cultures grown for 2 days, while the M9+NH4+ culture was grown for only 4 h. Nitrate concentrations of leaf washings measured using ion chromatography (IC) varied from about 1 to over 20 μg/g. The nitrogen fertilization regime of the plant did not directly relate to the concentration of nitrate to which the plants were treated (Table 3). Thus, substantial amounts of nitrate were found by both methods on plants, largely irrespective of how the plants were propagated.
Fig 4.
Ice nucleation activity of Enterobacter cloacae EcCT501R(pNICE) (A and C) and Pseudomonas syringae LK2(pSKNas4) (B and D) grown in vitro on bean leaf exudate (A and B) or on plants (C and D). Inocula of the bioreporter strains were grown for 2 days in M9 medium containing NH4Cl as the nitrogen source. The washed cells were then transferred to M9 medium without nitrogen but containing bean leaf exudates amended with various nitrogen compounds or onto plants grown with different nitrogen fertilization regimes. Ice nucleation activity was measured at −5°C, four (A, C, and D) or six (C) hours after inoculation into bean leaf exudates (A and B) or onto plants (C and D). The vertical bars represent the standard errors of the determinations of the mean, cell-normalized, ice nucleation activity with 3 replications. No replications were performed for panels A and B.
Table 3.
Quantification of nitrate levels in leaf exudates of bean plants grown with different forms of nitrogen fertilization as determined by ion chromatography
| Fertilizer applied to plants | Nitrate recovered in exudate |
|
|---|---|---|
| μg NO3−/g dry leaf | μM/leafa | |
| Ammonium | 2.22 | 7.1 |
| Nitrate | 1.2 | 3.9 |
| 50% nitrate | 4.13 | 13.3 |
| Hoagland's solution | 20.65 ± 8.8 | 60.35 ± 28.8 |
Concentration of nitrate in dew assuming that 1 ml of exudate is present on each leaf.
To determine the local concentrations of nitrate accessible to bacteria on bean leaves, the ice nucleation activity of EcCT501R(pNICE) that was sprayed directly onto plants was assessed (Fig. 4C). While the ice nucleation activity of the bioreporter was from 1,700 to 12,000-fold higher on the leaves than that in overnight cultures in a nitrate-free culture medium, only small differences in ice nucleation activity were seen on leaves of plants grown with different fertilization regimes (Fig. 4C). These results suggested that bacteria experienced substantial nitrate on all plants, although, as expected, the smallest amount of ice nucleation activity was seen on plants that had not received nitrogen fertilization.
Bacterial nitrate assimilation occurs on leaves.
Given that nitrate was detected in significant amounts on bean leaves, the nitrate assimilation bioreporter, LK2(pSKNas4), was used to determine if P. syringae expressed this trait on leaves. To determine whether nitrate reduction could occur at the level of the whole leaf, the activity of the reporter in leaf washings was determined. LK2(pSKNas4) incubated in a minimal medium containing both glucose and bean leaf exudates from leaves of plants grown with different nitrogen fertilization regimes expressed from 105- to 106-fold more ice nucleation activity in the leaf exudates than that grown in medium without added leaf exudate (Fig. 4B). The apparent induction of nitrate reductase was somewhat variable and largely independent of the plant nitrogen fertilization regimes, similar to that seen for the abundance of nitrate in the leaf washings (Fig. 4A).
The ice nucleation activity of LK2(pSKNas4) applied directly to leaves was assessed to determine if conditions that would enable nitrate assimilation occur on the leaf surface. LK2(pSKNas4) exhibited 15-fold more ice nucleation activity in planta than in the inoculum grown in the absence of nitrate (P = 0.01) (Fig. 4D). No such induction was observed when ammonium was applied with the bioreporter. Adding additional nitrate (10 μM) to the inoculum caused an increased ice nucleation activity of about 6-fold compared to that of cells on unamended plants, but this difference was not statistically significant (P = 0.19). Thus, at the population level, at least some expression of nasB occurs in cells of P. syringae on leaf surfaces.
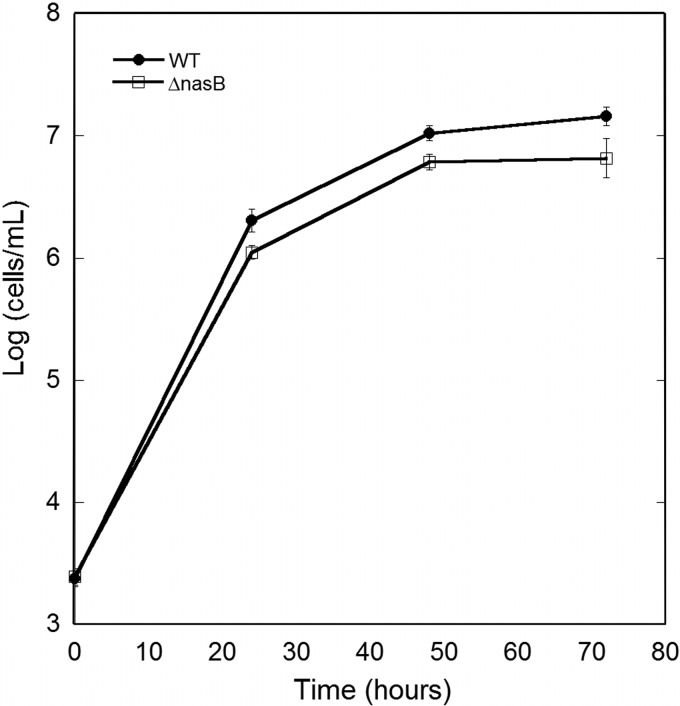
P. syringae ΔnasB does not exhibit reduced growth on bean leaves.
Given that nasB is expressed to some extent in P. syringae while on plants, we determined if the process of nitrate reduction contributes substantially to its growth on bean leaves. The population sizes of the wild-type strain and the ΔnasB mutant were similar on bean leaves at all of the time points tested (Fig. 5). While the population size of P. syringae ΔnasB was often lower than that of the wild-type strain at a given time, multivariate analysis of variance (ANOVA) revealed no differences in population sizes of the two strains over replicate experiments (P = 0.55).
Fig 5.
Growth of wild-type Pseudomonas syringae strain B728a and a ΔnasB mutant on bean primary leaves harvested at various times after inoculation. The vertical bars represent the standard errors of the determination of the mean log-transformed population size with 3 replications.
Nitrate assimilation is heterogeneous on the leaf surface.
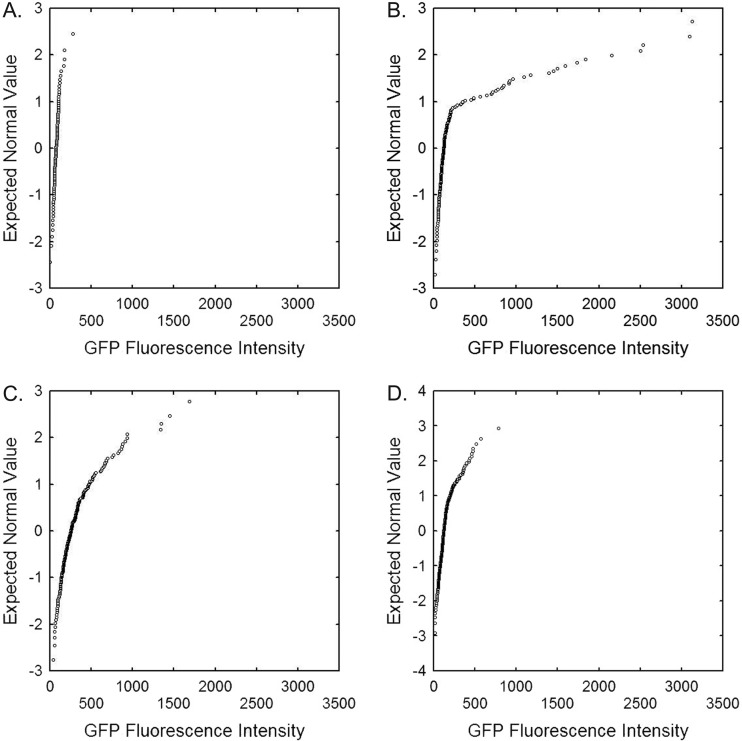
The lack of a contribution of nitrate assimilation to the growth of P. syringae on leaves was somewhat unexpected given the presence of nitrate on leaves (Fig. 4C) and the apparent expression of nasB on leaves (Fig. 4D). Both assessments were made using a sensitive ice nucleation reporter that provides population-level estimates of transcription. We considered that the bacterial population as a whole might not be exposed to nitrate and thus might not act as nitrate reducers. Hence, only a subset of the population might exhibit nitrate-dependent growth. That is, the heterogeneous nature of the leaf surface (27, 28), with respect to nitrate availability and perhaps also to ammonium and amino acid availability, might restrict nitrate assimilation to only a subset of the cells on a leaf. To test this hypothesis, a GFP-based nitrate reductase bioreporter, B728a(pOTNas4), was used to measure the expression of nasB in individual cells in a population of P. syringae on the leaf surface (Fig. 6 and Table 4). Nearly all cells of the inoculum of B728a(pOTNas4) grown in culture medium without added nitrate, as well as cells applied to plants in the presence of ammonium chloride (19 μM), exhibited relatively low GFP fluorescence (pixel intensity, <200 units), indicative of a lack of expression of nasB (Fig. 6A). We thus designated cells with such low relative fluorescence (<200 units) as uninduced for nasB. About 6% of the cells of B728a(pOTNas4) expressed nasB at a level above this baseline on the leaf surface (Fig. 6D). Such cells presumably perceived sufficient nitrate to induce nasB but not enough ammonium or other preferred nitrogen source to repress it. This percentage of induced cells increased to 17.6% or 63.5% in positive-control samples in which the cells were applied in solutions of 0.1% nitrate or preinduced by growth with nitrate (10 μM) in culture media before application to the plant, respectively (Fig. 6B and C). The local environment of those cells applied with added nitrate that exhibited GFP fluorescence presumably lacked sufficient ammonium or amino acids to suppress nasB. Thus, nitrate appears to be present in about 30% of the sites in which ammonium or other nitrogen sources are absent. The cumulative normal distribution plot of the GFP fluorescence of the cells recovered from plant samples reveals that those with low fluorescence intensities (<200 units) are not sampled from a population having a normal distribution of GFP fluorescence intensities. That is, cells appear to be either induced or not, with the induced cells being described by a normal distribution of fluorescence intensities (Fig. 6).
Fig 6.
Cumulative normal probability distribution plots of GFP fluorescence exhibited by individual cells of GFP-based nitrate bioreporter Pseudomonas syringae B728a(pOTNas4) harvested from bean plants 24 h after inoculation. (A and B) NH4Cl (A) or KNO3 (B) at 0.1% was added to the inoculum. (C) B728a(pOTNas4) was preincubated in 0.1% KNO3 overnight before inoculation onto plants. (D) B728a(pOTNas4) was inoculated onto the plant in buffer only. GFP fluorescence intensity was corrected for the intensity of the background in each slide. Each symbol represents the GFP fluorescence intensity of an individual cell harvested from the leaves.
Table 4.
Pseudomonas syringae B728a(pOTNas4) intensity
| Treatment | % intensitya at time (h) after inoculation |
|
|---|---|---|
| 0 | 24 | |
| Applied in water | 0 | 6 |
| Applied with NO3− | 42.8 | 17.6 |
| Applied with NH4+ | 0.5 | 0.9 |
| Pretreated with NO3− | 54.5 | 63.5 |
Percentage of cells of Pseudomonas syringae B728a(pOTNas4) having a GFP intensity above 200 units after sampling from bean plants at the time points indicated.
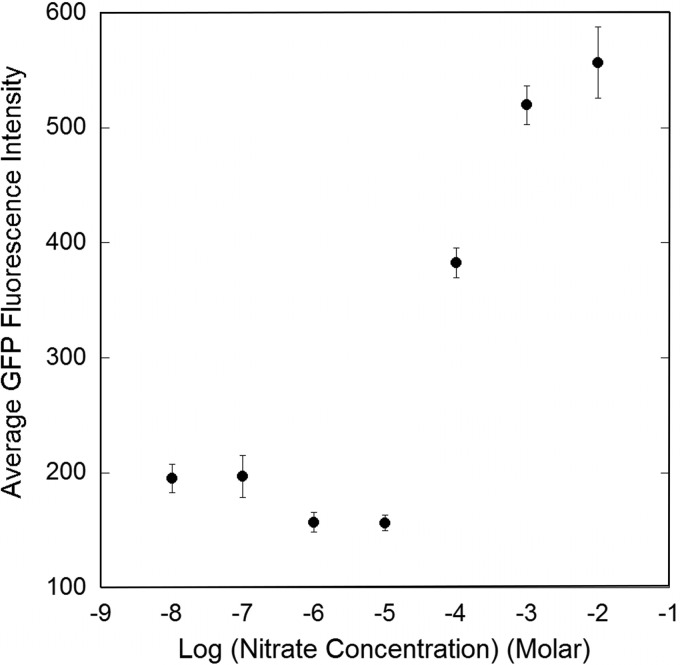
The relationship between GFP fluorescence exhibited by B728a(pOTNas4) and the concentration of nitrate in culture media was determined to aid in estimating the concentration of nitrate at a given location on leaves (Fig. 7). The intensity of GFP fluorescence of individual cells decreased steadily with decreasing nitrate concentrations to about 10−5 M, at which point GFP fluorescence could no longer be detected. It thus appears that cells of B728a(pOTNas4) that exhibited detectable GFP fluorescence on leaves must have experienced a local concentration of nitrate of at least 10−5 M, and some may have experienced more than 10−4 M.
Fig 7.
Mean GFP fluorescence of individual cells of Pseudomonas syringae B728a(pOTNas4) recovered from M9 medium containing various concentrations of nitrate. The vertical bars represent the standard errors of the determination of mean GFP fluorescence. The fluorescence intensity of approximately 100 cells was measured for each nitrate concentration.
Disruption of nasB causes a growth deficiency in bean leaf exudates in a carbon-rich environment.
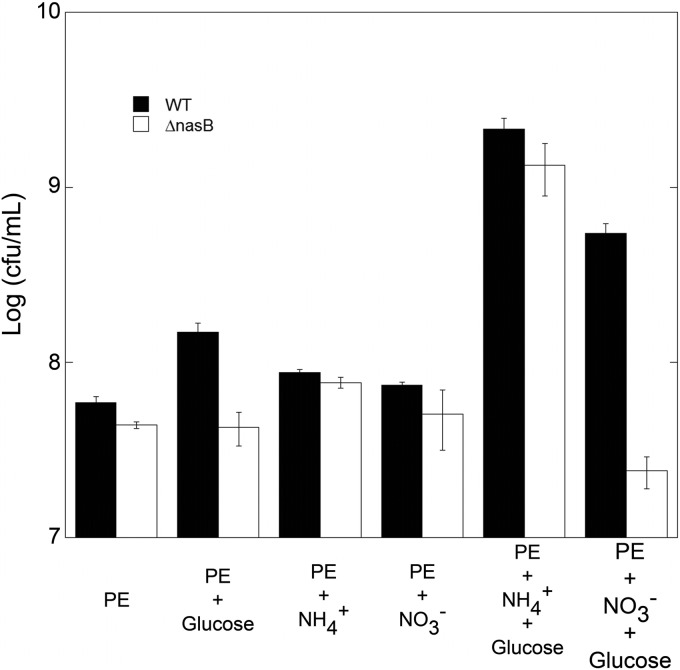
We hypothesized that the lack of contribution of nasB to growth of P. syringae on leaves was due to the fact that carbon availability may limit the growth of bacteria in most sites on leaves. In those sites where other factors, such as nitrogen, were limiting, only a small subset of cells experienced relatively high nitrate levels but not also high levels of other nitrogenous compounds. In such a situation, nitrate assimilation would lead to small relative increases in population sizes that would be hard to measure given the relatively large variation in the apparent carrying capacity of individual leaves (29). To minimize the variations in bacterial population assessments associated with leaf-to-leaf variability and to ensure that growth was not carbon limited, we assessed the growth of both a wild-type strain and the ΔnasB mutant in leaf exudates amended with either (i) potassium phosphate buffer only, (ii) 0.1% KNO3, (iii) 0.1% NH4Cl, (iv) 0.4% glucose, (v) 0.1% KNO3 and 0.4% glucose, or (vi) 0.1% NH4Cl and 0.4% glucose. P. syringae clearly exhibited carbon-limited growth in the bean exudate, since there was a 2.5-fold increase in the population size of the wild-type strain in the glucose-amended medium compared to that in unamended medium (P = 0.008, t test). While plant exudate was primarily carbon limited, nitrogen availability was also secondarily limiting, since the addition of both glucose and either nitrate or ammonium to the exudates increased the growth of P. syringae more than the addition of either alone (Fig. 8). No additional growth of the wild-type strain was seen in the exudates amended with nitrate or ammonium relative to that in exudates amended with buffer only. Importantly, the ΔnasB mutant did not exhibit an increase in growth upon addition of glucose alone or when nitrate was added to exudates. The ΔnasB mutant attained only a slightly smaller population size than the wild-type strain when cultured in exudate alone (P = 0.05). These results support the model that under conditions where carbon limitation is removed, nitrate is a significant nitrogen source for growth on leaves.
Fig 8.
Growth of wild-type Pseudomonas syringae strain B728a and a ΔnasB mutant in bean leaf exudates 48 h after inoculation at a final concentration of 105 cells/ml. The strains were grown on plant exudates alone (PE), PE containing 0.4% glucose, PE containing 0.1% NH4Cl, PE containing 0.1% KNO3, PE containing 0.4% glucose and 0.1% NH4Cl, or PE containing 0.4% glucose and 0.1% KNO3. The vertical bars represent the standard errors of mean log-transformed population size with 3 replications.
DISCUSSION
Nitrate is apparently a substantial source of nitrogen for bacteria on bean leaves. Both whole-cell nitrate biosensors and ion chromatography reveal substantial nitrate (4 μM to 60 μM) on moistened leaves (Table 3). It was interesting to note that both methods detected nitrate in exudate even from beans fertilized only with ammonium. This is likely due to nitrification by soil microbes that yielded nitrate that was taken up by the plant. Nitrification is a widespread process that most likely results in the common occurrence of at least some nitrate uptake and thus exudation onto leaves by most plants. Various factors can affect nitrate accumulation in plant tissue and subsequent leakage of nutrients onto the plant surface (17). It is thus likely that the availability of nitrate on leaves will vary substantially between plant species and under different environmental conditions. Despite this, nitrate is likely to be common on most plants. However, metaproteomics of epiphytic bacterial communities on soybean, clover, and Arabidopsis thaliana did not reveal the presence of nitrate reductases (30). Instead, many amino acid transporters were expressed in all the communities analyzed. This suggests that despite the presence of nitrate there is an abundance of amino acids on the leaf surface. Additionally, assessment of the transcriptome of P. syringae within bean pods, in macerated beans, and in apoplastic fluids found that a nitrate reductase was expressed only in cells grown in apoplastic fluid and not under the other conditions (31). Given that we find that nasB is expressed only in a small subpopulation of cells of P. syringae on leaves (Fig. 6D), it is likely that the abundance of the nitrate reductase itself or of its transcripts will be relatively low, and hence they might easily have been overlooked in these studies. Our study has shown that studying bacterial processes in individual cells in a heterogeneous environment like the leaf surface better enables the assessment of the contribution of processes that do not occur in all cells. It is quite likely, that as we have shown here for assimilatory nitrate reduction, a subset of the bacterial population may be conducting different processes than the rest of the population. Because of this, the contribution of many traits in bacteria in these heterogeneous environments may be overlooked or underestimated.
It was perhaps not surprising to find nitrate present on the leaf surface despite the fact that it had not been previously described. While earlier studies have not directly measured levels of nitrate on leaves, studies profiling the metabolites on several plant species revealed the presence of inorganic nitrogen as well as amino acids (6). Guttation fluid can contribute to the leachate found on leaves (6, 32) and guttation fluids from rye, wheat, and barley contain about 1 mg/liter (15.9 μM) nitrate (33). Such a concentration is within the range of what we measured in bean leaf exudates (Table 3). Clearly, the local concentration of any resource such as nitrate will be influenced by the amount of water into which it is dissolved on the leaf.
While detection of nitrate on the leaf surface was supportive of our hypothesis that it is a potential resource for bacteria, the importance of nitrate assimilation in the epiphytic growth of P. syringae on beans required us to demonstrate a difference in the fitness of strains differing in its consumption. While blockage of the nitrate reductase gene of P. syringae did not result in a measurable growth deficiency when bacteria were allowed to colonize bean plants, we saw a deficiency of growth of the ΔnasB mutant compared to the wild-type strain in plant exudates. This growth difference is exaggerated when the exudate was supplemented with glucose. This suggests that the growth of P. syringae on the bean leaf surface is most limited by carbon and only secondarily limited by nitrogen, consistent with previous studies that examined the epiphytic growth of P. syringae on potatoes (9). A growth difference between the wild-type strain and the ΔnasB mutant on plants was probably not observed, since most sites on leaves have an excess of other preferred nitrogen sources such as ammonium and amino acids relative to the amount of carbon available at those sites. It is known that the leaf surface is a heterogeneous environment where there are microsites where nutrients such as fructose, sucrose, and iron are spatially variable (20, 28, 34). The population of bacteria on leaves has been described as the sum of those subpopulations in relatively isolated microsites on leaves that differ in availability of carbon (35). A contribution of nitrate to overall population size would thus not likely be seen until the bacteria consumed the preferred nitrogen sources before consuming the available nitrate, and only then at sites where carbon was relatively high in proportion to the available nitrogen compounds. The proportion of these later cells that expressed nitrate assimilation was estimated directly with the whole-cell P. syringae nitrate reductase bioreporter; while many cells in the population do not express nasB on the leaf surface, a small proportion (about 6%) of cells do (Table 4 and Fig. 6D). We propose that the bean leaf contains microsites with variable concentrations of nitrogenous compounds in the forms of ammonium, amino acids, and nitrate. It is likely that only a portion of the population on a leaf will be found in locations either where more preferred forms of nitrogen are low, allowing for the consumption of nitrate, or where nitrogen rather than carbon sources were the most limiting resources on the leaf. However, because only a minority of the cells appeared to be capable of expressing nitrate reductase on the leaf, the contribution of nitrate to the overall bacterial population size was relatively small. The variations in leaf-to-leaf population sizes of bacteria such as P. syringae are relatively great (36), presumably due to intrinsic differences in the amount of resources that leach onto the leaf surface. Additionally, there is potentially a constant flux of nutrients at a given location on the leaf surface driven both by the consumption of the nutrients by microorganisms and production by the plants. As such, the relatively small differences in population sizes of P. syringae that are expected to be contributed by consumption of nitrate (2-fold or less) (Fig. 8) are difficult to measure. Nonetheless, such a contribution of nitrate to bacterial population sizes would be ecologically significant. It is also likely that the contribution of nitrate to the nitrogen budget of epiphytic bacteria will be quite context dependent. Plants apparently vary substantially in the amount and forms of nitrogen that they contain and hence the forms of nitrogen that will be exuded onto their leaves (37, 38). Likewise, the greenhouse-grown plants studied here received substantial nitrogen fertilization and hence probably differed greatly from plants grown in natural habitats in their content of nitrogen. It seems likely that plants in natural settings will have leaf surfaces that will be nitrogen limited rather than carbon limited because of the lesser amounts of available nitrogen to which they have access. Likewise, because of the prominence of nitrification in natural habitats (39–41), much of the nitrogen would be expected to be in the form of nitrate. It thus should be fruitful to assess the relative fitness of the ΔnasB mutant in such natural settings. While nitrate assimilation is apparently not essential for the growth of P. syringae on bean leaves, it contributes to the success of at least some cells even in this habitat. Nitrate assimilation is likely to be important for the growth of P. syringae on plants, other than bean, that may be more limited for nitrogen than carbon or for plants that store nitrate, such as spinach and lettuce. Assimilatory nitrate reduction thus probably facilitates epiphytic fitness in the multiple environments in which P. syringae is likely to be found.
ACKNOWLEDGMENTS
The research described in this paper has been funded in part by the United States Environmental Protection Agency (EPA) under the Science to Achieve Results (STAR) Graduate Fellowship Program. EPA has not officially endorsed this publication, and the views expressed herein may not reflect the views of the EPA.
Footnotes
Published ahead of print 16 November 2012
REFERENCES
- 1. Allen AE, Booth MG, Verity PG, Frischer ME. 2005. Influence of nitrate availability on the distribution and abundance of heterotrophic bacterial nitrate assimilation genes in the Barents Sea during summer. Aquat. Microb. Ecol. 39:247–255 [Google Scholar]
- 2. DeAngelis KM, Ji PS, Firestone MK, Lindow SE. 2005. Two novel bacterial biosensors for detection of nitrate availability in the rhizosphere. Appl. Environ. Microbiol. 71:8537–8547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Myrold DD, Posavatz NR. 2007. Potential importance of bacteria and fungi in nitrate assimilation in soil. Soil Biol. Biochem. 39:1737–1743 [Google Scholar]
- 4. Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morgan JV, Tukey HB. 1964. Characterization of leachate from plant foliage. Plant Physiol. 39:590–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tukey HB. 1966. Leaching of metabolites from above-ground plant parts and its implications. Bull. Torrey Bot. Club 93:385–401 [Google Scholar]
- 7. Sinhababu A, Kushari D. 1984. Effect of leaf leachates of Polyalthia longifolia on the growth and nitrogen fixation of Azolla pinnata. Aquat. Ecol. 18:103–108 [Google Scholar]
- 8. Benner R, Peele ER, Hodson RE. 1986. Microbial utilization of dissolved organic matter from leaves of the red mangrove, Rhizophora mangle, in the Fresh Creek estuary, Bahamas. Estuar. Coast. Shelf Sci. 23:607–619 [Google Scholar]
- 9. Wilson M, Lindow SE. 1994. Ecological similarity and coexistence of epiphytic ice-nucleating (Ice+) Pseudomonas syringae strains and a non-ice-nucleating (Ice−) biological control agent. Appl. Environ. Microbiol. 60:3128–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brandl MT, Amundson R. 2008. Leaf age as a risk factor in contamination of lettuce with Escherichia coli O157:H7 and Salmonella enterica. Appl. Environ. Microbiol. 74:2298–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hirano SS, Upper CD. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morris CE, Kinkel LL, Xiao K, Prior P, Sands DC. 2007. Surprising niche for the plant pathogen Pseudomonas syringae. Infect. Genet. Evol. 7:84–92 [DOI] [PubMed] [Google Scholar]
- 13. Loper JE, Lindow SE. 1987. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77:1449–1454 [Google Scholar]
- 14. Andersen GL, Beattie GA, Lindow SE. 1998. Molecular characterization and sequence of a methionine biosynthetic locus from Pseudomonas syringae. J. Bacteriol. 180:4497–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nelson EB. 1988. Biological control of pythium seed rot and preemergence damping-off of cotton with Enterobacter cloacae and Erwinia herbicola applied as seed treatments. Plant Dis. 72:140–142 [Google Scholar]
- 16. Loper JE, Lindow SE. 1997. Reporter gene systems useful in evaluating in situ gene expression by soil-and plant-associated bacteria, p 482–492 In Hurst CJ, Knudsen GR, McInerney MJ, Stetzenbach LD, Walter MV. (ed), Manual of environmental microbiology. ASM Press, Washington, DC [Google Scholar]
- 17. Maynard DN, Barker AV, Minotti PL, Peck NH. 1976. Nitrate accumulation in vegetables. Adv. Agron. 28:71–118 [Google Scholar]
- 18. Marco ML, Legac J, Lindow SE. 2005. Pseudomonas syringae genes induced during colonization of leaf surfaces. Environ. Microbiol. 7:1379–1391 [DOI] [PubMed] [Google Scholar]
- 19. Miller WG, Leveau JHJ, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13:1243–1250 [DOI] [PubMed] [Google Scholar]
- 20. Hoagland DR, Arnon DI. 1950. The water-culture method for growing plants without soil. California Agricultural Experiment Station, circular 347. University of California, Berkeley, CA [Google Scholar]
- 21. Van Beusichem ML, Kirkby EA, Baas R. 1988. Influence of nitrate and ammonium nutrition on the uptake, assimilation, and distribution of nutrients in Ricinus communis. Plant Physiol. 86:914–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramos F, Blanco G, Gutierrez JC, Luque F, Tortolero M. 1993. Identification of an operon involved in the assimilatory nitrate-reducing system of Azotobacter vinelandii. Mol. Microbiol. 8:1145–1153 [DOI] [PubMed] [Google Scholar]
- 23. Gutierrez J-C, Ramos F, Ortner L, Tortolero M. 1995. NasST, two genes involved in the induction of the assimilatory nitrite-nitrate reductase operon (nasAB) of Azotobacter vinelandii. Mol. Microbiol. 18:579–591 [DOI] [PubMed] [Google Scholar]
- 24. Lin JT, Stewart V. 1998. Nitrate assimilation by bacteria. Adv. Microb. Physiol. 39:1–30 [DOI] [PubMed] [Google Scholar]
- 25. Chen BM, Wang ZH, Li SX, Wang GX, Song HX, Xi-Na W. 2004. Effects of nitrate supply on plant growth, nitrate accumulation, metabolic nitrate concentration and nitrate reductase activity in three leafy vegetables. Plant Sci. 167:635–643 [Google Scholar]
- 26. Neely HL, Koenig RT, Miles CA, Koenig TC, Karlsson MG. 2010. Diurnal fluctuation in tissue nitrate concentration of field-grown leafy greens at two latitudes. HortScience 45:1815–1818 [Google Scholar]
- 27. Joyner DC, Lindow SE. 2000. Heterogeneity of iron bioavailability on plants assessed with a whole-cell GFP-based bacterial biosensor. Microbiology 146:2435–2445 [DOI] [PubMed] [Google Scholar]
- 28. Leveau JHJ, Lindow SE. 2001. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. U. S. A. 98:3446–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kinkel LL, Wilson M, Lindow SE. 1995. Effect of sampling scale on the assessment of epiphytic bacterial populations. Microb. Ecol. 29:283–297 [DOI] [PubMed] [Google Scholar]
- 30. Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA. 2009. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:16428–16433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hernandez-Morales A, De la Torre-Zavala S, Ibarra-Laclette E, Hernandez-Flores JL, Jofre-Garfias AE, Martinez-Antonio A, Alvarez-Morales A. 2009. Transcriptional profile of Pseudomonas syringae pv. phaseolicola NPS3121 in response to tissue extracts from a susceptible Phaseolus vulgaris L. cultivar. BMC Microbiol. 9:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carlson RM, Cabrera RI, Paul JL, Quick J, Evans RY. 1990. Rapid direct determination of ammonium and nitrate in soil and plant-tissue extracts. Commun. Soil Sci. Plant Anal. 21:1519–1529 [Google Scholar]
- 33. Goatley JL, Lewis RW. 1966. Composition of guttation fluid from rye wheat and barley seedlings. Plant Physiol. 41:373–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Willis DK, Hrabak EM, Rich JJ, Barta TM, Lindow SE, Panopoulos NJ. 1990. Isolation and characterization of a pseudomonas syringae pv. syringae mutant deficient in lesion formation on bean. Mol. Plant Microbe Interact. 3:149–156 [Google Scholar]
- 35. Remus-Emsermann MNP, Tecon R, Kowalchuk GA, Leveau JHJ. 2012. Variation in local carrying capacity and the individual fate of bacterial colonizers in the phyllosphere. ISME J. 6:756–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wilson M, Hirano SS, Lindow SE. 1999. Location and survival of leaf-associated bacteria in relation to pathogenicity and potential for growth within the leaf. Appl. Environ. Microbiol. 65:1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Durzan DJ. 1968. Nitrogen metabolism of Picea glauca. I. Seasonal changes of free amino acids in buds, shoot apices, and leaves, and the metabolism of uniformly labelled 14C-l-arginine by buds during the onset of dormancy. Can. J. Bot. 46:909–919 [Google Scholar]
- 38. Mattson WJ. 1980. Herbivory in relation to plant nitrogen content. Annu. Rev. Ecol. Syst. 11:119–161 [Google Scholar]
- 39. Booth MS, Stark JM, Rastetter E. 2005. Controls on nitrogen cycling in terrestrial ecosystems: a synthetic analysis of literature data. Ecol. Monogr. 75:139–157 [Google Scholar]
- 40. Stark JM, Hart SC. 1997. High rates of nitrification and nitrate turnover in undisturbed coniferous forests. Nature 385:61–64 [Google Scholar]
- 41. Stevenson FJ. 1982. Nitrogen in agricultural soils. American Society of Agronomy, Madison, WI [Google Scholar]